Antimicrobial resistance (AMR) is ranked among the top ten worldwide public health problems by the World Health Organization (WHO) [1,2]. According to the WHO Priority Pathogens List, Gram-negative bacteria (such as Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa) urgently require novel treatment options [3,4]. Broad-spectrum antibiotics abuse unconsciously fueling the AMR pathogens increase, multi-drug-resistant micro-organisms have capability to adapt through multiple AMR mechanisms when facing antibiotic treatment [5,6]. In order to prevent the emergence of a post-antibiotic era, the impending challenge of antibiotic resistance necessitates multifaceted strategy studies of antibiotic susceptibility testing (AST), AMR markers identification, and AMR detection, which may help the development of better medications and provide insight into bacterial persistence and resistance mechanisms [1,7].
The key issues in AMR detection include employing simple, low-cost technologies to provide credible results rapidly (in minutes or hours, as opposed to days with traditional approaches). Automated systems have aided in efforts to reduce analysis times during AST deployment, and several approaches and methodologies on the basis of optical microscopy imaging have been established to estimate the growth of bacteria [8-10]. The traditional and improved AST techniques provide AST magnetic, mass, and mechanical (bio) sensors, AST optical (bio) sensors, and chemical sensors for the monitoring of phenotypic AMR. Additionally included are the AST electrochemical (bio) sensors, chemical sensors for the monitoring of genotypic AMR. To address issues with low starting pathogen numbers and contaminated sample matrices, a variety of approaches must be integrated. Future iterations of these technologies will be one step closer to a clinical diagnosis that enables data-driven antimicrobial prescribing, decreases costs, and mitigates the issue of growing antibiotic resistance if these advances are successfully incorporated [11,12]. As a result, the markers for AMR monitoring have been widely explored. Until now, several reviews have summarized the advances in AMR monitoring using sensors and biosensors [11,13,14]. Thus, an all-round yet timely review of the markers family for AMR monitoring is of urgence [11,14]. The general idea behind the AMR markers monitoring technology is to target AMR biomarkers or pathways with an imaging contrast agent, like a radionuclide or fluorophore. Due to the specificity of these tracers, these approaches have the potential to provide a dynamic, rapid and real-time assessment of the AMR microenvironment (such as the presence of particular enzymes or the pH of the surrounding tissue) and host-pathogen interactions.
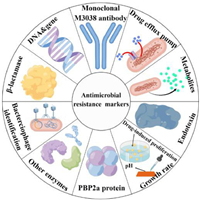
Herein, as shown in Fig. 1, this review represents an effort to provide current research events in the AMR monitoring areas and an attempt to meld these results into an overall and detailed list of the progress of AMR markers testing, followed by details on emerging drug-induced proliferation (DIP) quantification, pH of bacterial growth sensing, genotypic determination, enzymes (such as β-lactamases, carbapenemase and prolyl aminopeptidase) or metabolites detection, endotoxins detection, bacteriophage identification, drug efflux pump efficiency determination, monoclonal M3038 antibody labeling, staphyloferrin labeling, PBP2a protein labeling. This review will contribute to the further research for full use of novel markers detection and do promoting effects on humans' health and technical advancement in AMR monitoring technology worldwide.
|
Download:
|
| Fig. 1. An overview of markers for antimicrobial resistance monitoring. Produced by Figdraw (www.figdraw.com). | |
It is essential to appropriately quantify drug-induced proliferation (DIP) in order to detect partial AMR [15]. The turbidity test is useful for tracking the development of bacteria, but it also has a limited sensitivity and a high experimental error rate. In a high-throughput (HTS) method, the drug-induced proliferation rate may be precisely quantified by combining bacteria, an inhibitor, and EZMTT dye [16,17]. Hu and colleagues were able to generate reliable IC50 values using the EZMTT approach on clinically obtained pathogenic bacteria in 4h [18]. The test is also sensitive enough to spot variations in IC50 in response to shifting incubation durations or cell densities. The EZMTT-dye can be used to detect of AMR instances with 30% growth (5% growth in the bacteriostatic condition, which would have gone undetected under conventional clinical recommendations. HTS mode DIP rate can realize 10-fold better sensitivity than the turbidity assay. As a consequence, lengthy medical treatment can be avoided and the development of microorganisms resistant to antibiotics is prevented.
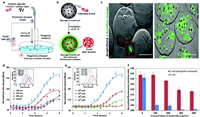
2.2. pH of bacterial growth sensingWhile fixed standards are necessary to link the biochemical parameters and bacterial growth rates, indirect approaches like biochemical activity assessment through concentration measuring of molecules such as O2 [19], CO2 and ATP can be used to estimate bacterial growth rates [20,21]. Since bacterial growth will change the pH of the medium, many research groups have used pH detection as an indicator of growth. In the majority of cases, the pH of the culture becomes acidic over time. Due to this, a number of technologies that use fluorescence as a pH-sensitive readout have been established, however, the majority of them have photobleaching or instability issues with their sensing probes in growth media [22]. To create sensors for a series of parameters, carbon-based nanoprobes with adjustable characteristics and established photobleaching stability have been utilized [23,24]. As Fig. 2 presented, Chandra and colleagues developed a system to detect bacterial growth by observing pH changes through changes in fluorescence [25]. In this study, they described how micron-sized alginate hydrogels (microgels) containing pH-sensitive carbon dots can be used to measure the pH of bacterial life. The platform can also monitor the microorganism's medication AMR profile within 4–6h, which is presently impossible using any other clinically employed approaches. The pH ratiometric platform ensures the detection of real-time pH changes in the bacterial population boom point, can achieve sensitively detection of bacteria densities as low as ~104 CFUs. Microgels culture platform offers multiplexing possibilities. The sensor can be applied to high-throughput evaluation of samples for AST with various antibiotics, quickly offering assessment on the most feasible high-quality antibiotic combination.
|
Download:
|
| Fig. 2. (a) Diagram of the equipment used to create alginate microgels. (b) Diagram illustrating how the growth of encapsulated E. coli increases fluorescence intensity as a result of a pH change. (c) Fluorescence microscopy photos of live E. coli upon encapsulation (scale bar: 100µm). (d) Fluorescence intensity ratios that are pH-dependent and change over time for different sizes of E. microgels containing E. coli. (e) The ratio of pH-dependent fluorescence intensity changes with E. coli. (f) The difference between non-resistant and ampicillin-resistant E. coli in terms of the fluorescence intensity ratio when ampicillin is present. Reproduced with permission [25]. Copyright 2018, Royal Society of Chemistry. | |
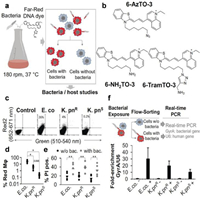
The use of green fluorescent protein (GFP) derivatives as fluorescent reporters is increasingly common in research on biological processes in cells and at the molecular level these days [26]. For the case of AMR infection, Valdivia and colleagues applied GFP to fluorescently label bacteria to learn about host-pathogen interactions, achieving unparalleled sensitivity down to single cell resolution [27]. However, some strains may be resistant to antibiotics applied to select transformants carrying transgenic fluorogenic protein. Therefore, the investigation of host cell interactions with drug-resistant bacteria using fluorogenic labeling has lagged. Schulte and colleagues synthesized and studied a new far-red fluorescent cyanine derivative: 6-TramTO-3, which is a fluorescent probe used for tracking live bacterial cells (Fig. 3) [28]. In the presence of double strain DNA, the signal increases 27 times, and has no significant effect on bacterial activity. Therefore, their exemplary study of pathogenic microorganisms revealed different interaction patterns between multidrug-resistant and antibiotic-sensitive Klebsiella and macrophages.
|
Download:
|
| Fig. 3. (a) Bacterial labeling that is compatible with growth is represented schematically. (b) Chemical structure of the synthesized and tested far-red dyes. (c) On example dot plots of human macrophages exposed to 6-TramTO-3-labeled bacteria, a movement toward the red channel indicates bacterial uptake. (d) Macrophages with red shifts are quantified. (e) Measuring the number of macrophages that have been propidium iodide (PI) positive with or without bacteria. (f) An experimental plan and qRT-PCR using RNA from both fractions to compare the relative abundance of bacterial housekeeping RNA GyrA to human U6 snRNA. Reproduced with permission [28]. Copyright 2018, Wiley-VCH. | |
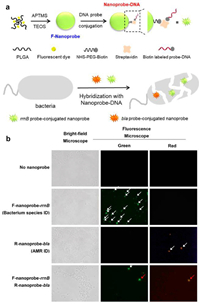
As shown in Fig. 4, Lee and colleagues stated the improvement of a new fluorescent in situ hybridization nanoprobe (FISH) and its utility in the identification of Escherichia coli that is ampicillin-resistant [29]. They developed a new fluorescent nanoprobe for FISH technology and successfully applied it to the identification of microorganisms resistant to antibiotics. The stable nanoprobe was chemically prepared by improved sol-gel and consisted of fluorogenic dye supported poly(d, L-lactide-co-glycolide) (PLGA) and silica nanoparticles (NPs). The fluorescent dyes that are highly loaded in the nanoprobe provide a high-powered fluorescent signal, and silica nanoparticles enable us to modify single-stranded DNA simply. This is the first study using the FISH method and a nanoprobe to identify bacteria that are resistant to antibiotics. They predicted that FISH method based on nanoprobe would be used to detect various pathogens and diagnose the emergence of infectious drug-resistant bacteria.
|
Download:
|
| Fig. 4. (a) Illustration of a schematic for creating stable DNA nanoprobe for FISH. (b) Using the nanoprobe-mixed FISH approach, bright-field images and fluorescence images of E. coli are used to identify the bacterial species and AMR. Reproduced with permission [29]. Copyright 2019, under the terms of the Creative Commons Attribution License (CC BY). Correspondence: Jinyoung Jeong, jyjeong@kribb.re.kr. | |
A novel low-density microarray for commercial use was evaluated to identify typical wide spectrum of carbapenemase (blaNDM and blaKPC) genes and cephalosporinase genes mediated by lactamase plasmids. In general, by using the reference technique of bla gene sequencing, the specificity and sensitivity of microarray testing of plasmid-mediated blaAmpC, blaKPC and blaNDM genes can be 100%. Due to its fast and accurate diagnostic performance, the microarray seems to be very suitable for infection or epidemiological research, in which a large number of isolates need to be characterized. In most countries, the low prevalence between carbapenemase- and pAmpC-producing strains, and the long testing process (7h) still hamper the routine use of this technology. Bogaerts and colleagues realized that additional targets can be added and verified at any time as needed, such as oxacillin enzyme coding-genes, especially for non-fermenting bacteria, secondary ESBLs (BEL, VEB, PER and GES) and carbapenem oxygenase [30].
4. Enzyme or metabolites detection 4.1. β-Lactamases detectionIn a high-throughput method, Chan and colleagues used the E166Cf (a β-lactamase mutant labeled by fluorescein) mutant which is a fluorogenic device to display the β-lactamases of bacteria mentioned above in relation to penicillins and cephalosporins [31]. They further demonstrate that the effects of the E166Cf mutant's fluorescence emission alterations consistent with the bacterial β-lactamases' reactive behavior. When antibiotics that are β-lactamase resistant and bacterial β-lactamases are present together, the E166Cf mutant maintains its fluorescence emission signals. On the contrary, the fluorescence emission signals of the E166Cf mutant were diminished in the attendance of drugs that are both bacterial β-lactamases and β-lactamase-sensitive. The fluorescence markers from the E166Cf mutant permit an explicit distinction of β-lactamase-resistant drugs from β-lactamase sensitive ones in the monitoring of bacterial β-lactamases against a series of antibiotic drugs. This straightforward method may also provide a tool for selecting robust β-lactam antibiotics toward the AMR infection treatment.
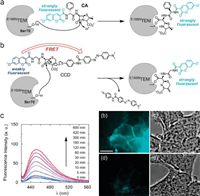
Fig. 5 showed a unique protein labeling method created by Mizukami and colleagues combined a genetically altered β-lactamase with small organic fluorescent β-lactam probes [32]. It was designed according to the FRET principle, the genetically modified hydrolytic enzyme was covalently bonded to a well-crafted fluorogenic probe. They labeled proteins with a delicate fluorescent dye for in vitro or living cells application. The E166NTEM tag protein can also be utilized to accurately identify proteins in higher eukaryotes because it is not present in mammalian cells.
|
Download:
|
| Fig. 5. (a) Fluorescent probe structure and CA labeling mechanisms. (b) Fluorescent probe structure and CCD labeling mechanisms. (c) CCD Time-dependent fluorescence emission spectra in the existence of E166NTEM and optical microscopy images of E166NTEM-EGFR, EGFR-expressing HEK293T and cells that were labeled with CCD (scale bar: 10µm). Reproduced with permission [32]. Copyright 2009, American Chemical Society. | |
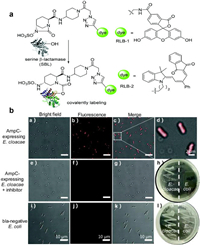
Chen and colleagues published a groundbreaking probe derived from relebactam for the covalent tagging of each serine β-lactamases as well as β-lactamases (Fig. 6) [33]. This imaging agent showed exceptional selectivity against several proteins identified in tilted bacteria. Additionally, the binding of NIR (near-infrared) P-Mero4 with serine β-lactamases produced a sensitively fluorescent response, and the fluorescence intensity may increase by over 300 times. The wash-free imaging for expression of serine β-lactamase in bacterial microorganisms has been used to demonstrate the utility and advantage of this reagent. One way for determining AMR is the identification of certain enzymes or metabolites [34-36]. As depicted in Fig. 7, Zhang and colleagues innovatively synthesized C-2 as a fluorogenic β-lactamase probe [37]. When excited at 440nm, the fluorescent molecule C-2 showed fluorescent emission at 590nm. This is brought on by the F-1 and F-2 fluorophores' effective FRET in its composition. When C-2 is broken down by β-lactamase, the FRET fluorescence will diminish as one fluorophore molecule leaves away. As C-2 degrades, the 590nm fluorescence intensity at declines while the fluorescence intensity at 520nm rises. The molecule has a methoxyimino chain, triggering its resistance to the TEM-1 enzyme, yet it should be vulnerable to the overexpressed AmpC and NDM-1 enzymes. In fact, when C-2 was incubated with highly pure TEM-1, there was very little degradation. Contrarily, under identical conditions, its counterpart (C-1) without the large methoxyimino group was instantly destroyed. Therefore, C-2 is able to discriminate between β-lactamases that have the ability to hydrolyze methoxyimino cephalosporins and those that do not. This assay would promptly offer helpful guidance on antibiotic choice and discourage the overuse of broad-spectrum antimicrobials.
|
Download:
|
| Fig. 6. (a) Design of serine β-lactamase covalent labeling probes. (b) RLB-2, AmpC-expressing Enterobacter cloacae was photographed using fluorescence microscope (scale bar: 10µm). Reproduced with permission [33]. Copyright 2019, Royal Society of Chemistry. | |

|
Download:
|
| Fig. 7. (a) C-1, C-2, and the hydrolyzed byproduct structures and quantitative measuring the β-lactamases activity of several bacterial species. (b) C-2 time-dependent fluorescence change when was treated with E. cloacae cell lysate. (c) Temporal response of the β-lactamase activity from Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, and the inducible AmpC strain of Enterobacter cloacae. RR (relative ratio)=(F590/F520)/(F590/F520). (d) A series of serial dilution studies were used to determine the smallest number of cells needed to detect β-lactamase activity. (e) The profiles of β-lactamase inhibition against various inhibitors for distinct types of β-lactamases. Reproduced with permission [37]. Copyright 2012, Wiley-VCH. | |
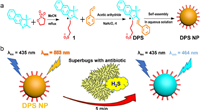
The TEM-1 fluorescent probe design, which uses methoxyimino-substituted cephalosporin as the enzymatic recognition moiety and has bulky substituents on β-lactam rings to prevent hydrolysis by narrow-spectrum β-lactamases, was inspired by the distinctive extended-spectrum β-lactam antibiotics structures [37-39]. With the core structure of cefotaxime serving as the enzymatic recognition moiety, Mao and colleagues had created the first-ever ratiometric fluorescent probe CTX-HN for the selective testing of extended-spectrum β-lactamase (ESBL) activity [40]. Because of the probe concentration is independent from environmental factors, the fluorescence intensity ratio (I563 nm/I452 nm) of this probe, which rises over 2500 times following hydrolysis by ESBLs, is especially appealing in the monitoring of AMR. The detection limits of CTX-HN for NDM-1 β-lactamase within 30min were 0.4pmol/L with high sensitivity. The speedy identification of extended-spectrum antibiotic-resistant microorganisms from a series of clinically enormous bacteria has additionally confirmed the probe's utility. Our group created the novel self-assembled nano fluorescent probe DPS NP, which was proposed for a specific diagnostic approach for bacterial β-lactam antibiotic resistance (Fig. 8) [41]. It is capable of sensitively and selectively measuring changes in H2S concentration for imaging and screening of H2S-related AMR, as well as using colorimetric and ratiometric fluorogenic visualization to measure the concentration of H2S. Direct confirmation of the existence of H2S-related AMR pathways may be made by screening assays against methicillin-resistant S. aureus (MRSA) with four different antibiotics displayed by DPS NP. Additionally, effective microscopy fluorescence imaging for bacterial H2S suggests DPS NP can be employed to investigate the mechanism of AMR. DPS NP can significantly increase the effectiveness and visualize monitoring accuracy for H2S-related hazardous biochemical environments because of its quick, sensitive, and non-invasive qualities.
|
Download:
|
| Fig. 8. (a) Synthetic route of H2S probe DPS NP. (b) Schematic representation of ratiometric fluorescence monitoring of superbugs released H2S. Copied with permission [41]. Copyright 2022, Elsevier B.V. | |
Mao and colleagues created a reagent that is fluorogenic and immobilizable to precisely identify bacteria that are antibiotic-resistant [42]. A monofluoromethyl (CFC-1) or difluoromethyl (CFC-2) substituted coumarin was used as an activatable fluorophore in the β-lactamase probes [43]. These probes are selectively activated by β-lactamase to produce quinone methides [44], which act as extraordinarily reactive Michael acceptors to react with nearby proteins or potential β-lactamase nucleophiles (such as the thiol of Cys, the hydroxyl of Tyr-or Ser, the amine of Lys), covalently linking the proteins around the factor of interest. The antibiotic-hydrolyzing β-lactamase is used as an activator for selectively labeling antibiotic-resistant bacteria and activating their fluorescence emission. In spite of the fact that its blue fluorescence emission might limit the sensitivity of detection because of its relatively weaker fluorescence or autofluorescence background interference, CFC-2 stops the activated fluorophore from diffusing away from the point of interest once it has been activated by enzymatic reaction. Due to its fluorescence emission, it is perfect for cell-labeling investigations that enable quick screening for bacteria that are resistant to antibiotics. This probe has many advantages, such as simple structural design, great labeling efficiency, and high sensitivity. Thai and colleagues developed a fluorogenic β-lactamase substrate used as a probe that incorporates the pyridinium moiety of ceftazidime (CAZ) in the chemical structure of the BODIPY-based fluorescent dye and the oxyimino moiety of cefotaxime (CTX) in order to detect bacteria that are resistant to antibiotics [38]. The probe fluorescence emission signal was increased with enzyme, suggesting that it would be useful for testing drug-resistant enzymes and determining the kinetic properties of enzymes. It is demonstrated that the fluorogenic cephalosporin analogue can provide detection signals in response to bacterial CAZ- and/or CTX-resistance.
By fabricating a substrate of penicillin G on the surface of a pH-responsive fluorophore-doped mesoporous silica nanoparticle [45], Tummala and colleagues provided a quick and sensitive monitoring approach for the β-lactamase activity assessment in bacteria resistant to antibiotics [46]. Fluorescein isothiocyanate (FITC), a water-soluble fluorescent molecule with a high quantum yield, was chemically integrated into the particle surface. Then, the substrate of β-lactamase (penicillin G, PenG) was loaded with fluorescent mesoporous particles Penicilloic acid was created when the substrate and β-lactamase interacted, and the environmental pH reduction contributed to the particle fluorescence quenching. The strains both sensitive and resistant to antibiotics of 25 clinical samples of bacteria were examined. In less than an hour, the suggested approach might identify the existence of β-lactamases in clinically important tests. Additionally, the β-lactamase activity detection limit was 7.8×10−4U/mL, which is very low.
4.2. Carbapenemase detectionFluorescent β-lactamase probes have received a lot of interest in this field due to their high sensitivity, simplicity of use, and affordability [38,47,48]. However, most of the current β-lactamase probes are on the basis of the core cephalosporins structure, which can be hydrolyzed by mainstream β-lactamases and lack specificity for carbapenemases. A stereochemically changed cephalosporin served as the enzyme recognition motif for the beautifully constructed carbapenemase-selective probes that Shi and colleagues recently realized [49]. Though structurally distinct from carbapenems, these probes are selective to particular carbapenemases due to various substituents (R1) on their surfaces. As shown in Fig. 9, using carbapenemase activity as a target, Mao and colleagues created a highly particular probe containing a core carbapenem structure [50]. The carbapenemase core structure was used as the enzymatic recognition moiety in the probe design. Moreover, the activatable fluorophore in this study was an alkenyl-linked boron dipyrromethene (BODIPY) dye, since most carbapenems lack leaving groups. Fluorescent reagents can reproduce carbapenem hydrolysis profiles, increasing fluorescence intensity by over 200 folds after carbapenemases activation; the detection limit of CB-1 for IMP-1 within 30min was 1.1pmol/L. Because CB-1 emits green fluorescence, it reduces potential interference from bacteria's autofluorescence or culture medium's fluorescence. Additionally, this research has shown that the usage of caged enamine-BODIPY fluorophore switches with fluorescence emission turn-on activation, may serve as an inspiration for a general approach to the development of fluorogenic imaging reagents based on BODIPY.

|
Download:
|
| Fig. 9. (a) CB-1 fluorescence in reaction to various β-lactamases. (b) CB-1 fluorescence change after incubated with the designated β-lactamases. (c) Fluorescence intensity change of CB-1 with a variety of β-lactamases. (d) Variations in fluorescence intensity of CB-1 after incubated with the appropriate carbapenemases in the inhibitors existence. (e) Time dependency of the CB-1 fluorescence intensity when different β-lactamases are present. (f) Time dependence of imipenem absorbance when various β-lactamases are present. Reproduced with permission [50]. Copyright 2017, Wiley-VCH. | |
The most significant AMR clinically mechanism in carbapenem-resistant pathogens, aside from practically all other β-lactams, is the production of carbapenemases, microbial enzymes that catalyze the carbapenems hydrolytic degradation [51]. Kim and colleagues employed benzyl ether to both improve the probe's interaction with the active sites of several carbapenemase types and to generate a cascade reaction with the probe following enzymatic hydrolysis for the testing of a variety of carbapenemase-producing Enterobacteriaceae (CPE) [52]. After carbapenemase hydrolyzes the β-lactam ring of probe 1, a cascade reaction occurs that releases hydroxy benzyl alcohol 3 and umbelliferone 4 in an anionic form, leading to intense fluorescence emission. They also examined the fluorescent assay performance using the pellet obtained from blood cultures which are positive, had directly evaluated with matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and tests for antibiotic sensitivity are performed using Vitek 2. This is the first demonstration of a fluorescence assay for detecting carbapenemases directly from positive blood cultures as well as bacterial isolates.
Kim and colleagues described a fluorescent probe library based on carbapenem, utilizing hydroxymethyl carbapenem 7a or hydroxyalkyl carbapenem 7b as crucial intermediates [53]. They have synthesized a set of compound materials with three various sorts of connectors (e.g., benzyl ethers, carbamate, and amines), used to detect bacteria that produce carbapenemase. The fluorescence emission signal was improved after several types of carbapenemases hydrolyzed compound 1b with a benzyl ether linker, proving that the active junction is essential for the stability of the probes and its capacity to increase activity. In particular, clinical sample screening showed that probe 1B had better selectivity to carbapenemase-producing organisms (CPOs) than other kind β-lactamase or non-carbapenemase-producing bacteria and could be used for the quick and precise identification of CPOs for quick identification and treatment.
Feng and colleagues developed β-LEAF (β-lactamase triggered fluorescence probe) for β-lactamase fluorescence test [54]. When broken by β-lactamases such as penicillinases, ESBL, AmpC β-lactamases, and carbapenemases, the β-LEAF fluorescence emission turns on. They show that carbapenemases generation in bacteria may be quickly and precisely identified in only one step in 10min by means of fluorescence identification of β-lactamase activity (FIBA). The increase rate, sometimes referred to the rate of fluorescence intensity rise, is a measure of the bacterial β-lactamase activity that declines as the activity is blocked. The addition of imipenem, which binds to the enzyme active site and prevents β-LEAF access, will lessen the rising rate for a noncarbapenemase β-lactamase. In contrast, the addition of imipenem did not have any effect on the increase of carbapenemase since carbapenemase can rapidly cleave imipenem and release inhibition. Accordingly, the growth rate with and without imipenem may be compared to identify bacteria that generate carbapenemases.
Song and colleagues described a brand-new caging approach for creating fluorogenic probes and how it may be used to monitor β-lactamase activity. The typical technique is caging the fluorophore chemically using a group that the analyte can selectively remove [55]. The 'off-state' probe may be provided by the caging section, which could be connected to the fluorophore directly. If a direct sealing method cannot be used, or if a directly sealed fluorescence intensity detector is used, poor performance will result, for example, because of the steric effect of a large number of fluorophores, so, for design convenience, it is often designed to connect two components connector. In their caging method, a thiophenyl linker is attached to a fluorescent dye that is caged by dinitrophenyl, an excellent sensing group. To increase intramolecular reaction speed and subsequently fluorescence activation rate, the linker's length has been examined and improved. On this basis, they developed a new green fluorescent probe Cephalosporin-arylated Tokyo green (CAT) and tested the selectivity of this probe for distinguishing Metallo-carbapenemases (NDM-1, IMP-1, VIM-27) in carbapenem Enterobacter (CRE).
Using alkenyl-linked boron dipyrromethene (BODIPY) dye as an activatable fluorescent dye and a carbapenem with a carbon-carbon double bond attached as an enzyme recognition motif, Wang and colleagues combined carbapenem and umbelliferone to create the fluorescent probe CPC-1 [56]. All carbapenemases can selectively activate probes, and can significantly increase fluorescence intensity, but for other β-lactamases, even extended spectrum β-lactamase, have no effect.
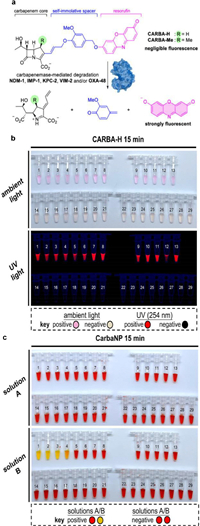
The first potent pan-carbapenemase reporter, CARBA-H, was reported by Ma and colleagues (Fig. 10) [34]. CARBA-H targets the top five applicable carbapenemases, including OXA-48, whose detection limit is picomolar. One of the most clinically significant carbapenemases is OXA-48, yet none of the chemical probes in use could handle the difficulty of detecting it. A number of mechanistic investigations have shown that the 1β-methyl group significantly alters the OXA-48 enzyme's hydrolytic kinetics. When the 1-methyl substituent is absent, pancarbapenemase can be recognized and strains that encode carbapenemase, such as OXA-48, OXA-181 and OXA-232, can be identified. The instrument's chromogenic and fluorescent readouts are easily distinguishable within 15min of current detection technology. In addition, CARBA-H can also be used to measure carbapenemase activity in urine samples of labeled CPE.
|
Download:
|
| Fig. 10. (a) CARBA-H design and (b) colorimetric and fluorogenic reaction of CARBA-H. (c) Regarding medical isolates in accordance with the CLSI protocol, CarbaNP solution A and solution B. PCR was used to identify the carbapenemases encoded in the clinical pathogens isolates. Reproduced with permission [34]. Copyright 2021, under the terms of the Creative Commons Attribution License (CC BY-NC-ND 4.0). Correspondence: Dan Yang, yangdan@hku.hk. | |
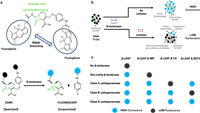
As depicted in Fig. 11, Feng and colleagues used β-lactam activity (FIBA) for fluorescence identification to rapidly detect and classify bacterial carbapenemases [57]. Within 10min, by combining the β-lactamase substrate β-LEAF (β-lactamase-activated fluorophore) with isolates of pathogens and corresponding inhibitors (imipenem, for non-carbapenem enzyme β-lactamase, clavulanic acid for type A carbapenemase, EDTA for type B carbapenemase), the sensitivity is 95%−100%. FIBA is about $1/test and consists of only one mixing procedure. Future research should focus on expanding FIBA testing to include additional medical isolates, especially those that are not currently included in the test plate and those together with weaker hydrolytic characteristics and lower carbapenem MICs (such as OXA-48, VIM, and SME). It is important to highlight that the diversity of isolates examined includes 147 isolates from 19 different species, including 18 carbapenemase subtypes and 31 noncarbapenemase β-lactamases, indicating the applicability of this unique technique.
|
Download:
|
| Fig. 11. The concept of carbapenemase identification and categorization with FIBA is illustrated schematically. (a) A cleavable β-lactam core coupled to two fluorophores makes up the β-LEAF probe. (b) β-lactamase inhibitor lowers β-LEAF β-lactamase cleavage and fluorescence emission. (c) IMP (imipenem) is the noncarbapenemase (non-carba) inhibitor. CA (clavulanic corrosive) is the class A carbapenemase inhibitor and EDTA is the class B carbapenemase inhibitor. Copied with permission [57]. Copyright 2021, American Society for Microbiology. | |
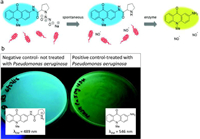
Prior studies have demonstrated that the enzymatic detection of bacteria is possible by affixing amino acids to derivatives of 2-aminoacridones [58]. Furthermore, Hibbard and colleagues chose 2-aminoacridone derivative as the fluorogenic reporter to pathogens monitoring, they developed materials which can both recognize and eliminate the dangerous lethal multi-drug resistant microorganism stress Pseudomonas aeruginosa (Fig. 12) [59]. They do this by synthesizing a nitric oxide donor connected to a fluorogenic molecule. A synthetic method is created to join proline to Pro/Fluoro 6, a fluorescent amino acridone molecule. In the existence of Pseudomonas aeruginosa, a new prolyl aminopeptidase-specific indicator, experiments demonstrate a clear color change from blue to yellow under UV light. These findings show that Pro/Fluoro/NO 8 is a remarkably adaptable molecule that has two distinct functions: it can fluorescently identify P. aeruginosa and destroy it by producing NO.
|
Download:
|
| Fig. 12. (a) Diagram displaying the emission of the color shift of indicator with nitric oxide. (b) Representative agar plates displaying the anticipated color shift when Pseudomonas aeruginosa is present. Reproduced with permission [59]. Copyright 2019, Royal Society of Chemistry. | |
The Limulus Amebocyte Lysate (LAL) test [60,61], electrochemical and colorimetric detection [62], surface-enhanced Raman scattering (SERS) [63], and peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) [64-66] are the most common methods used to identify free endotoxin, a significant pyrogen whose defilement in medications and drugs is severely controlled by administration. As shown in Fig. 13, Gupta and colleagues designed a series of water-soluble metal-based aggregation-induced emission luminogens (AIEgens). Cyclometalated iridium(Ⅲ) polypyridine complexes [Ir(PQ)2(N^N)]Cl (1−3), where PQ is 2-phenyl quinoline and N^N is 2, 2′-bipyridine derivatives, exhibit dual functionality for monitoring and removing of drug-resistant micro-organism in aqueous solutions [67]. Ir(PQ)2(N^N)]Cl (1−3) exhibited a distinct luminescence response to target these naturally biomarker on the outer wall/membrane of the microbe in water samples within 5min, resulting in unprecedentedly detection limits as low as 1.2 CFU/mL. It is remarkable that microorganism may be detected with naked eye at a high concentration. Additionally, toward highly antibiotic-resistant pathogens such as MRSA and carbapenem-resistant A. baumannii (CRAB), the complexes showed strong antibacterial activity.

|
Download:
|
| Fig. 13. Chemical structures of (a) Lipopolysaccharide (LPS) and (b) Lipoteichoic acid (LTA). (c) Proposed model cyclometalated Ir(Ⅲ) complex-mediated theranosis. (d and e) ROS production was quantified using DCFDA dye. (f–i) Time-resolved fluorescence spectroscopy results were acquired using DiSC3(5) and PI dyes, separately. Reproduced with permission [67]. Copyright 2020, American Chemical Society. | |
TM4 mycobacteriophage phAE87:: hsp60-EGFP (EGFP-phage) include the improved GFP gene was designed and evaluated for identifying drug-resistant Mycobacterial tuberculosis strains [68]. Similar to the luciferase phage, EGFP-phage is used to infect M. tuberculosis that is being grown in the presence of antibiotics. After these strains have been inactivated, they are spotted onto glass slides, and the fluorescent bacilli that signify AMR are then imaged by using a fluorescence microscope. Rondón and colleagues described the principal check of the EGFP-phage for AMR identifying in naturally occurring M. tuberculosis [69]. The consequences obtained are superior to the majority of the D29 phage replication assay, while the luciferase phage has been claimed to have 100 percentage responsiveness and particularity. Improved results have additionally been accounted by the microscopic observation drug susceptibility (MODS) approach [70], as well as more current Xpert methods [71]. The EGFP-phage approach sensitivity can be raised to 98% for use with scientific samples, and the protocol can be made simpler, it might be helpful as a quick and affordable method to detect M. tuberculosis strains that are extensively or multidrug resistant in environments with limited resources and infrastructure [69].
Further developed diagnostics and medication treatment for Mycobacterial tuberculosis are desperately required [72,73]. Fluorescence-activated cell sorting (FACS) allows for quick counting of metabolically active bacteria following phage infection [74]. O'Donnell and colleagues contrasted the correspondent phage examine to Quality Xpert MTB/RIF for sputum analysis for M. tuberculosis and rifampin (RIF) blockage [75]. Metabolically active bacteria in sputum from patients were directly imaged using fluorescence microscopy or FACS. These results represent a significant improvement over earlier-generation phages reporter and are encouraging the reporter for identification of phenotypic AMR to RIF.
7. Drug efflux pump efficiency determinationBlair and colleagues examined a variety of techniques, including minimum inhibitory concentration (MIC) assays, to assess the drug efflux pump efficiency of transporters [76]. It is a challenge to make it certain that no drug translocation with comparable MICs has occurred [77]. Lu and colleagues used MALDI-TOF MS in their investigation to evaluate drug efflux from a few drug transporters and compared the result to those of more consequences [78]. E. coli drug transporter AcrB (RND family) was firstly assessed using the MIC method, and this investigation revealed that the E. coli with overexpression of AcrB show a variety of antibiotic and dye resistance. The research which was based on fluorescence emission revealed that AcrB in E. coli may reduce intracellular dye accumulation and exhibit a range of constants for diverse dyes' efflux rates, indicating the efflux efficiency of AcrB. In addition, the MS data revealed that E. coli with overexpression of AcrB resulted in enhanced abundance of different drugs and dyes at varying rates over time in the extracellular space, using AcrB to realize continuous substrate efflux. The study is aimed to develop a quick, accurate method for measuring drug and dye efflux by drug transporters using MALDI-TOF MS. E. coli's secondary drug transporter AcrB was selected as the permeability model to calculate drug efflux (erythromycin and rifampicin) and dyes (Hoechst 33342, Nile red, and EtBr) over time. The advantages of using MALDI-TOF MS for drug efflux assessment are minimum sample preparation procedures, quick manipulation, and accurate analysis.
Stone and colleagues synthesized and characterized two novel fluorescent derivatives of erythromycin [79]. Erythromycin was modified site-selectively to add an azide substituent, which was then used in azide-alkyne 'click' chemistry to link two different color fluorophores with an alkyne substituent to create two new fluorescent roxithromycin derivatives. Fluorescence emission was seen to be localized within the efflux-inhibited Gram-negative bacteria, showing significantly higher absorption. On the contrary, efflux increased Gram-positive bacteria did not exhibit reduced uptake. Single-cell microfluidics were employed to analyze the kinetics of the accumulation of these probes, and the results showed significant population heterogeneity that may be related to persistence and/or resistance. This exploratory study demonstrates the potential value of macrolide fluorescence probes in optical imaging of the interactions between bacteria and antibiotics and offers a fresh understanding of AMR mechanisms. Future applications include quantifying macrolide AMR pathways and identifying variations in antibiotic sensitivity among bacterial populations.
8. Protein labeling 8.1. Monoclonal M3038 antibody labelingIt is one of today's greatest challenges in addressing the problem of multiple drug-resistant bacteria (MDRBs) Salmonella enterica serovar Typhimurium definitive type 104 (DT104) [80,81]. Ronholm et al. have demonstrated that by using particular monoclinic antibodies for Salmonella DT104's O: 4 antigen, one may identify the bacteria with minimum to no cross-reactivity to other relative pathogens [82]. Khan and colleagues published a M3038 antibody-conjugated popcorn-shaped gold nanoparticle-based SERS assay for the first time, with remarkable selectivity for various drug-resistant Salmonella DT104 pathogens (ATCC 700408) at the 10 colony-forming unit (CFU)/mL threshold from contaminated water and romaine vegetables [83]. It has been demonstrated that this colorimetric test has a strong selectivity for MDRB Salmonella typhimurium DT104 with an outstanding detection limit (103 CFU/g) when used to identify the pathogen in water and lettuce samples within 5min.
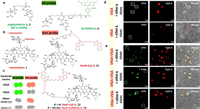
8.2. Targeted protein labelingThe primary method by which S. aureus gets iron in scarce conditions is siderophore-mediated iron absorption. Because of this, the siderophore-derived probe was able to distinguish S. aureus from other germs [84]. It was able to specifically target vancomycin-sensitive S. aureus (VSSA), further separating vancomycin-resistant S. aureus (VRSA) and VSSA, through perfecting the vancomycin-derived probes [85]. Wang and colleagues had established that siderophore-based probes are potential bacterial targeting agents. The two main siderophores for S. aureus are staphyloferrin A (SA) and B (SB). The fluorescent probes based on Vancomycin (Van) have been utilized to visualize kinetics of bacterial cell walls and identify Gram-positive microorganism as diagnostic agents for the identification of Van resistance (Fig. 14) [86,87]. The majority of applications depended on the affinity between Van and the mobile wall d-Ala-d-Ala-residues in cell walls. They effectively targeted and distinguished between VRSA and VSSA in complex pathogens mixture using the siderophore-derivative probe SA-DORS-FL in combination with the vancomycin-derivative probe VanC-SulfoCy5.5. The MECA gene, which produces the PBP2a protein and causes β-lactam to become resistant to antibiotics, causes methicillin AMR [88]. Verifying the corresponding relationship between protein expression and gene vectors and quickening the diagnosis of methicillin AMR are very important for improving the management of antibiotics and reducing the spread of AMR [89]. Silvwstri and colleagues examined the utility of immunofluorescence toward PBP2a protein to discover a novel potential technique for methicillin AMR monitoring [90]. They proved that PBP2a protein can be identified via immunofluorescence in Staphylococcus pseudintermedius (SP) isolates, so as to methicillin-resistant bacteria. To hasten the detection of methicillin AMR in SP, it is worthwhile to examine its effectiveness on cytological samples.
|
Download:
|
| Fig. 14. (a) Staphyloferrin A (SA or DORS) and its derivative probe, SA-DORS-FL chemical structures. (b) Van and its derivative probes, VanC-SulfoCy5.5, VanC-Cy5.5, and VanN-Cy5.5, have the following structures. (c) Detection mechanism of probes VanCSulfoCy5.5 and SA-DORS-FL. (d, e) probes 6 and 13 were used to treat non-Staphylococcus bacteria, E. coli, A. baumannii, V. cholera, and B. subtilis with DP, VRSA or VSSA. The channels FTIC/Cy5.5, Cy5.5, and FTIC are each represented severally (scale bar: 5µm). Reproduced with permission [87]. Copyright 2021, American Chemical Society. | |
Enzymes, genes, and proteins are excellent models of chemo- and bio-markers that are the subject of AMR monitoring research. These markers will aid in increasing selectivity and, in turn, bacterial identification, which is a vital step for enhancing the validity of the (bio)sensing method of AMR surveillance. Antimicrobial peptides, for example, are currently regarded to be intriguing biorecognition components for the identification and detection of bacteria. These peptides have the ability to interact with bacteria, enfeebled them and preventing their further development. Antimicrobial peptides can recognize many different types of bacteria, but few can realize AMR monitoring. The ability to interact with a variety of bacterial species can be potentially used for AMR monitoring probes creation. The creation of enhanced and quicker biosensor-based pathogenic risk detection methods are, therefore, unquestionably the most attractive aims in the context of the global growth of AMR. This naturally implies the future development of more effective markers as well as sufficient biorecognition components to enable the precise identification of diverse AMR.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThis work was financially supported by the National Natural Science Foundation of China (No. 82202221), the Natural Science Foundation of Anhui Province (No. 2208085QB39), and College Students' Innovative Entrepreneurial Training Plan Program (No. 202110359071).
| [1] |
X. Lai, M.L. Han, Y. Ding, et al., Nat. Commun. 13 (2022) 343. DOI:10.1038/s41467-022-28012-5 |
| [2] |
S. Santajit, N. Indrawattana, BioMed Res. Int. 2016 (2016) 2475067. |
| [3] |
S. McGuire, Adv. Nutr. 7 (2016) 418-419. DOI:10.3945/an.116.012211 |
| [4] |
A.P. Zavascki, L.Z. Goldani, J. Li, R.L. Nation, J. Antimicrob. Chemother. 60 (2007) 1206-1215. DOI:10.1093/jac/dkm357 |
| [5] |
M. Boolchandani, A.W. D'Souza, G. Dantas, Nat. Rev. Genet. 20 (2019) 356-370. |
| [6] |
G.G. Rao, N.S. Ly, J.B. Bulitta, et al., J. Antimicrob. Chemother. 71 (2016) 3148-3156. DOI:10.1093/jac/dkw293 |
| [7] |
M. Krishnamurthy, M.M. Lemmon, E.M. Falcinelli, et al., Infect. Drug Resist. 12 (2019) 1393. DOI:10.2147/idr.s196874 |
| [8] |
J. Choi, J. Yoo, M. Lee, et al., Sci. Transl. Med. 267ra174 (2014). |
| [9] |
S. Metzger, R.A. Frobel, W.M. Dunne, Jr., Diagn. Microbiol. Infect. Dis. 79 (2014) 160-165. DOI:10.1016/j.diagmicrobio.2013.11.029 |
| [10] |
M. Fredborg, K.R. Andersen, E. Jørgensen, et al., J. Clin. Microbiol. 51 (2013) 2047-2053. DOI:10.1128/JCM.00440-13 |
| [11] |
E.C. Reynoso, S. Laschi, I. Palchetti, et al., Chemosensors 9 (2021) 232. DOI:10.3390/chemosensors9080232 |
| [12] |
Y.W. Lin, N.A. Rahim, J. Zhao, et al., Antimicrob. Agents Chemother. 63 (2019) e02176-18. DOI:10.1128/AAC.02176-18 |
| [13] |
B.J.C. Walsh, D.P. Giedroc, J. Biol. Chem. 295 (2020) 13150-13168. DOI:10.1074/jbc.rev120.011304 |
| [14] |
K. Hsieh, K.E. Mach, P. Zhang, et al., Acc. Chem. Res. 55 (2022) 123-133. DOI:10.1021/acs.accounts.1c00462 |
| [15] |
L.A. Harris, P.L. Frick, S.P. Garbett, et al., Nat. Methods 13 (2016) 497-500. DOI:10.1038/nmeth.3852 |
| [16] |
W. Zhang, M. Zhu, F. Wang, et al., Anal. Biochem. 509 (2016) 33-40. DOI:10.12928/telkomnika.v14i3A.4426 |
| [17] |
Z. Chen, D. Li, N. Xu, et al., J. Med. Chem. 62 (2019) 589-603. DOI:10.1021/acs.jmedchem.8b01198 |
| [18] |
Q. Hu, Y. Yu, D. Gu, et al., ACS Infect. Dis. 5 (2019) 1252-1263. DOI:10.1021/acsinfecdis.9b00132 |
| [19] |
J. Cao, S. Nagl, E. Kothe, et al., Microchim. Acta 182 (2015) 385-394. DOI:10.1007/s00604-014-1341-3 |
| [20] |
M. Gottfredsson, H. Erlendsdottir, S. Gudmundsson, Antimicrob. Agents Chemother. 35 (1991) 2658-2661. DOI:10.1128/AAC.35.12.2658 |
| [21] |
M. Stemler, G. Stemke, J. Robertson, J. Clin. Microbiol. 25 (1987) 427-429. DOI:10.1128/jcm.25.2.427-429.1987 |
| [22] |
N.J. Cira, J.Y. Ho, M.E. Dueck, et al., Lab Chip 12 (2012) 1052-1059. DOI:10.1039/C2LC20887C |
| [23] |
P. Zuo, X. Lu, Z. Sun, et al., Microchim. Acta 183 (2016) 519-542. DOI:10.1007/s00604-015-1705-3 |
| [24] |
A. Chandra, N. Singh, ACS Biomater. Sci. Eng. 3 (2017) 3620-3627. DOI:10.1021/acsbiomaterials.7b00740 |
| [25] |
A. Chandra, N. Singh, Chem. Commun. 54 (2018) 1643-1646. DOI:10.1039/c7cc08678d |
| [26] |
O. Shimomura, Angew. Chem. Int. Ed. 48 (2009) 5590-5602. DOI:10.1002/anie.200902240 |
| [27] |
R.H. Valdivia, A.E. Hromockyj, D. Monack, et al., Gene 173 (1996) 47-52. DOI:10.1016/0378-1119(95)00706-7 |
| [28] |
L.N. Schulte, B. Heinrich, H. Janga, et al., Angew. Chem. Int. Ed. 57 (2018) 11564-11568. DOI:10.1002/anie.201804090 |
| [29] |
W.S. Lee, S. Lee, T. Kang, et al., Nanomaterials 9 (2019) 750. DOI:10.3390/nano9050750 |
| [30] |
P. Bogaerts, A.M. Hujer, T. Naas, et al., Antimicrob. Agents Chemother. 55 (2011) 4457-4460. DOI:10.1128/AAC.00353-11 |
| [31] |
P.H. Chan, K.C. Chan, H.B. Liu, et al., Anal. Chem. 77 (2005) 5268-5276. DOI:10.1021/ac0502605 |
| [32] |
S. Mizukami, S. Watanabe, Y. Hori, et al., J. Am. Chem. Soc. 131 (2009) 5016-5017. DOI:10.1021/ja8082285 |
| [33] |
Y. Chen, M. Xu, W. Xu, et al., Chem. Commun. 55 (2019) 9919-9922. DOI:10.1039/c9cc04533c |
| [34] |
C.W. Ma, K.K.H. Ng, B.H.C. Yam, et al., J. Am. Chem. Soc. 143 (2021) 6886-6894. DOI:10.1021/jacs.1c00462 |
| [35] |
S.P. Gholap, C. Yao, O. Green, et al., Bioconjugate Chem. 32 (2021) 991-1000. DOI:10.1021/acs.bioconjchem.1c00149 |
| [36] |
H.L. Chan, L. Lyu, J. Aw, et al., ACS Chem. Biol. 13 (2018) 1890-1896. DOI:10.1021/acschembio.8b00172 |
| [37] |
J. Zhang, Y. Shen, S.L. May, et al., Angew. Chem. Int. Ed. 51 (2012) 1865-1868. DOI:10.1002/anie.201107810 |
| [38] |
H.B.D. Thai, J.K. Yu, B.S. Park, et al., Biosens. Bioelectron. 77 (2016) 1026-1031. DOI:10.1016/j.bios.2015.10.081 |
| [39] |
J. Aw, F. Widjaja, Y. Ding, et al., Chem. Commun. 53 (2017) 3330-3333. DOI:10.1039/C6CC09296A |
| [40] |
W. Mao, X. Qian, J. Zhang, et al., ChemBioChem 18 (2017) 1990-1994. DOI:10.1002/cbic.201700447 |
| [41] |
D. Gong, X. Huang, Z. Yi, et al., Mater. Today Commun. 32 (2022) 103959. DOI:10.1016/j.mtcomm.2022.103959 |
| [42] |
W. Mao, L. Xia, Y. Wang, et al., Chem. Asian J. 11 (2016) 3493-3497. DOI:10.1002/asia.201601344 |
| [43] |
J. Ge, L. Li, S.Q. Yao, Chem. Commun. 47 (2011) 10939-10941. DOI:10.1039/c1cc14653j |
| [44] |
D.H. Kwan, H.M. Chen, K. Ratananikom, et al., Angew. Chem. 123 (2011) 314-317. DOI:10.1002/ange.201005705 |
| [45] |
M.L. Vera, A. Cánneva, C. Huck-Iriart, et al., J. Colloid Interface Sci. 496 (2017) 456-464. DOI:10.1016/j.jcis.2017.02.040 |
| [46] |
S. Tummala, W.A. Huang, B.H. Wu, et al., ChemistryOpen 9 (2020) 1074-1081. DOI:10.1002/open.202000221 |
| [47] |
Y. Cheng, H. Xie, P. Sule, et al., Angew. Chem. Int. Ed. 53 (2014) 9360-9364. DOI:10.1002/anie.201405243 |
| [48] |
L. Li, Z. Li, W. Shi, et al., Anal. Chem. 86 (2014) 6115-6120. DOI:10.1021/ac501288e |
| [49] |
H. Shi, Y. Cheng, K.H. Lee, et al., Angew. Chem. 126 (2014) 8251-8254. DOI:10.1002/ange.201402012 |
| [50] |
W. Mao, L. Xia, H. Xie, Angew. Chem. Int. Ed. 56 (2017) 4468-4472. DOI:10.1002/anie.201612495 |
| [51] |
K. Bush, Antimicrob. Agents Chemother. 62 (2018)e01076-18. |
| [52] |
H.S. Kim, J.O. Kim, J.E. Lee, et al., J. Clin. Microbiol. 58 (2019)e01026-19. |
| [53] |
J. Kim, Y. Kim, A.Z. Abdelazem, et al., Bioorg. Chem. 94 (2020) 103405. DOI:10.1016/j.bioorg.2019.103405 |
| [54] |
Y. Feng, A. Palanisami, J. Kuriakose, et al., Emerg. Infect. Dis. 26 (2020) 793. DOI:10.3201/eid2604.181655 |
| [55] |
A. Song, Y. Cheng, J. Xie, et al., Chem. Sci. 8 (2017) 7669-7674. DOI:10.1039/C7SC02416A |
| [56] |
J. Wang, W. Xu, S. Xue, et al., Org. Biomol. Chem. 18 (2020) 4029-4033. DOI:10.1039/d0ob00114g |
| [57] |
Y. Feng, J.W. Swain, A. Palanisami, et al., J. Clin. Microbiol. 59 (2021) e02517-e02520. |
| [58] |
M. Cellier, A.L. James, S. Orenga, et al., PLoS One 11 (2016) e0158378. DOI:10.1371/journal.pone.0158378 |
| [59] |
H.A.J. Hibbard, M.M. Reynolds, J. Mater. Chem. B 7 (2019) 2009-2018. DOI:10.1039/c8tb02552e |
| [60] |
P.F. Roslansky, T.J. Novitsky, J. Clin. Microbiol. 29 (1991) 2477-2483. DOI:10.1128/jcm.29.11.2477-2483.1991 |
| [61] |
G.H. Zhang, L. Baek, P.E. Nielsen, et al., J. Clin. Microbiol. 32 (1994) 416-422. DOI:10.1128/jcm.32.2.416-422.1994 |
| [62] |
J. Sun, A.R. Warden, J. Huang, et al., Anal. Chem. 91 (2019) 7524-7530. DOI:10.1021/acs.analchem.8b04997 |
| [63] |
W. Gao, B. Li, R. Yao, et al., Anal. Chem. 89 (2017) 9836-9842. DOI:10.1021/acs.analchem.7b01813 |
| [64] |
M. Kang, C. Zhou, S. Wu, et al., J. Am. Chem. Soc. 141 (2019) 16781-16789. DOI:10.1021/jacs.9b07162 |
| [65] |
Y. Li, Z. Zhao, J. Zhang, et al., Adv. Funct. Mater. 28 (2018) 1804632. DOI:10.1002/adfm.201804632 |
| [66] |
G. Jiang, J. Wang, Y. Yang, et al., Biosens. Bioelectron. 85 (2016) 62-67. DOI:10.1016/j.bios.2016.04.071 |
| [67] |
A. Gupta, P. Prasad, S. Gupta, et al., ACS Appl. Mater. Interfaces 12 (2020) 35967-35976. DOI:10.1021/acsami.0c11161 |
| [68] |
M. Piuri, L. Rondón, E. Urdániz, et al., Appl. Environ. Microbiol. 79 (2013) 5608-5615. DOI:10.1128/AEM.01016-13 |
| [69] |
L. Rondón, M. Piuri, W.R. Jacobs, et al., J. Clin. Microbiol. 49 (2011) 1838-1842. DOI:10.1128/JCM.02476-10 |
| [70] |
J. Minion, E. Leung, D. Menzies, et al., Lancet Infect. Dis. 10 (2010) 688-698. DOI:10.1016/S1473-3099(10)70165-1 |
| [71] |
H.H. El-Hajj, S.A.E. Marras, S. Tyagi, et al., J. Clin. Microbiol. 39 (2001) 4131-4137. DOI:10.1128/JCM.39.11.4131-4137.2001 |
| [72] |
C.R. Horsburgh Jr., J. Am. Med. Assoc. 283 (2000) 2575-2576. DOI:10.1001/jama.283.19.2575 |
| [73] |
W.H. Organization, World Health Organization Global Tuberculosis Report 2013, WHO Press, Geneva, 2014.
|
| [74] |
P. Jain, T.E. Hartman, N. Eisenberg, et al., J. Clin. Microbiol. 50 (2012) 1362-1369. DOI:10.1128/JCM.06192-11 |
| [75] |
M.R. O'Donnell, A. Pym, P. Jain, et al., J. Clin. Microbiol. 53 (2015) 2188-2194. DOI:10.1128/JCM.03530-14 |
| [76] |
J.M. Blair, L.J. Piddock, MBio 7 (2016) e00840-00816. |
| [77] |
R. Iyer, A.L. Erwin, Res. Microbiol. 166 (2015) 516-524. DOI:10.1016/j.resmic.2015.06.006 |
| [78] |
W.J. Lu, H.J. Lin, P.H. Hsu, et al., Antibiotics 9 (2020) 639. DOI:10.3390/antibiotics9100639 |
| [79] |
M.R.L. Stone, U. Łapińska, S. Pagliara, et al., RSC Chem. Biol. 1 (2020) 395-404. DOI:10.1039/d0cb00118j |
| [80] |
D. Anderson, et al., Nat. Rev. Microbiol. 8 (2010) 260-271. DOI:10.2134/jeq2009.0093 |
| [81] |
H.K. Allen, J. Donato, H.H. Wang, et al., Nat. Rev. Microbiol. 8 (2010) 251-259. DOI:10.1038/nrmicro2312 |
| [82] |
J. Ronholm, Z. Zhang, X. Cao, et al., Hybridoma 30 (2011) 43-52. DOI:10.1089/hyb.2010.0066 |
| [83] |
S.A. Khan, A.K. Singh, D. Senapati, et al., Chem. Commun. 47 (2011) 9444-9446. DOI:10.1039/c1cc13199k |
| [84] |
E.D. Weinberg, Microbiol. Rev. 42 (1978) 45-66. DOI:10.1128/mr.42.1.45-66.1978 |
| [85] |
M. van Oosten, T. Schäfer, J.A. Gazendam, et al., Nat. Commun. 4 (2013) 2584. DOI:10.1038/ncomms3584 |
| [86] |
H.Y. Park, S.D. Zoller, V. Hegde, et al., Sci. Rep. 11 (2021) 1622. DOI:10.1038/s41598-020-78362-7 |
| [87] |
T.S.A. Wang, P.L. Chen, Y.C.S. Chen, et al., ACS Infect. Dis. 7 (2021) 2584-2590. DOI:10.1021/acsinfecdis.1c00235 |
| [88] |
B. Ballhausen, A. Kriegeskorte, N. Schleimer, et al., Antimicrob. Agents Chemother. 58 (2014) 3791-3798. DOI:10.1128/AAC.02731-13 |
| [89] |
A. Rohde, J.A. Hammerl, S. Al Dahouk, Ann. Clin. Microbiol. Antimicrob. 15 (2016) 55. DOI:10.1186/s12941-016-0167-8 |
| [90] |
S. Silvestri, E. Rampacci, V. Stefanetti, et al., Front. Vet. Sci. 8 (2021) 740934. DOI:10.3389/fvets.2021.740934 |
 2023, Vol. 34
2023, Vol. 34 

