b Clinical Metabolomics Center, China Pharmaceutical University, Nanjing 211198, China;
c Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai 200032, China;
d College of Chinese Materia Medica and Food Engineering, Shanxi University of Chinese Medicine, Jinzhong 030619, China
Small peptides, particularly dipeptides and tripeptides, have recently attracted much attention due to their nutritional value and diverse biological functions. These peptides have demonstrated various beneficial effects, such as anti-hypertensive [1], vasorelaxant [2], and antioxidant properties [3]. Some dipeptides have also been proven to be involved in disease progression and hold potential to serve as biomarkers in clinics [4–7]. Additionally, dipeptides are essential ingredients that contribute to unique tastes of many fermented foods, including alcoholic beverages [8,9], soy sauce [10,11], and cheese [12]. Given the significance of small peptides, efforts have been advanced in their qualitative and quantitative analyses. However, because of the complexity arising from numerous structural isomers and the low abundance present in matrices, achieving rapid separation and precise quantification still remains a challenge.
Several mass spectrometry (MS)-based methods in combination with different separation techniques have been reported for the direct analysis of di/tripeptides, including reverse-phase liquid chromatography (RPLC) [13], hydrophilic interaction liquid chromatography (HILIC) [14], and capillary electrophoresis (CE) [15]. RPLC usually results in poor separation due to the strong polarity of di/tripeptides. HILIC chromatography allows for better retention of these polar compounds [16] but suffers from possible peak drifting and tailing. In a recent study, Hitoshi et al. combined two detection instruments, LC-MS and CE-MS, which enabled the quantitative analysis of 335 dipeptides [17]. Additionally, matrix-assisted laser desorption/ionization (MALDI) has emerged as an in-situ analytical method for di/tripeptides when coupled with MS [18–20]. Nevertheless, the absence of chromatographic separation and the interference caused by the matrix and complex background factors limit its application for high coverage of a large number of small peptides with low abundance.
Compared to direct analysis, chemical derivatization offers an alternative approach for the determination of small peptides through enhancing their detection sensitivity and retention on reversed-phase columns [21]. Several pre-column derivatization reagents, including phenyl isothiocyanate [22], 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate [8,23], and 2,4,6-trinitrobenzene sulfonate [24,25], which could rapidly react with the amino group, have been employed in the analyses of di/tripeptides. However, many of these studies have limitations in terms of the number of the small peptide species analyzed, with the majority of them only focusing on no more than 10 types of dipeptides. This limited coverage can be attributed to the differential derivatization efficiency of these reagents for different small peptides. Recently, some LC-MS methods have been reported for the analysis of di/tripeptides using chemical derivatization via dansylation, a method that targets the amine groups of di/tripeptides [26–28]. These approaches employed a commonly used reagent, namely dansyl chloride, for relative quantification of the di/tripeptides or annotation from biological samples with high coverages of up to 400 dipeptides. Despite these advancements, there is still a need to develop novel derivatization strategies to achieve absolute quantification of small peptides.
A recent study has reported that amines could react with ethoxy methylidine derivatives of β-keto amides under mild conditions to produce amide-substituted β-aminoenones with high (Z)-stereoselectivity, atomic efficiency and excellent yield [29]. To the best of our knowledge, quantitative LC-MS/MS analysis of amino compounds based on this mechanism has not yet been documented. Herein, we designed a three-step synthetic route to prepare an ethoxy methylidine derivative of β-keto amide named 4-(2-(ethoxymethylene)-3-oxobutanamido)-N,N,N-trimethylbenzenaminium iodide (EOTMBA). The synthetic method was easy to operate and was cost-effective. Notably, the effectiveness of chemical derivatization using EOTMBA was evaluated, and our findings demonstrated that the 97 small peptides (89 dipeptides and 8 tripeptides) (Table S1 in Supporting information) could be efficiently derivatized in methanol to produce stable amide-substituted β-aminoenones without the use of any catalyst. The EOTMBA labeling significantly enhanced the detection sensitivity of di/tripeptides, improved their retention, and facilitated the separation of isomers in the RPLC-MS/MS analysis. Generally, accurate quantifications of analytes largely depend on stable isotope standards. Stable isotope chemical tagging strategies have been widely employed to minimize complex matrix effects, enabling accurate determination of target analytes [30–37]. In this work, we also synthesized a paired isotope deuterium-labeled reagent d-EOTMBA for the first time. This reagent allowed for the generation of “one to one” internal standard for all di/tripeptides. Our developed chemical isotope labeling-assisted LC-MS/MS method achieved high coverage, high sensitivity and absolute quantification for 97 small peptides within a relatively short analytical time of 22 min. The established method was successfully applied to accurately quantify small peptides in various biological samples, including plasma, urine, and bile samples from both healthy individuals and patients with biliary tract diseases as well as extracts of the animal-derived medicinal material Eupolyphaga sinensis Walker.
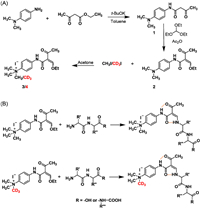
We firstly prepared the derivatization reagents EOTMBA and its paired isotope deuterium-labeled d-EOTMBA through three-step synthetic routes (Fig. 1A). NMR analyses were performed to characterize the intermediates 1, 2 as well as EOTMBA and d-EOTMBA. The 1H and 13C spectra are provided in the Supporting Information (Figs. S1-S8 in Supporting information). In addition, high-resolution mass spectrometry was applied to characterize EOTMBA and d-EOTMBA. The results showed that EOTMBA produced a molecular ion peak at m/z 291.1699 (M+, theoretical value 291.1703) and a fragment ion of m/z 276.1463 ([M-CH3]+, theoretical value 276.1468) in the positive ion mode (Fig. S9A in Supporting information). Similarly, d-EOTMBA generated a molecular ion peak at m/z of 294.1884 (M+, theoretical value 294.1891), a fragment ion at m/z 276.1463 ([M-CD3]+, theoretical value of 276.1468) and a fragment ion at 279.1652 ([M-CH3]+, theoretical value 279.1657) (Fig. S9B in Supporting information). These findings together provided evidence of the successful synthesis of the derivatization reagents EOTMBA and d-EOTMBA.
|
Download:
|
| Fig. 1. Reaction schemes for (A) synthesis of EOTMBA as well as its deuterium-labeled isotope reagent d3-EOTMBA and (B) di/tripeptides derivatized by EOTMBA or d3-EOTMBA. | |
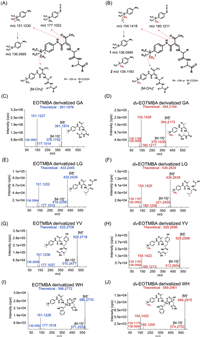
The di/tripeptides were derivatized by EOTMBA or d-EOTMBA to form stable amide-substituted β-aminoenones through aminolysis (Fig. 1B). This process is known to lead to the production of a highly (Z) selective product due to the existence of dual intramolecular hydrogen bonding through C=O···H-N interactions [29]. The confirmation of single chromatographic peaks for the derivatives in our analysis further validated this observation. Subsequently, we identified the derivatives and examined their fragmentation patterns by high-resolution mass spectrometry. In the positive ion mode, all the EOTMBA derivatives produced a product ion of [M-15]+ due to the loss of a methyl group from the quaternary ammonium moiety of the molecular ion (M+) (Fig. 2A). In addition, a product ion with m/z 177.1022 was generated, which could be assigned to the [4-isocyanato-N,N,N-trimethylbenzenaminium]+ (C10H13N2O) (Fig. 2A). Of note, after C-N bond cleavage, another product ion corresponding to [4-amino-N,N,N-trimethylbenzenaminium]+ (C9H15N2) exhibited the highest abundance at m/z 151.1230 (Fig. 2A). This product ion could further dissociate into a fragment ion of m/z 136.0995 (Fig. 2A). It is worth noting that di/tripeptides derivatized with d-EOTMBA showed similar fragmentation behaviors to those derivatized with EOTMBA. Specifically, product ions at m/z 180.1211, m/z 154.1418 and m/z 139.1183 were observed (Fig. 2B). The fragmentation patterns of EOTMBA- and d-EOTMBA-derivatized products of four typical dipeptides, including GA (containing the straight-chain amino acid glycine), LG (containing the branched-chain amino acid leucine), YV (containing the aromatic amino acid tyrosine), and WH (containing the heterocyclic amino acid tryptophan), are presented in Figs. 2C-J. Obviously, the precursor and product ions of di/tripeptides labeled with d-EOTMBA demonstrated a 3 Da increase compared to those labeled with EOTMBA. Based on the fragmentation patterns, the MRM parameters of the EOTMBA- and d-EOTMBA-derivatized products, including MRM transitions, Q1 values, collision energy, and Q3 values, were optimized and summarized in Table S2 (Supporting information).
|
Download:
|
| Fig. 2. Schematic MS/MS fragmentation patterns of (A) EOTMBA- and (B) d3-EOTMBA-derivatized di/tripeptides, and MS/MS spectra of fragment ions generated by EOTMBA-derivatized (C) GA, (E) LG, (G) YV and (I) WH as well as d3-EOTMBA-derivatized (D) GA, (F) LG, (H) YV and (J) WH. | |
To obtain the best reaction efficiency, we optimized the derivatization conditions, including the reaction temperature, molar ratio and reaction time. As shown in Fig. S10A (Supporting information), the peak areas of 97 di/tripeptides derivatives exhibited a significant increase as the temperature was raised from 20 ℃ to 60 ℃. Considering the potential volatility of methanol, we selected 60 ℃ as the optimal derivatization temperature. For the molar ratio, we investigated different ratios of substrate to EOTMBA, including 1:20, 1:50, 1:100, 1:150 and 1:200. The results showed that a molar ratio of 1:100 yielded the highest abundance of derivatives (Fig. S10B in Supporting information). We also considered the impact of reaction time on the labeling efficiency. As shown in Fig. S10C (Supporting information), the reaction time had no obvious influence on the peak areas of the derivatives and we finally selected 1.5 h as the suitable derivatization time. Taken together, the derivatization reaction of the di/tripeptides with EOTMBA was efficiently carried out at a molar ratio of 1:100 under 60 ℃ for 1.5 h. Under this optimized condition, we then examined the derivatization efficiencies by comparing the peak areas of dipeptide substrates (GA, LG, YV and WH) without and after EOTMBA derivatization. The results showed that the small peptides could be quickly converted to their derivatized products with conversion rates more than 90% (Figs. S11A-D in Supporting information).
The derivatization reagent EOTMBA contains a benzene ring in its structure, which could remarkably reduce hydrophilicity and increase the retention of the derivatized products on the reversed phase HSS T3 column. Prior to derivatization, we observed that most of the native di/tripeptides (91 out of 97) were coeluted within 4.5 min (Fig. 3A). After derivatization with EOTMBA, their chromatographic behavior was significantly improved, with elution times ranging from 1.6 min to 19.2 min (Fig. 3B and Table S3 in Supporting information). Of note, the structural isomers such as IA, LA and AI (Fig. S12A in Supporting information), LD, γ-EV and DL (Fig. S12C in Supporting information), IW and LW (Fig. S12E in Supporting information) were well retained and separated after EOTMBA labeling (Figs. S12B, D, and F in Supporting information). Most of di/tripeptides derivatives achieved baseline separation within a retention time of 22 min by LC-MS/MS analysis (Fig. 3B). Furthermore, the introduction of a quaternary amine moiety via EOTMBA derivatization significantly enhanced the ionization efficiency of the di/tripeptides derivatives. We determined the limit of quantification (LOQ) using a signal-to-noise ratio (S/N) close to 10, and the range was found to vary from 0.25 fmol/L to 5 nmol/L (Table S3). The sensitivity of our method was compared with previously reported approaches (Table S3). A previous work used 2,4,6-trinitrobenzene sulfonate as derivatization reagent for LC-MS/MS detection of dipeptides with LOQs of 0.25 nmol/L and 1.17 nmol/L for VY and MY, respectively (Table S3). In contrast, our method achieved LOQs of VY at 10 fmol/L and MY at 1 fmol/L, demonstrating a remarkable increase in sensitivity by 4–6 orders of magnitude (Table S3). Several other MS-based quantification methods for dipeptides reported limits of detections ranging from tens of pmol/L to hundreds of nmol/L (Table S3). Obviously, our method exhibited significantly enhanced detection sensitivities and this makes it possible to quantitatively analyze di/tripeptides in biological samples at ultra-trace levels.

|
Download:
|
| Fig. 3. Typical LC-MS/MS chromatograms of (A) underivatized di/tripeptides and (B) EOTMBA-derivatized di/tripeptides. | |
The developed methodology was then fully validated for linearity, accuracy, precision, recovery, matrix effect and stability. Calibration curves were established for 97 di/tripeptides over three consecutive days at 10 different concentrations. As shown in Table S4 (Supporting information), all calibration curves had good linearity with coefficient of determination (R2) greater than 0.99. The accuracy and precision were determined by analyzing mixed standards solution at low, medium and high concentration levels. The results showed that the intra-day accuracy was 86.3%–113.0% with coefficient of variation (CV) values at 0.6%-14.5% and the inter-day accuracy was 87.6%–109.8% with CV values ranging from 1.3% to 11.3%, suggesting good accuracy and precision were achieved (Table S5 in Supporting information). Recovery and matrix effects were assessed in different types of biological samples, including plasma, urine, bile and extract of Eupolyphaga sinensis Walker. As shown in Table S6 (Supporting information), the recoveries at 50%, 100% and 150% spiked levels were 70.1%–129.2% with CV values of 0.5%-13.9% in plasma, 70.8%–130.0% with CV values of 0.9%–15.0% in urine, 70.3%–113.4% with CV values of 0.4%-14.7% in bile, and 85.0%–115.0% with CV values of 0.4%–14.7% in extract of Eupolyphaga sinensis Walker. The matrix effect ranged from 74.7% to 130.7% with CV values less than 18.3% (Table S7 in Supporting information). In addition, the stability was also examined. The 24 h autosampler stability was determined by analyzing derivatized standard solutions and quality control (QC) samples with CV values below 12.8% (Table S8 in Supporting information). The long-term stability of 97 di/tripeptides standards (14 days at -80 ℃) ranged from 1.4% to 11.6% (Table S8). The CV values of short-term stability of derivatization reagents EOTMBA and d-EOTMBA (6 h and 12 h at room temperature as well as 6 h and 12 h at 4 ℃) were <6.4% (Table S9 in Supporting information). Overall, our method gave reliable and reproducible results in the quantification of di/tripeptides in complex biological samples.
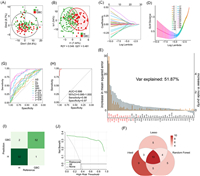
Biliary tract cancers, mainly including gallbladder cancer and cholangiocarcinoma, are highly aggressive tumors with poor prognosis [38]. Identification of reliable biomarkers to aid in their early diagnosis is therefore crucial. We utilized our method to quantitatively analyze di/tripeptides in 147 plasma, 49 urine and 46 bile samples collected from healthy individuals and patients with biliary tract diseases to identify potential biomarkers. All the participants signed the informed consent forms for the examination of their samples. All experiments with the clinical samples were conducted in accordance with the guidelines of Helsinki Declaration and approved by the Ethics Committee of Zhongshan Hospital (Shanghai, China). For the plasma samples, typical total ion chromatograms of the di/tripeptides generated from direct analysis and EOTMBA derivatization are presented in Fig. S13 (Supporting information). After labeling, the separation and MS response of the di/tripeptides were remarkably improved. A total of 83 di/tripeptides (78 dipeptides and 5 tripeptides) were accurately quantified (Table S10 in Supporting information). To determine the differences in di/tripeptides profiles in plasma between healthy individuals (n = 94) and patients with gallbladder cancer (n = 53), unsupervised principal component analysis (PCA) and supervised orthogonal partial least squares discriminant analysis (OPLS-DA) analyses were performed (Figs. 4A and B). The OPLS-DA model showed a significant distinction between the two groups with R2Y of 0.546 and Q2Y of 0.481(Fig. 4B). Lasso regression analysis (Figs. 4C and D), random forest (Fig. 4E) and Student’s t-test were applied to identify the differential di/tripeptides. A Venn diagram analysis revealed that 10 di/tripeptides were consistently identified as differential markers using all the three statistical screening methods (Fig. 4F and Table S11 in Supporting information). Among them, the levels of γ-EM, γ-EW, γ-EA, AS, β-AH, ST and γ-EIG in patients with gallbladder cancer were significantly lower relative to the healthy group (Table S11). Receiver operating characteristic (ROC) curves were constructed to evaluate the diagnostic performance of the 10 potential di/tripeptides biomarkers, of which 5 had a calculated area under the curve (AUC) value greater than 70%, with good confidence intervals, sensitivity and specificity (Fig. 4G and Table S12 in Supporting information). We optimized the biomarker panel of di/tripeptides with enumeration method, and found that the combination of 9 dipeptides, γ-EM, γ-EW, γ-EA, AS, β-AH, ST, WH, LL and WE, provided the best diagnostic performance with an AUC of 0.996 (Fig. 4H). The confusion matrix of the logistic regression model indicated that out of 94 healthy individuals, 92 were correctly predicted, while out of 53 patients with gallbladder cancer, 52 were correctly predicted (Fig. 4I). Decision curve analysis demonstrated that the combined model of 9 dipeptides had a good net benefit in predicting gallbladder cancer (Fig. 4J).
|
Download:
|
| Fig. 4. Di/tripeptides profiling of plasma samples for clinical diagnosis of patients with gallbladder cancer. Score plots of (A) principal component analysis (PCA) and (B) orthogonal partial least squares discriminant analysis (OPLS-DA). (C) Screening of the potential differential di/tripeptides by lasso regression of machine learning method. (D) Cross-validation of the constructed signature. (E) Screening of the potential differential di/tripeptides by random forest. The di/tripeptides marked red represent the potential differential biomarkers with P values less than 0.05. (F) Venn diagram of the differential di/tripeptides screened by lasso regression, random forest and Student’s t-test. Receiver operating characteristic (ROC) curve analyses of (G) the 10 di/tripeptides biomarker candidates and (H) the combination of 9 differential dipeptides for differentiating patients with gallbladder cancer from healthy individuals. (I) Illustrative confusion matrix for the disease prediction model. (J) Decision curve analysis (DCA) for predicting gallbladder cancer. H, heathy; GBC, gallbladder cancer. | |
For the urine samples, improved separation and MS response of di/tripeptides were achieved through EOTMBA derivatization (Fig. S14 in Supporting information). A total of 87 di/tripeptides were quantified (Table S13 in Supporting information). The PCA and OPLS-DA analyses demonstrated a limited distinction between the healthy individuals (n = 32) and patients with biliary tract cancer (n = 17) (Figs. S15A and B in Supporting information). The differential di/tripeptides between the two groups were screened by lasso regression analysis (Figs. S15C and D in Supporting information), random forest (Fig. S15E in Supporting information), Student’s t-test, and Venn diagram analysis (Fig. S15F in Supporting information). A total of 5 differential di/tripeptides, namely, γ-EIG, DL, β-AH, IE and DF, were identified as potential biomarkers (Table S14 in Supporting information). Remarkably, all 5 di/tripeptides demonstrated calculated AUC values higher than 70% for the diagnosis of patients with biliary tract cancer (Fig. S15G and Table S15 in Supporting information) and their combination exhibited an optimal diagnostic performance with an AUC of 0.893 (Fig. S15H). The confusion matrix correctly predicted 31 out of 32 healthy individuals and 12 out of 17 patients with biliary tract cancer (Fig. S15I in Supporting information). Decision curve analysis confirmed the net benefit of prediction model based on the 5 di/tripeptides for predicting patients with biliary tract cancer (Fig. S15J in Supporting information).
Additionally, our method was applied to profile the di/tripeptides in bile samples collected from 27 patients with benign biliary conditions and 19 patients with biliary tract cancer. Obviously, EOTMBA derivatization resulted in improved separation and increased MS response of di/tripeptides in bile (Fig. S16 in Supporting information). In total, 68 di/tripeptides were quantified (Table S16 in Supporting information). The PCA and OPLS-DA analyses showed no significant distinction between the two groups (Figs. S17A and B in Supporting information). Three dipeptides, including WN, GN and EF, were identified as potential biomarkers using the three statistical screening methods (Figs. S17C–E in Supporting information) and Venn diagram analysis (Fig. S17F in Supporting information). As indicated in Table S17 (Supporting information), all the 3 dipeptides were significantly increased in patients with biliary tract cancer compared with patients with benign biliary conditions. The calculated AUC values of the 3 dipeptides were found to be greater than 70% (Fig. S17G and Table S18 in Supporting information). The optimized panel consisted of 2 dipeptides, WN and GN, provided an AUC of 0.870, indicating a good diagnostic performance (Fig. S17H in Supporting information). The confusion matrix for the diagnostic model indicated a higher prediction probability. Specifically, it correctly predicted 12 out of 13 patients with benign biliary conditions and 15 out of 19 patients with biliary tract cancer (Fig. S17I in Supporting information). Additionally, the decision curve analysis provided further support for the combined diagnostic model, demonstrating its favorable net benefit in accurately predicting biliary tract cancer (Fig. S17J in Supporting information). All these results together demonstrate that our method significantly improved the detection of di/tripeptides in complex biological samples and highlight its immense promise in the early clinical diagnosis of patients with biliary tract cancer.
As a type of animal-derived traditional Chinese medicine, Eupolyphaga sinensis Walker contains proteins, polypeptides, and amino acids. There have been limited reports on the presence of di/tripeptides in Eupolyphaga sinensis Walker. We employed our developed method to analyze di/tripeptides in 11 batches of Eupolyphaga sinensis Walker. The application of EOTMBA derivatization significantly enhanced the detection coverage of di/tripeptides and improved their chromatographic behavior, as depicted in Fig. S18 (Supporting information). A total of 81 di/tripeptides were successfully identified and quantified, with their respective contents listed in Table S19 (Supporting information). The total content of di/tripeptides in Eupolyphaga sinensis Walker extracts ranged from 194.751 to 664.698 μg/g. It is worthy of note that, this is the first report detailing the characterization of di/tripeptides in Eupolyphaga sinensis Walker. These findings provide insights into the profiling of di/tripeptides in Eupolyphaga sinensis Walker, which can serve as a foundation for further characterization and content analysis related to this traditional Chinese medicine.
In summary, we developed a chemical isotope-labeled quantitative strategy to simultaneously analyze 97 di/tripeptides by LC-MS/MS. We synthesized EOTMBA and its paired d-EOTMBA as novel derivatization reagents. The derivatization with EOTMBA significantly improved the chromatographic separation of the di/tripeptides and remarkably enhanced their detection sensitivities with LOQs ranging from 0.25 fmol/L to 5 nmol/L. Using this method, we quantified di/tripeptides in clinical plasma, urine and bile samples, and identified some di/tripeptides as potential diagnostic biomarkers for patients with biliary tract cancer. Furthermore, we successfully applied this method to quantify di/tripeptides in the extract of Eupolyphaga sinensis Walker. Overall, our analytical method enabled sensitive and accurate determination of di/tripeptides in various biological samples with complex matrices. The ultra-sensitivity of our method also highlights the possibility of utilizing it to detect previously unreported di/tripeptides, showing great promise for potential application in disease diagnosis and quality control of traditional Chinese medicines.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsWe are thankful for Hai-Tao Meng at Shimadzu (China) Co., Ltd for providing technical support in mass spectrometry work. This work was financially supported in part by the National Natural Science Fund of China for Distinguished Young Scholars (No. 81825023), National Natural Science Foundation of China (No. 82003979), and Natural Science Foundation of Shanghai (No. 23ZR1459100).
Supplementary materialsSupplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2024.109670.
| [1] |
M. Sasai, X. Sun, C. Okuda, et al., Biochem. Biophys. Res. Commun. 503 (2018) 1070-1074. DOI:10.1016/j.bbrc.2018.06.118 |
| [2] |
M. Tanaka, M. Tokuyasu, T. Matsui, K. Matsumoto, Life Sci. 82 (2008) 869-875. DOI:10.1016/j.lfs.2008.02.001 |
| [3] |
L. Zheng, Y. Zhao, H. Dong, G. Su, M. Zhao, J. Funct. Foods 21 (2016) 485-496. DOI:10.1016/j.jff.2015.12.003 |
| [4] |
T. Soga, M. Sugimoto, M. Honma, et al., J. Hepatol. 55 (2011) 896-905. DOI:10.1016/j.jhep.2011.01.031 |
| [5] |
M. Saoi, K. Sasaki, H. Sagawa, et al., J. Proteome Res. 19 (2020) 2689-2699. DOI:10.1021/acs.jproteome.9b00405 |
| [6] |
Z. Yang, D. Wu, S. Lu, et al., Proc. Natl. Acad. Sci. U. S. A. 119 (2022) e2117089119. DOI:10.1073/pnas.2117089119 |
| [7] |
J. Li, J. Li, H. Wang, et al., Gastroenterology 157 (2019) 257-259.e5. DOI:10.1053/j.gastro.2019.03.020 |
| [8] |
K. Takahashi, M. Tokuoka, H. Kohno, et al., J. Chromatogr. A 1242 (2012) 17-25. DOI:10.1016/j.chroma.2012.03.076 |
| [9] |
E. Sherman, M. Coe, C. Grose, D. Martin, D.R. Greenwood, J. Agric. Food Chem. 68 (2020) 13380-13396. DOI:10.1021/acs.jafc.0c04095 |
| [10] |
S. Yamamoto, K. Shiga, Y. Kodama, et al., J. Biosci. Bioeng. 118 (2014) 56-63. DOI:10.1016/j.jbiosc.2013.12.019 |
| [11] |
M. Jünger, V.K. Mittermeier-Kleßinger, A. Farrenkopf, et al., J. Agric. Food Chem. 70 (2022) 6503-6518. DOI:10.1021/acs.jafc.2c01688 |
| [12] |
S. Toelstede, A. Dunkel, T. Hofmann, J. Agric. Food Chem. 57 (2009) 1440-1448. DOI:10.1021/jf803376d |
| [13] |
X. Li, C. Yao, Y. Li, et al., J. Pharm. Anal. 12 (2022) 263-269. DOI:10.1016/j.jpha.2021.07.007 |
| [14] |
M. Gallego, F. Toldrá, L. Mora, Food Chem. 370 (2022) 130977. DOI:10.1016/j.foodchem.2021.130977 |
| [15] |
A. Hirayama, K. Igarashi, M. Tomita, T. Soga, J. Chromatogr. A 1369 (2014) 161-169. DOI:10.1016/j.chroma.2014.10.007 |
| [16] |
Y. Tang, R. Li, G. Lin, L. Li, Anal. Chem. 86 (2014) 3568-3574. DOI:10.1021/ac500109y |
| [17] |
H. Ozawa, A. Hirayama, T. Ishikawa, et al., Anal. Chem. 92 (2020) 9799-9806. DOI:10.1021/acs.analchem.0c01258 |
| [18] |
A. Heres, C. Saldaña, F. Toldrá, L. Mora, Food Chem. Mol. Sci. 3 (2021) 100048. DOI:10.1016/j.fochms.2021.100048 |
| [19] |
K. Li, S. Guo, W. Tang, B. Li, Chem. Commun. 57 (2021) 12460-12463. DOI:10.1039/D1CC05026E |
| [20] |
V.V. Ilyushenkova, M.E. Zimens, N.Y. Polovkov, et al., Talanta 253 (2023) 123922. DOI:10.1016/j.talanta.2022.123922 |
| [21] |
B.L. Qi, P. Liu, Q.Y. Wang, et al., Trends Analyt. Chem. 59 (2014) 121-132. DOI:10.1016/j.trac.2014.03.013 |
| [22] |
M. Aito-Inoue, K. Ohtsuki, Y. Nakamura, et al., J. Agric. Food Chem. 54 (2006) 5261-5266. DOI:10.1021/jf060531s |
| [23] |
A.D. Askretkov, A.A. Klishin, D.I. Zybin, et al., J. Anal. Chem. 75 (2020) 1038-1045. DOI:10.1134/S1061934820080031 |
| [24] |
E.M.N. Nakashima, H.Q. Qing, M. Tanaka, T. Matsui, Biosci. Biotechnol. Biochem. 77 (2013) 2094-2099. DOI:10.1271/bbb.130448 |
| [25] |
V.T. Hanh, Y. Kobayashi, M. Maebuchi, et al., Food Chem. 190 (2016) 345-350. DOI:10.1016/j.foodchem.2015.05.053 |
| [26] |
Z. Cheng, L. Li, Anal. Chem. 95 (2023) 6629-6636. DOI:10.1021/acs.analchem.2c05796 |
| [27] |
S. Tang, P. Zhang, M. Gao, et al., Anal. Chim. Acta. 1274 (2023) 341570. DOI:10.1016/j.aca.2023.341570 |
| [28] |
X. Lu, P. Dou, C. Li, et al., J. Proteome. Res. 22 (2023) 1896-1907. DOI:10.1021/acs.jproteome.3c00002 |
| [29] |
P. Subramaniam, C. Ramasubbu, S. Athiramu, Green Chem. 19 (2017) 2541-2545. DOI:10.1039/C7GC00909G |
| [30] |
F.L. Liu, T.T. Ye, J.H. Ding, et al., Anal. Chem. 93 (2021) 6848-6856. DOI:10.1021/acs.analchem.1c00915 |
| [31] |
S. Li, F.L. Liu, Z. Zhang, et al., Anal. Chem. 94 (2022) 4866-4873. DOI:10.1021/acs.analchem.2c00346 |
| [32] |
S.L. Wang, Y. Wang, L. Wu, et al., Anal. Chem. 94 (2022) 3590-3599. DOI:10.1021/acs.analchem.1c04924 |
| [33] |
S. Zhao, L. Li, Anal. Chem. 90 (2018) 13514-13522. DOI:10.1021/acs.analchem.8b03435 |
| [34] |
C.F. Wang, L. Li, Anal. Chem. 94 (2022) 11650-11658. DOI:10.1021/acs.analchem.2c02220 |
| [35] |
Y. Hu, X. Hong, Z. Yuan, et al., Chin. Chem. Lett. 34 (2023) 108023. DOI:10.1016/j.cclet.2022.108023 |
| [36] |
H. Zhao, J. Li, X. Ma, et al., Chin. Chem. Lett. 29 (2018) 102-106. DOI:10.1016/j.cclet.2017.06.013 |
| [37] |
H. Xiao, P. Liu, S. Zheng, et al., Chin. Chem. Lett. 31 (2020) 2423-2427. DOI:10.1016/j.cclet.2020.03.003 |
| [38] |
J.W. Valle, R.K. Kelley, B. Nervi, D.Y. Oh, A.X. Zhu, Lancet 397 (2021) 428-444. DOI:10.1016/S0140-6736(21)00153-7 |
 2024, Vol. 35
2024, Vol. 35 

