2. 中国科学院大学, 北京 100049;
3. 复旦大学 生命科学学院, 上海 200438
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. School of life Sciences, Fudan University, Shanghai 200438, China
在高等植物及微生物体内,莽草酸代谢途径为芳香族氨基酸的合成提供底物.对于植物而言,此途径还为多种次生代谢物的合成提供前体物质.在植物正常生长的过程中,大约有20%的碳流量经莽草酸代谢途径合成芳香族氨基酸、木质素、黄酮类、生物碱及植物激素等[1, 2].这些莽草酸途径介导的代谢物不仅具有植物生长发育调控、信号传导、抗病抗逆等重要生理功能[3, 4],而且它们可以介导人体与肠道菌群的共代谢进而影响人体健康[5-7],甚至具有一定的药理作用[8, 9].
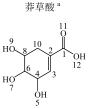
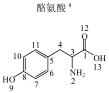
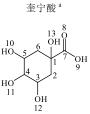
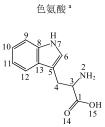
莽草酸由赤藓糖-4-磷酸与磷酸烯醇式丙酮酸经一系列酶的催化代谢而成,它进一步被代谢为分支酸,进而形成色氨酸、苯丙氨酸及酪氨酸等3种芳香类必需氨基酸[10-12].莽草酸也是奎宁酸的代谢前体及奥塞米韦等抗病毒药物的中间体.色氨酸代谢会产生吲哚乙酸、5-羟基吲哚乙酸及吲哚乳酸等生物碱[13].吲哚乙酸这种植物激素生长素是由吲哚丙酮酸脱羧酶(IPDC)催化吲哚丙酮酸合成[14].苯丙氨酸可进一步通过羟基化而代谢为酪氨酸,这两种氨基酸再经过苯丙氨酸解氨酶(PAL)催化而形成肉桂酸和对羟基肉桂酸,进而合成植物激素水杨酸及一系列多酚酸等植物次生代谢物.上述代谢物还可以再通过哺乳动物与肠道微生物的共代谢而产生一批芳香类代谢物[5, 7, 15-17].
为此,植物莽草酸代谢途径中部分代谢物的定量分析颇为重要.研究发现丹参中多酚酸具有环境及品种依赖性[3, 4],莽草酸途径代谢物存在于绿豆种子[18, 19].研究还发现具有抗肿瘤及抗雄激素活性的杨梅醇是经4-香豆酸转化为4-羟基苯丙酸而实现其生物合成[20];玛咖中类黄酮木质素具有抗炎及抗癌的活性,4-香豆酸也具有一定的生理活性[21].其次,多酚酸是植物抗逆抗病的重要次生代谢物,它们对丹参根茎抵抗脱水有显著应答[4],莽草酸代谢途径介导的次生代谢物在植物代谢组应答盐胁迫过程中有潜在重要功能[22].再次,水稻对褐飞虱的抗性与其莽草酸途经介导的次生代谢相关[23],奎宁酸及绿原酸含量与桃子抵抗害虫攻击的抗性呈正相关[24].此外,吲哚乙酸与水杨酸都是重要的植物激素,其功能的不可或缺性显而易见.因此,“次生代谢物”这个传统名称已无法准确反映此类代谢物的重要性.
如此重要代谢物组的核磁共振(NMR)分析显然需要完备的NMR数据与归属.虽然大部分的代谢物结构的NMR数据归属已被报道[13, 20, 24-36],但测定NMR数据的溶剂系统较多,代谢组学研究常使用水或乙腈作为溶剂.另外,前述文献中5-羟基吲哚乙酸及吲哚乳酸等莽草酸代谢途径介导代谢物的NMR基础数据及信号归属不甚完善,一些季碳NMR数据未见报道.因此,针对代谢组学研究中常用水与乙腈作为溶剂的特点,本文使用1D 1H NMR与2D 1H-13C HMBC NMR谱图对该途径介导的26种代谢物进行了系统的分析研究,对它们的所有1H与13C NMR信号进行了归属,为基于NMR的相关代谢组学等研究提供了基础数据.
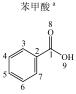
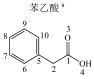
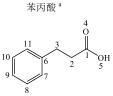
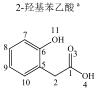
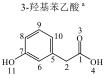
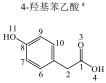
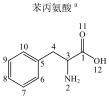
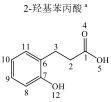
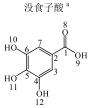
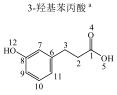
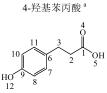
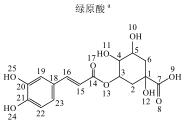
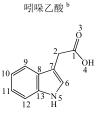
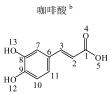
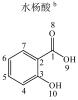
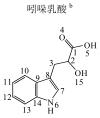
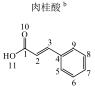
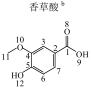
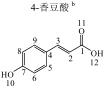
1 实验部分 1.1 仪器及试剂水杨酸、2-羟基苯乙酸、3-羟基苯乙酸、4-羟基苯乙酸、2-羟基苯丙酸、4-羟基苯丙酸及吲哚乳酸购自上海Sigma-Aldrich公司.肉桂酸、4-香豆酸、莽草酸、咖啡酸、奎宁酸、芥子酸及绿原酸购自上海阿拉丁化学试剂有限公司.酪氨酸、色氨酸购自上海如吉生物科技有限公司.5-羟基吲哚乙酸、吲哚乙酸购自北京百灵威化学试剂有限公司.3-羟基苯丙酸购自天津阿法埃莎试剂公司.没食子酸、香草酸、阿魏酸购自南京泽朗医药科技公司.苯甲酸、苯乙酸、苯丙酸、苯丙氨酸、三水合磷酸氢二钾(K2HPO4·3H2O)、二水合磷酸二氢钠(NaH2PO4·2H2O)购自上海国药集团化学试剂有限公司.分析纯叠氮钠(NaN3)购于天津福晨化学试剂厂.氘代乙腈(CD3CN,99.8%氘代)、重水(D2O,99.9%氘代)、用于定标的2, 2, 3, 3-氘代三甲基硅烷丙酸钠(TSP)购自Cambridge Isotope Laboratories公司.配制溶液的超纯水来自Elix Advantage System(默克Millipore,德国)纯水系统(电阻率大于18.2 MΩ·cm-1).26种代谢物的化学结构及代谢途径见图 1所示.
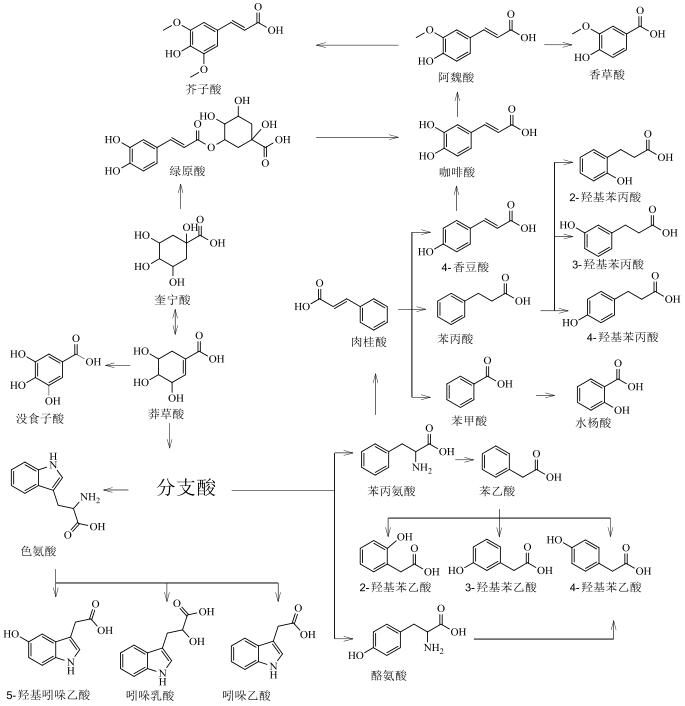
|
图 1 26种代谢物的结构及代谢途径 Figure 1 Structure and metabolic pathway of the 26 metabolites |
酪氨酸与绿原酸分别使用D2O配制的磷酸盐缓冲溶液(0.1 mol/L,pD = 7.4)作为溶剂,其余代谢物直接溶于D2O(含TSP 0.5 mg/mL)或CD3CN中(见表 1).溶于CD3CN溶剂的样品以溶剂中残留质子信号定标.溶于D2O的样品以TSP的甲基信号定标(δH 0.00,δC0.00).1D 1H NMR和2D 1H-13C HMBC图谱均在Bruker AVIII 600 MHz NMR谱仪(Bruker BioSpin)上采集,1H NMR和13C NMR的共振频率分别为600.13 MHz和150.90 MHz,实验温度为298 K.1D 1H NMR谱的参数设置如下:脉冲序列为Noesygppr1d,采样点数(TD)为32 k,累加次数(NS)为16,空扫次数(DS)为4,等待时间(D1)为2 s,谱宽(SW)设置为12 000 Hz,90˚脉宽(P1)约为10 ms.2D 1H-13C HMBC谱的参数设置如下:脉冲序列为HMBCgppr,F2维(1H)的采样点数为2 048,F1维(13C)采样点数为200,NS为8,DS为16,D1为2 s,2-羟基苯丙酸、4-羟基苯丙酸、苯丙氨酸的SW分别设为6 000 Hz(1H)与30 000 Hz(13C),其余样品的SW为6 000 Hz和33 000 Hz.所有代谢物的分析均使用直径为5 mm的高质量NMR样品管.
| 表 1 26种莽草酸途径介导代谢物NMR信号归属 Table 1 Assignments of NMR signals of the 26 shikimate pathway mediated metabolites |
本文研究的代谢物存在于植物等细胞时,其所在环境pH值有一定分布,但研究中常使用不同的溶剂进行提取.经典的植物化学研究中常使用有机溶剂的水溶液作为NMR检测溶剂,而代谢组学研究中也常使用水与乙腈作为溶剂.因此,本文根据代谢物的溶解性重点选择了D2O与CD3CN作为溶剂进行溶液配制,代谢物的信号归属采用与文献中所报道的相似策略[19, 37-39].众所周知,溶剂化效应使得不同溶剂对这些代谢物的1H与13C核的化学位移有不同的影响(特别是可置换活泼氢),文献[19, 37-39]中使用的氘代溶剂包括氯仿、二甲亚砜、甲醇、丙酮、水及其它们的不同比例混合物,故此文不准备与文献中如此复杂的多种溶剂系统中所得数据进行简单对比.鉴于乙腈是植物代谢组学研究常用的提取溶剂,也是代谢物结构确定所用方法——液相色谱-固相卒取-核磁共振-质谱联用技术(LC-SPE-NMR-MS)的关键溶剂,但文献中没有本文所关注代谢物群的乙腈溶液的NMR数据.因此本文所得信息库是对文献已有数据的补充,也是LC-SPE-NMR等相关研究的参比基础数据.本文中使用的代谢物均是纯品,因此1D 1H NMR与2D 1H-13C HMBC谱图的结合分析足以实现所有1H和13C NMR信号的归属.
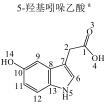
此文对26种莽草酸代谢途径介导的代谢物进行了谱图归属.这里以5-羟基吲哚乙酸为例,对其1D 1H NMR谱(图 2)和2D 1H-13C HMBC谱(图 3)进行了解析讨论.首先,1H NMR谱中脂肪区只有一个单峰δH 3.78归属为H-2,芳香区唯一的单峰δH 7.26归属为H-6.芳香区出现的ABX耦合系统分别归属为苯环的H-12(δH 7.38)、H-11(δH 6.84)及H-10(δH 7.03).其次,直接联接质子的13C核的化学位移均通过1H-13C HMBC谱中对应的卫星峰确定,C-2、C-6、C-9、C-11与C-12的化学位移分别归属为δC 33.8、128.6、105.6、114.6与115.9.羧基的13C核信号一般在δC 165~185之间,因此谱图中唯一的一个羧基信号δC180.9归属为C-1.
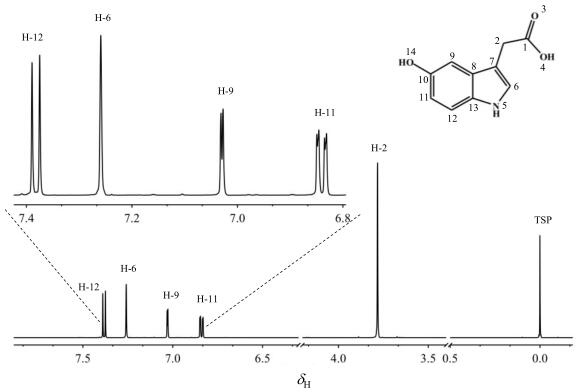
|
图 2 5-羟基吲哚乙酸的1H NMR谱(D2O) Figure 2 1H NMR spectrum of 5-hydroxyindoleacetic acid (D2O) |
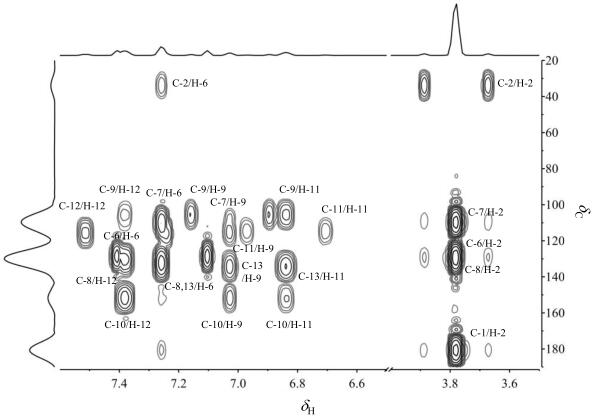
|
图 3 5-羟基吲哚乙酸的1H-13C HMBC谱(D2O) Figure 3 1H-13C HMBC spectrum of 5-hydroxyindoleacetic acid (D2O) |
上述归属还可以使用1H-13C HMBC谱中的远程耦合关系进行再确认.C-1与H-2及H-6均有远程耦合.H-6与C-2、C-7、C-8及C-13均有远程耦合,同时H-2与C-7、C-8有远程耦合,由此可确定C-7、C-8与C-13的化学位移分别为δC109.8、129.5及134.4.这些还可以通过C-8/H-12与C-13/H-11的耦合进一步确认.另一个季碳C-10的化学位移可以通过它与H-12、H-11、H-9之间的耦合关系确定为δC151.6.由于氧原子的电负性大于氮原子,而碳原子电负性均弱于前二者,C-10的化学位移大于C-13且C-7与C-8化学位移均小于前二者也比较合理.综上所述,5-羟基吲哚乙酸的所有1H与13C NMR信号则可以得到归属.其余代谢物NMR谱中信号均按照上述解析方法进行了归属,其结果罗列于表 1中.
3 结论根据1H NMR谱的自旋耦合,及1H-13C HMBC谱中的耦合信息,本文对莽草酸途径介导的26种代谢物的1H与13C NMR信号进行了系统归属,对文献中已有报道的多个代谢物水溶液和乙腈溶液的NMR数据进行补充,对文献中未见报道的5-羟基吲哚乙酸及吲哚乳酸等代谢物季碳的化学位移信息进行了完善,这些代谢物NMR数据库将为基于NMR的代谢组学等研究提供基础数据.需要指出的是,这些代谢物的化学位移会有一定的溶剂依赖性,因此使用文中所报道的化学位移进行植物提取物中代谢物归属时,需要考虑溶剂及代谢物之间相互作用等因素的影响.生物代谢组中此类物质的浓度一般处于经典稀溶液的范畴,代谢物间相互作用的影响较小.本文中的NMR数据,特别是代谢物的J耦合常数及1H-13C HMBC谱图提供的原子间远程耦合关系对代谢物的指认有一定帮助.
| [1] | HERRMANN K M. The shikimate pathway:early steps in the biosynthesis of aromatic-compounds[J]. Plant Cell, 1995, 7(7): 907-919. DOI: 10.1105/tpc.7.7.907. |
| [2] | MAEDA H, DUDAREVA N. Annual review of plant biology[M]. Palo Alto: Annual Reviews, 2012. |
| [3] | DAI H, XIAO C N, LIU H B, et al. Combined NMR and LC-DAD-MS analysis reveals comprehensive metabonomic variations for three phenotypic cultivars of Salvia miltiorrhiza bunge[J]. J Proteome Res, 2010, 9(3): 1565-1578. DOI: 10.1021/pr901045c. |
| [4] | DAI H, XIAO C N, LIU H B, et al. Combined NMR and LC-MS analysis reveals the metabonomic changes in Salvia miltiorrhiza bunge induced by water depletion[J]. J Proteome Res, 2010, 9(3): 1460-1475. DOI: 10.1021/pr900995m. |
| [5] | WANG Y L, TANG H R, NICHOLSON J K, et al. A metabonomic strategy for the detection of the metabolic effects of chamomile (Matricaria recutita L.) ingestion[J]. J Agric Food Chem, 2005, 53(2): 191-196. DOI: 10.1021/jf0403282. |
| [6] | SHI X H, XIAO C N, WANG Y L, et al. Gallic acid intake induces alterations to systems metabolism in rats[J]. J Proteome Res, 2013, 12(2): 991-1006. DOI: 10.1021/pr301041k. |
| [7] | NICHOLSON J K, HOLMES E, WILSON I D. Gut microorganisms, mammalian metabolism and personalized health care[J]. Nat Rev Microbiol, 2005, 3(5): 431-438. DOI: 10.1038/nrmicro1152. |
| [8] | CHEN C, TANG H R, SUTCLIFFE L H, et al. Green tea polyphenols react with 1, 1-diphenyl-2-picrylhydrazyl free radicals in the bilayer of liposomes:Direct evidence from electron spin resonance studies[J]. J Agric Food Chem, 2000, 48(11): 5710-5714. DOI: 10.1021/jf000807a. |
| [9] | LU Z B, NIE G J, BELTON P S, et al. Structure-activity relationship analysis of antioxidant ability and neuroprotective effect of gallic acid derivatives[J]. Neurochem Int, 2006, 48(4): 263-274. DOI: 10.1016/j.neuint.2005.10.010. |
| [10] | MIR R, JALLU S, SINGH T P. The shikimate pathway:Review of amino acid sequence, function and three-dimensional structures of the enzymes[J]. Crit Rev Microbiol, 2015, 41(2): 172-189. DOI: 10.3109/1040841X.2013.813901. |
| [11] | WEAVER L M, HERRMANN K M. Dynamics of the shikimate pathway in plants[J]. Trends Plant Sci, 1997, 2(9): 346-351. DOI: 10.1016/S1360-1385(97)84622-5. |
| [12] | WILSON D J, PATTON S, FLOROVA G, et al. The shikimic acid pathway and polyketide biosynthesis[J]. J Ind Microbiol Biotechnol, 1998, 20(5): 299-303. DOI: 10.1038/sj.jim.2900527. |
| [13] | NIGOVIC B, ANTOLIC S, KOJIC-PRODIC B, et al. Correlation of structural and physico-chemical parameters with the bioactivity of alkylated derivatives of indole-3-acetic acid, a phytohormone (auxin)[J]. Acta Crystallogr, 2000, 56(1): 94-111. DOI: 10.1107/S0108768199006199. |
| [14] | SCHUTZ A, GOLBIK R, KONIG S, et al. Intermediates and transition states in thiamin diphosphate-dependent decarboxylases. A kinetic and NMR study on wild-type indolepyruvate decarboxylase and variants using indolepyruvate, benzoylformate, and pyruvate as substrates[J]. Biochemistry, 2005, 44(16): 6164-6179. DOI: 10.1021/bi0473354. |
| [15] | TIAN Y, ZHANG L M, WANG Y L, et al. Age-related topographical metabolic signatures for the rat gastrointestinal contents[J]. J Proteome Res, 2012, 11(2): 1397-1411. DOI: 10.1021/pr2011507. |
| [16] | ZHAO Y, WU J F, LI J V, et al. Gut microbiota composition modifies fecal metabolic profiles in mice[J]. J Proteome Res, 2013, 12(6): 2987-2999. DOI: 10.1021/pr400263n. |
| [17] | LIN H, AN Y P, HAO F H, et al. Correlations of fecal metabonomic and microbiomic changes induced by high-fat diet in the pre-obesity state[J]. Sci Rep, . DOI: 10.1038/srep21618. |
| [18] | WU X Y, LI N, LI H D, et al. An optimized method for NMR-based plant seed metabolomic analysis with maximized polar metabolite extraction efficiency, signal-to-noise ratio, and chemical shift consistency[J]. Analyst, 2014, 139(7): 1769-1778. DOI: 10.1039/C3AN02100A. |
| [19] |
WU X Y, LI N, TANG H R. Quantitative analysis of metabolites in Mungbean (Vigna Radiata) extracts using NMR techniques[J].
Chinese J Magn Reson, 2014, 31(4): 548-563.
吴香玉, 李宁, 唐惠儒. 绿豆(Vigna Radiata)代谢物组成的核磁共振定量分析[J]. 波谱学杂志, 2014, 31(4): 548-563. DOI: 10.11938/cjmr20140409. |
| [20] | KAWAI S, NAKATA K, ICHIZAWA H, et al. 3-(4-Hydroxyphenyl)propionic acid is involved in the biosynthesis of myricanol in Myrica rubra[J]. J Wood Sci, 2010, 56(2): 148-153. DOI: 10.1007/s10086-009-1082-9. |
| [21] | BAI N S, HE K, ROLLER M, et al. Flavonolignans and other constituents from lepidium meyenii with activities in anti-inflammation and human cancer cell lines[J]. J Agric Food Chem, 2015, 63(9): 2458-2463. DOI: 10.1021/acs.jafc.5b00219. |
| [22] | ZHANG J T, ZHANG Y, DU Y Y, et al. Dynamic metabonomic responses of tobacco (Nicotiana tabacum) plants to salt stress[J]. J Proteome Res, 2011, 10(4): 1904-1914. DOI: 10.1021/pr101140n. |
| [23] | LIU C X, HAO F H, HU J, et al. Revealing different systems responses to brown planthopper infestation for pest susceptible and resistant rice plants with the combined metabonomic and gene-expression analysis[J]. J Proteome Res, 2010, 9(12): 6774-6785. DOI: 10.1021/pr100970q. |
| [24] | CAPITANI D, SOBOLEV A P, TOMASSINI A, et al. Peach fruit:Metabolic comparative analysis of two varieties with different resistances toinsect attacks by NMR spectroscopy[J]. J Agric Food Chem, 2013, 61(8): 1718-1726. DOI: 10.1021/jf303248z. |
| [25] |
HUANG Y, SHAO H K, LI K, et al. Chemical analysis and activity evaluation of anti-inflammatory constituents of Fi-cus microcarpa L. f[J].
Chinese Traditional Patent Medicine, 2014, 36: 1227-1233.
黄洋, 邵慧凯, 李康, 等. 小叶榕叶抗炎成分分析及活性评价[J]. 中成药, 2014, 36: 1227-1233. DOI: 10.3969/j.issn.1001-1528.2014.06.025. |
| [26] | FORINO M, TARTAGLIONE L, DELL'AVERSANO C, et al. NMR-based identification of the phenolic profile of fruits of Lycium barbarum (goji berries). Isolation and structural determination of a novel N-feruloyl tyramine dimer as the most abundant antioxidant polyphenol of goji berries[J]. Food Chem, 2016, 194:1254-1259. |
| [27] | SCOTT K N. NMR parameters of biologically important aromatic acids. 2.phenylacetic acid and derivatives[J]. J Magn Reson, 1972, 6(1): 55-73. |
| [28] | SWISLOCKA R, PIEKUT J, LEWANDOWSKI W. The relationship between molecular structure and biological activity of alkali metal salts of vanillic acid:spectroscopic, theoretical and microbiological studies[J]. Spectroc Acta A-Mol Biomol Spectrosc, 2013, 100: 31-40. DOI: 10.1016/j.saa.2012.01.044. |
| [29] | WALKER T S, BAIS H P, HALLIGAN K M, et al. Metabolic profiling of root exudates of arabidopsis thaliana[J]. J Agric Food Chem, 2009, 57(19): 9346-9346. DOI: 10.1021/jf902670b. |
| [30] | GOTTLIEB H E, KUMAR S, SAHAI M, et al. Ethyl brevifolin carboxylate from Flueggea microcarpa[J]. Phytochemistry, 1991, 30(7): 2435-2438. DOI: 10.1016/0031-9422(91)83676-C. |
| [31] | VINOD K S, PERIANDY S, GOVINDARAJAN M. Spectroscopic analysis of cinnamic acid using quantum chemical calculations[J]. Spectroc Acta A-Mol Biomol Spectrose, 2015, 136: 808-817. DOI: 10.1016/j.saa.2014.09.098. |
| [32] | KINROSS J M, ALKHAMESI N, BARTON R H, et al. Global metabolic phenotyping in an experimental laparotomy model of surgical trauma[J]. J Proteome Res, 2011, 10(1): 277-287. DOI: 10.1021/pr1003278. |
| [33] | CAI R, ARNTFIELD S D, CHARLTON J L. Structural changes of sinapic acid and sinapine bisulfate during autoclaving with respect to the development of colored substances[J]. J Am Oil Chem Soc, 1999, 76(4): 433-441. DOI: 10.1007/s11746-999-0021-7. |
| [34] | AMIN R P, KUNAPARAJU N, KUMAR S, et al. Structure elucidation and inhibitory effects on human platelet aggregation of chlorogenic acid from Wrightia tinctoria[J]. J Complemen Integr Med, 2013, 10(1): 97-104. |
| [35] | |
| [36] | CONNELLY J C, CONNOR S C, MONTE S, et al. Application of directly coupled high performance liquid chromatography-NMR-mass spectometry and H-1 NMR spectroscopic studies to the investigation of 2, 3-benzofuran metabolism in Sprague-Dawley rats[J]. Drug Metab Dispos, 2002, 30(12): 1357-1363. DOI: 10.1124/dmd.30.12.1357. |
| [37] |
AN Y P, YANG X Y, LI H D, et al. NMR analysis of nicotinamide N-oxide and pseudouridine in rat urine[J].
Chinese J Magn Reson, 2014, 31(2): 232-242.
安艳捧, 杨晓艳, 李洪德, 等. 大鼠尿液中N-氧化烟酰胺和伪尿嘧啶核苷的NMR分析[J]. 波谱学杂志, 2014, 31(2): 232-242. |
| [38] |
TIAN Y, TANG H R. Identification and structural determination of saccharides in rat feces[J].
Chinese J Magn Reson, 2012, 29(3): 361-371.
田园, 唐惠儒. 大鼠粪样中几种糖类物质的结构确定[J]. 波谱学杂志, 2012, 29(3): 361-371. |
| [39] |
YANG X Y, WU X Y, AN Y P, et al. An NMR study on keto-enol tautomerism of indole-3-pyruvic acid[J].
Chinese J Magn Reson, 2014, 31(1): 81-90.
杨晓艳, 吴香玉, 安艳捧, 等. 吲哚丙酮酸的酮-烯醇互变异构化的NMR研究[J]. 波谱学杂志, 2014, 31(1): 81-90. |
 2017, Vol. 34
2017, Vol. 34