b School of Pharmacy, Shandong Academy of Sciences, Qilu University of Technology, Ji'nan 250353, China;
c College of Chemical Engineering, North China University of Science and Technology, Tangshan 063210, China
Riding the wave of human genome decryption [1], nucleic acid-derived therapeutics have emerged as a new paradigm in the arsenal of modern medicine for addressing various diseases at genetic level. Meanwhile, localized treatment with small interfering RNA (siRNA), such as nasal siRNA delivery [2], is still in the early stages of clinical trials. The nasal mucosa, as the first line defense against pathogens and aeroallergens, forms a continuous barrier comprised of tight junctions, adherens junctions, desmosomes and hemidesmosomes [3], which restricted the passage of molecules larger than 1 kDa [4]. Additionally, the proteases and nucleases in the nasal cavity potentially undermine the integrity of biological drug molecules [5].
Herein, we report, for the first time, the combined use of ionic liquids (ILs) and cationic lipids to achieve in situ formation of cationic lipid nanocomplexes (CLNs), with the dual purposes of enhancing siRNA mucosa penetration and improving gene transfection. (2,3-Dioleoyloxy-propyl)-trimethylammonium-chloride (DOTAP) was primarily chosen as the cationic lipid for our formulation to enhance gene transfection [6]. Firstly, our research utilized the superior solubilizing ability of ILs [7] to load the lipophilic DOTAP and the hydrophilic siRNA, forming siRNA-DOTAP-ILs (siRNA-DILs) formulation. Next, as well-reported penetration enhancer [8,9], ILs were expected to enhance the penetration of siRNA-DILs components across the nasal mucosa. During penetration, the internal structure of water-miscible ILs would gradually break down, allowing dynamic in situ self-assembly of CLNs from siRNA-DILs to load siRNA for enhanced transfection.
Experimental section in this study was detailed in Supporting information. We synthesized seven choline-based ILs (Table S1 in Supporting information) [10] to assess their compatibility of forming CLNs with DOTAP. The ILs were characterized by proton magnetic resonance (1H NMR) (Figs. S1–S7 in Supporting information). We prepared siRNA-DILs or blank DILs as described in Fig. 1A, successfully dispersing up to 50% of DOTAP in the ILs without visible stratification. Upon hydration, [Ch][Ger] with 1%–50% of DOTAP could form CLNs with sizes ranging from 100 nm to 200 nm and polydispersity index (PDI) below 0.3 (Fig. S8A in Supporting information). Both [Ch][Ole] and [Ch][Lin] were also capable of forming nanoparticles with 1%–30% of DOTAP (130–280 nm, PDI < 0.5). Furthermore, zeta potential measurement indicated that only [Ch][Ger] loaded with more than 10% of DOTAP could self-assemble into positively charged CLNs (Fig. S8B in Supporting information). Such results suggested that the ILs anions played a crucial role in governing the morphology and surface charges of CLNs. Anions derived from geranic acid, oleic acid and linoleic acid possessed backbone chains with more than 8 carbons and lacked hydrophilic groups, resulting in higher compatibility in forming CLNs with DOTAP. Furthermore, the difference in surface charges indicated that the ILs anions directly participated in CLNs formation, and the proportion of ILs anions in CLNs was inversely correlated with zeta potential. To some extent, the carbon chain of anions in [Ch][Ole] and [Ch][Lin] resembled that of DOTAP, leading to an excess of negatively charged ions in CLNs that could potentially impede siRNA transfection. Therefore, [Ch][Ger] was solely chosen as the ILs for our DILs formulation in the following investigations.
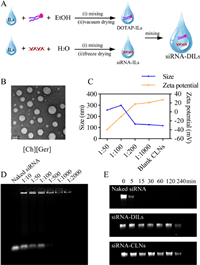
|
Download:
|
| Fig. 1. Characterization of self-assembled CLNs. (A) Illustration of siRNA-DILs preparation process. (B) TEM images of siRNA-CLNs sampled from the in vitro release experiment. Scale bar: 100 nm. (C) Sizes and zeta potentials of self-assembled siRNA-CLNs with different loading ratios. (D) Gel retarding assay to determine siRNA loading efficiency. (E) RNase degradation assay. | |
The in vitro release of siRNA from DILs revealed that the total siRNA release only reached 35%, 18% and 13% with 10%, 30% and 50% DOTAP, respectively (Fig. S8C in Supporting information). Such results indicated that exorbitant DOTAP contents increased the lipophilicity of DILs and trapped siRNA within the formulation, potentially impeding siRNA permeation in nasal mucosa. Therefore, [Ch][Ger] containing 10% DOTAP was chosen as our DILs formulation. The transmission electron microscopy (TEM) images of CLNs sampled from the in vitro release experiments demonstrated that siRNA loaded in [Ch][Ger] containing 10% DOTAP was able to form homogeneous sphere-like CLNs of approximately 100 nm after diffusing into the hydrated environment (Fig. 1B). Similar siRNA diffusion experiments were performed using other hydrophilic ILs as substitutes for [Ch][Ger], which ended up with unsatisfying lipid complexes (Fig. S8D in Supporting information).
The loading ratio (siRNA: DILs, w/w) could significantly affect the size and zeta potential of self-assembled CLNs. The incorporation of siRNA in CLNs led to an increase in particle size and a gain in negative charges (Fig. 1C). When the loading ratio was less than 1:200, the sizes and surface charges were comparable to blank CLNs (117 nm, 27.2 mV), which was desirable for efficient siRNA transfection. Regardless of the loading ratios, siRNA-CLNs achieved the smallest particle size at 1 h, indicating that the self-assembly of CLNs was a dynamic process (Fig. S8E in Supporting information). Furthermore, the gel retarding assay depicted that less siRNA migrated towards the positive pole with the increase in the use of DILs (Fig. 1D). The RNA quantification assay (Fig. S8F in Supporting information) also indicated that the encapsulation efficiency reached 90.0% when loading ratio was 1:500.
In addition, we performed RNase resistance assay to test our hypothesis that DILs formulation, as well as the formation of CLNs, could protect siRNA from degradation by RNase. Both DILs and self-assembled CLNs could effectively protect siRNA for at least 4 h, while naked siRNA was completely degraded by RNase within 15 min (Fig. 1E). Based on current literature [11–14], we suppose that CLNs could act as physical barriers to prevent RNase binding. Meanwhile, ILs contribute to siRNA stability owing to hydrophobic and polar interactions between ions and siRNA backbone, so that 2′-OH, the critical site of siRNA for degradation, was not recognized by RNase.
Molecular dynamics (MD) simulation was conducted to elucidate the self-assembly process of siRNA-CLNs at the molecular level (Fig. 2A). According to Fig. 2B, DOTAP cations aggregated around the terminal of siRNA. A portion of choline cations and geranic anions diffused into water. Additionally, a significant number of choline cations were found to intercalate with siRNA. Moreover, we spotted micelle-like structures around siRNA and in water, which were composed of 4–10 geranic anions (Fig. 2C). These geranic clumps were also observed to fill the gaps within DOTAP aggregated cluster, somewhat stabilizing the CLNs structure. This phenomenon supports our hypothesis that the anions of ILs engaged in the formation of CLNs. The unique ability of geranic clumps to support DOTAP clusters may be the critical key to explaining why [Ch][Ger] was the only suitable ILs in our study to form uniform and nano-sized CLNs with DOTAP. Concentration profile analysis of MD results also coincided with our observation above (Fig. S9 in Supporting information). Subsequently, radial distribution function analysis (RDF) of MD results suggested that, regarding the interaction with siRNA, the highest probability was calculated between the hydroxyl group of choline and the phosphate group of siRNA at a separation of 3.5 Å after hydration (Fig. 2D), presumably due to hydrogen bonding. Strong interaction was found between the hydroxyl group of choline and carboxyl group of geranic anion at 2.5 Å, which was reduced by half after hydration (Fig. 2E). The intermolecular interaction among the carboxyl groups of geranic ions at 4.0 Å became stronger after hydration (Fig. 2F), indicating the cluster formation of geranic anions.
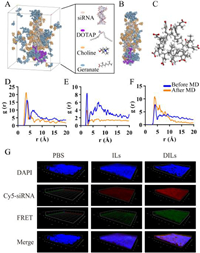
|
Download:
|
| Fig. 2. Investigation of CLNs self-assembly. (A) Snapshot of MD simulation unit with water hidden. (B) The formation of siRNA-loaded CLNs. (C) Micelle-like structure composed of 10 geranic anions after diffusing into water. (D) RDF analysis between siRNA phosphodiester groups and choline hydroxyl groups. (E) RDF analysis between choline hydroxyl groups and geranate carboxyl groups. (F) RDF analysis among intermolecular geranic carboxyl groups. (G) CLSM z-stack scanning of intact nasal mucosa for investigation of siRNA-CLNs in situ self-assembly from siRNA-DILs based on FRET effect. | |
Apart from in silico investigation, the fluorescence resonance energy transfer (FRET) technology [15] was employed to visualize the in situ self-assembly process of CLNs (Fig. S10A in Supporting information). When Cy3-siRNA and Cy5-siRNA were simultaneously loaded into self-assembled CLNs, the solution exhibited distinct emission peak at 665 nm under 552 nm excitation, which diminished when CLNs were dissembled by adding tetrahydrofuran (Fig. S10B in Supporting information). Hence, the FRET effect resulting from Cy3-siRNA and Cy5-siRNA assembly could be utilized as an indicator for in situ formation of CLNs. The z-stack confocal laser scanning microscopy (CLSM) images of rat mucosa penetration under the same parameter settings (Fig. 2G) revealed that only Cy3/Cy5-siRNA-DILs exhibited prominent FRET effect across the mucosal layer. In Cy3/Cy5-siRNA-ILs group, weak FRET effect was also spotted on the mucosa surface. We speculate the reason under such phenomenon was that ILs could temporarily form geranic micelles when diluted by nasal mucus to certain concentration, which could load Cy3/Cy5-siRNA to induce FRET effect but was highly unstable and broke down during mucosa penetration. Thus, a preliminary conclusion can be drawn that siRNA-CLNs were self-assembled from siRNA-DILs during the process of mucosa penetration. Nonetheless, advanced investigation is urgently required to further validate the in situ CLNs formation.
Efficient cellular uptake of self-assembled siRNA-CLNs is the prerequisite for siRNA to elicit gene silencing effects. In our study, three different cell lines, namely HNEpC, Calu-3 and P815, were incorporated to investigate the influence of loading ratios on cellular uptake efficiency. HNEpC and Calu-3 are human epithelial cell lines [16,17], while P815 belongs to mouse mast cell lineage [18]. The cytotoxicity of siRNA-CLNs was evaluated to determine the tolerated dose range (Fig. S11A in Supporting information). Cellular uptake efficiency was measured by flow cytometry (Figs. S11B and C in Supporting information) and visualized with CLSM (Fig. S11D in Supporting information). Generally, higher cellular uptake efficiency was achieved with higher loading ratios. For HNEpC and Calu-3 cells, when the loading ratio was set as 1:500, the siRNA delivery efficiency was comparable to that of lipofectamine 2000. For P815 cells, FAM-siRNA-CLNs with loading ratios of 1:500 and 1:1000 even outperformed lipofectamine 2000.
The in vivo mucosa penetration of FAM-siRNA was evaluated. Based on the CLSM images of mucosa sections (Fig. 3A), ILs enabled siRNA to be evenly distributed within the mucosa, while DILs enabled siRNA to be closely assembled around the cell nucleus, suggesting enhanced transfection efficiency. Moreover, the nasal retention of Cy5-siRNA following nasal administration was inspected using in vivo imaging system (IVIS) (Fig. S11E in Supporting information). Compared to naked siRNA, ILs significantly retained siRNA in the nasal cavity for up to 4 h, but only 20.3% of siRNA remained after 12 h (Fig. 3B), which might be down to immune clearance and siRNA degradation. DILs managed to retain over 40.0% siRNA after 12 h, allowing sufficient time to elicit gene silencing effects.
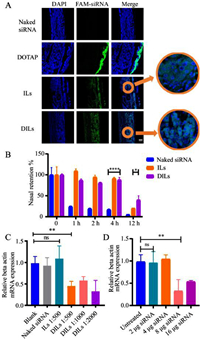
|
Download:
|
| Fig. 3. In vivo evaluation of siRNA-DILs nasal delivery. (A) CLSM images of siRNA penetration across nasal mucosa. Scale bar: 100 µm. (B) Nasal retention percentages of Cy5-siRNA delivered by phosphate buffer saline (PBS), ILs and DILs (n = 3). (C) Relative beta actin mRNA expression of rat mucosa treated with siRNA-DILs of different loading ratios (n = 6). (D) Relative beta actin mRNA expression of rat mucosa treated with siRNA-DILs of different siRNA dosages (n = 6). Data are represented as mean ± standard deviation (SD). *P < 0.05, **P < 0.01, ****P < 0.0001. ns, not significant. | |
To further validate the gene silencing effect of siRNA-DILs, the in vivo gene silencing efficacy was evaluated at the mRNA expression level using beta-actin-targeted siRNA on a rat model. Two days after nasal delivery, the nasal mucosa was sampled from the nasal septum and subjected to real-time polymerase chain reaction (PCR) analysis. Pure ILs did not enable siRNA to elicit significant gene silencing effects. A single dose of 16 µg siRNA in DILs resulted in reduction of beta-actin mRNA level by over 50% (Fig. 3C). However, in stark contrast to the cellular uptake results, siRNA-DILs with loading ratios of 1:500, 1:1000 and 1:2000 did not show a significant difference in gene silencing efficacy. It raises questions about the in vitro–in vivo correlation of CLNs self-assembly, which will be addressed in future investigation. Considering the proper volume of DILs to be evenly distributed in the rat nasal cavity, a loading ratio of 1:2000 was preliminarily set. Next, a range of siRNA doses were evaluated in rats (Fig. 3D). The results indicated that a mere 8 µg of siRNA in DILs could immensely reduce beta-actin mRNA expression by 68%. Therefore, 8 µg of siRNA in 16 mg DILs was eventually chosen as our siRNA-DILs formulation for the subsequent allergic rhinitis (AR) rat model. It should be noted that the gene silencing efficacy is also dependent on the siRNA sequence and therapeutic targets [19].
The therapeutic effects of siRNA-DILs were further investigated in rat AR model. In this study, the researchers targeted the calcium-activated K+ channel 3.1 (KCa3.1), which is upregulated in AR condition and plays a key role in allergic and inflammatory responses [20,21]. Recent evidence suggested that inhibition of KCa3.1 could ameliorate airway inflammation and hyperresponsiveness [18,22,23]. The AR model was induced with ovalbumin (OVA) and treated with KCa3.1-targeting siRNA (siKCa3.1) loaded in DILs (Fig. 4A). A group treated with non-coded siRNA (siNC) in DILs was included as a control to rule out non-specific effects. Treatments with siKC3.1-DILs resulted in attenuated AR symptoms (sneezes and nose scratches, Fig. 4B) as well as reduced levels of IgE (Fig. 4C), prostaglandin D2 (PGD2) (Fig. 4D) and histamine (Fig. 4E) in serum and nasal lavage fluid. In addition, the mRNA levels of KCa3.1 (Fig. 4F), MUC5AC (gel-forming mucin secreted by goblet cells [24], Fig. 4G) and tryptase (existed in secretory granules of mast cells [25], Fig. 4H) were elevated in the AR condition, and treatment with siKCa3.1-DILs significantly suppressed their expression. The mRNA levels of MUC5AC and tryptase positively correlated with KCa3.1 levels, indicating the importance of KCa3.1 in regulating the activity of mast cells and goblet cells. Histopathological examination of the nasal mucosa of AR rat confirmed the above findings (Fig. 4I). Specifically, Alcian blue and periodic acid Schiff (AB-PAS) staining revealed increased acidic mucus secretion in the AR progression, which was down-regulated with siKCa3.1-DILs treatment. The number of goblet cells did not show significant differences among the groups (Fig. 4J), possibly owing to mucosal destruction in the AR groups. Hematoxylin-eosin (H&E) staining revealed inflammatory cell infiltration and mucosal edema in untreated AR rats and siNC-DILs treated rats, while siKCa3.1-DILs treatment resulted in more orderly arranged epithelial cells with fewer inflammatory cells. Toluidine blue staining suggested mast cell infiltration (Fig. 4K) and increased mucosal thickness (Fig. 4L) in the untreated AR group, which were reduced in the siKCa3.1-DILs treated group. Immunohistochemical staining confirmed the reduced expression of KCa3.1 in the siKCa3.1-DILs treated group, as well as the restoration of zonula occludens protein 1 (ZO-1) levels (Fig. 4M). The latter represented the restoration of tight junctions in the nasal mucosa and suggested that the use of DILs as penetration enhancers did not cause irreversible damage to the tight junctions of the nasal mucosa.
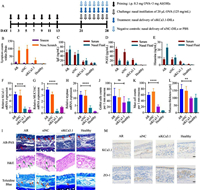
|
Download:
|
| Fig. 4. Evaluation of treating AR with siRNA-DILs (n = 10). (A) Time schedule of AR induction and treatment in rats. (B) Total counts of sneezes and nose scratches within 20 min. ELISA essay of IgE (C), PGD2 (D) and histamine (E) in rat serum and nasal lavage fluid. Relative mRNA expression of (F) KCa3.1, (G) MUC5AC and (H) tryptase measured by RT-qPCR. (I) Histopathological sections of rat nasal mucosa (mucus secretion, inflammatory cell infiltration and mast cell infiltration were marked with red, green and orange arrows, respectively). Scale bar: 40 µm. (J) Average number of goblet cells per 1 mm mucosa. (K) Average mucosa thickness. (L) Average number of mast cells per 1 mm mucosa. (M) KCa3.1 and ZO-1 immunohistochemical staining of nasal mucosa. Scale bar: 40 µm. Data are represented as mean ± SD. **P < 0.01, ****P < 0.0001. | |
It should be noted that all the animal experiments above were performed based on the concept of "reduction, replacement, and refinement", and the procedures were examined by the Institutional Animal Care and Use Committee at School of Pharmacy, Fudan University.
In conclusion, this study validated that [Ch][Ger] could support the in situ dynamic self-assembly of CLNs for localized siRNA treatment. Our formulation of DILs integrated the penetration ability of ILs and the gene transfection ability of cationic lipids, resulting in higher gene silencing effects at a lower dose. Our attempt to treat AR with siRNA-DILs yielded promising results, leading us to believe that such a platform could be applied to address malignant diseases like nasopharyngeal carcinoma. However, the concept of DILs is still in its preliminary stage and could be further explored as follows. Firstly, the ILs and lipid components could be optimized for higher biocompatibility. Secondly, delivery of other nucleic acid-based therapeutics could be investigated in DILs platforms, especially considering the immense demand for mRNA vaccine during the coronavirus disease (Covid-19) pandemic. Thirdly, the adverse effects associated with other siRNA delivery platforms should be thoroughly investigated with DILs, including the exacerbation of inflammation and the toxicity of excipients. Lastly, the long-term stability of siRNA in DILs should be examined.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentThe work was supported by National Natural Science Foundation of China (No. 82073801).
Supplementary materialsSupplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2023.109101.
| [1] |
M.J. Chaisson, J. Huddleston, M.Y. Dennis, et al., Nature 517 (2015) 608-611. DOI:10.1038/nature13907 |
| [2] |
J. Gottlieb, M.R. Zamora, T. Hodges, et al., J. Heart Lung Transpl. 35 (2016) 213-221. DOI:10.1016/j.healun.2015.08.012 |
| [3] |
S.M. Nur Husna, H.T. Tan, N. Md Shukri, N.S. Mohd Ashari, K.K. Wong, Front. Immunol. 12 (2021) 663626. DOI:10.3389/fimmu.2021.663626 |
| [4] |
J. Koizumi, T. Kojima, R. Kamekura, et al., J. Membr. Biol. 218 (2007) 1-7. DOI:10.1007/s00232-007-9029-9 |
| [5] |
Y. Ozsoy, S. Gungor, E. Cevher, Molecules 14 (2009) 3754-3779. DOI:10.3390/molecules14093754 |
| [6] |
R. Liu, C. Luo, Z. Pang, et al., Chin. Chem. Lett. 34 (2023) 107518. DOI:10.1016/j.cclet.2022.05.032 |
| [7] |
W. Huang, X. Wu, J. Qi, et al., Drug Discov. Today 25 (2020) 901-908. DOI:10.1016/j.drudis.2019.09.018 |
| [8] |
L. Zhang, Z. Dong, W. Liu, et al., Pharmaceuticals 15 (2022) 877. DOI:10.3390/ph15070877 |
| [9] |
Y. Zhang, C. Liu, J. Wang, et al., Chin. Chem. Lett. 34 (2023) 107631. DOI:10.1016/j.cclet.2022.06.054 |
| [10] |
W. Huang, Z. Fang, et al., Chin. Chem. Lett. 33 (2022) 4079-4083. DOI:10.1016/j.cclet.2022.01.043 |
| [11] |
A.A. Mokhtarieh, J. Lee, S. Kim, M.K. Lee, Biochim. Biophys. Acta Biomembr. 1860 (2018) 1318-1325. DOI:10.1016/j.bbamem.2018.02.027 |
| [12] |
M. Zhang, H. Jiang, L. Wu, et al., J. Control. Release 352 (2022) 422-437. DOI:10.1016/j.jconrel.2022.10.020 |
| [13] |
A. Mandal, N. Kumbhojkar, C. Reilly, et al., Sci. Adv. 6 (2020) eabb6049. DOI:10.1126/sciadv.abb6049 |
| [14] |
R.R. Mazid, U. Divisekera, W. Yang, et al., Chem. Commun. 50 (2014) 13457-13460. DOI:10.1039/C4CC05086J |
| [15] |
Z. Lv, M. Huang, P. Li, et al., Chin. Chem. Lett. 35 (2024) 108601. DOI:10.1016/j.cclet.2023.108601 |
| [16] |
K.A. Foster, M.L. Avery, M. Yazdanian, K.L. Audus, Int. J. Pharm. 208 (2000) 1-11. DOI:10.1016/S0378-5173(00)00452-X |
| [17] |
C.C. Liu, M. Xia, Y.J. Zhang, et al., Biochem. Bioph. Res. Commun. 500 (2018) 145-151. DOI:10.1016/j.bbrc.2018.03.204 |
| [18] |
H. Lin, C. Zheng, J. Li, C. Yang, L. Hu, Sci. Rep. 5 (2015) 13127. DOI:10.1038/srep13127 |
| [19] |
E. Fakhr, F. Zare, L. Teimoori-Toolabi, Cancer Gene Ther. 23 (2016) 73-82. DOI:10.1038/cgt.2016.4 |
| [20] |
A.R. Philp, F. Miranda, A. Gianotti, et al., Physiol. Genom. 54 (2022) 273-282. DOI:10.1152/physiolgenomics.00134.2021 |
| [21] |
C.C. Chou, C.A. Lunn, N.J. Murgolo, Expert Rev. Mol. Diagn. 8 (2008) 179-187. DOI:10.1586/14737159.8.2.179 |
| [22] |
Z.H. Yu, J.R. Xu, Y.X. Wang, et al., Am. J. Respir. Cell Mol. Biol. 48 (2013) 685-693. DOI:10.1165/rcmb.2012-0236OC |
| [23] |
H. Lin, C. Zheng, J. Li, C. Yang, L. Hu, Int. Immunopharmacol. 23 (2014) 642-648. DOI:10.1016/j.intimp.2014.10.017 |
| [24] |
J. Tong, Q. Gu, Curr. Allergy Asthma Rep. 20 (2020) 63. DOI:10.1007/s11882-020-00958-w |
| [25] |
J.J. Lyons, T. Yi, Curr. Opin. Immunol. 72 (2021) 94-106. DOI:10.1016/j.coi.2021.04.001 |
 2024, Vol. 35
2024, Vol. 35 

