b Department of Pharmaceutical Analysis, The School of Pharmacy, Fujian Medical University, Fuzhou 350108, China
The exosomes, secreted actively by cells, is extracellular vesicle with lipid bilayer membrane and diameter 40–160 nm (~100 nm on average) [1], carrying a lot of genetic information of mother cell, such as protein, lipid, RNA, DNA. It plays an important role in many physiological and pathological processes, including growth and migration of tumors, cell differentiation, antigen presentation, etc. [2-5]. Moreover, its unique bilayer membrane structure makes it more stable than commonly used biomarkers in liquid biopsy technology, such as circulating tumor cells (CTCs), circulating tumor DNA (ctDNA). It is reported that the blood stored in cold storage for 30 years can still be used to isolate exosomes successfully. At the same time, the bilayer membrane can effectively protect its contents from degradation of enzymes and enrich them properly to reduce the difficulty of detection [6, 7]. In addition, exosomes, which are widely existed in blood, saliva, urine, ascites, milk and other body fluids [8], are more abundant and easier to enrich than CTCs and ctDNA. In brief, exosomes and their contents [9], especially exosomal microRNAs [10-16], are more promising than traditional biomarkers in tracking the dynamic changes of tumors in real time and improving the efficiency of cancer screening and diagnosis.
At present, the most frequently used methods for detecting exosomal microRNA are northern blot hybridization [17] or reverse transcription-polymerase chain reaction (RT-PCR) [18]. However, these methods require either a large amount of samples and time, or complex operation and high cost, which hinder them from widely popularization. The electrochemical biosensor possessing the characteristics of fast analysis, low cost, low energy consumption, easy miniaturization and integration, is more in line with the requirements of point-of-care test (POCT) for large-scale early screening of cancer. Consequently, a large number of researchers have designed multiple electrochemical biosensors for the detection of exosomal miRNAs [19], For example, Boriachek et al. used complementary DNA-functionalized magnetic beads to capture the target miRNA sequence to form a hybrid double chain. The hybrid double chain releases heat and unwinds. The miRNA is detached and directly adsorbed on the surface of SPE-Au through the affinity of RNA-gold. Finally, the redox system of [Fe(CN)6]4−/3− was used to detect the level of exosomal miRNA on the surface of SPE-Au [20]. Zhang et al. based on a bipedal "DNA walker" (single-stranded DNA) and a toehold sequence-mediated DNA strand replacement reaction to construct a signal cascade amplified ratio electrochemical biosensor to achieve efficient detection of exosomes miR-21 [21]. Nevertheless, considering the extremely low level of exosomal microRNAs [22-23] and the complexity of clinical samples, it is of great demand to improve the sensitivity and specificity of the electrochemical biosensor for real applications in cancer diagnosis.
It is noteworthy that the rapid development of DNA self-assembly technology provides a new idea for the construction of ultra-sensitive electrochemical biosensors [24-30]. Through Watson-Crick base pairing, the rational designed DNA sequences can assemble into various shapes of nanoscale architectures with good thermal stability and enzyme tolerance. In recent years, scientists have proposed a variety of biosensors based on DNA self-assembly technology and applied them to the sensitive detection of microRNAs [31-35]. For example, Yuan et al. successfully built quite long DNA duplex on the surface of electrodes using target triggered hybridization chain reaction (HCR), and realized simultaneous detection of two kinds of microRNAs with increased sensitivity [36]. Yu et al. have fabricated a gold nanoparticles (AuNP) network on the electrode surface through integrating duplex-specific nuclease-assisted target recycling with catalytic hairpin assembly reaction for atto-molar (amol/L) detection of microRNA-141 [37]. These sensors above significantly improve the sensitivity of microRNAs detection. However, both the long DNA duplex and the AuNP network structure are not stable enough because they are connected to the electrode only through a single strand of DNA. Moreover, since both of the nanostructures adopt a vertical arrangement perpendicular to the electrode surface, a part of signal molecules on them are far away from the electrode, resulting in limited signal amplification efficiency. Fortunately, Helmig et al. have anchored a large number of DNA hairpin probes acted as "piers" on the DNA origami structure and then constructed a DNA "nano-bridge" over 200 nm long by utilizing HCR. The "nano-bridge" is extremely stable due to its numerous "piers". Furthermore, it is very conducive to the efficient amplification and transmission of signals because its transverse arrangement parallel to the assembly interface made all of the signal groups close to the electrode surface[38]. Based on the advantages of this kind of "nano-bridge", Chao et al. designed a DNA navigation system. By accurately "anchoring" the DNA hybridization chain reaction to the DNA origami structure, a DNA nanobridge "pier" was constructed to realize the design along the nano-interface the molecular cascade of pathways, combining this stable DNA molecular structure with intelligent computing, can provide new research directions for single-molecule intelligent sensing and disease diagnosis and treatment [39].
Inspired by the above research, the electrochemical sensing method based on DNA "nano-bridge" was developed to detect exosomal miR-21 which was always elevated when breast cancer was present. In short, two kinds of hairpins probes caught in a kinetic trap, one is modified on the (gold electrode) GE surface and the other is in solution, can coexist without an initiator. In the presence of target miR-21, the partially "caged" initiator sequence by the stem region of another hairpin probe is liberated and then triggers the cross-opening of the first two kinetically trapped hairpins, leading to the formation of DNA "nano-bridge" as well as the introduction of considerable signal reporters. Compared with the other DNA nano-structure vertical to the electrode surface in previous electrochemical methods, the proposed DNA "nano-bridge" spread on the electrode surface possesses plenty of "piers" and is more stable to ensure the reliability and repeatability of assay. Moreover, the signal groups labeled on the DNA "nano-bridge" is very close to the electrode surface, which increases the efficiency and dependability of signal transmission and lower false positive and false negative probability. In addition, DNA "nano-bridge" carrying substantial negative charges due to the negative charged DNA phosphate backbone, which may prevent the interference from the substance with the same negative charge in the real clinical samples, for example, DNA, protein, etc. To our knowledge, DNA nano-structure is first assembled horizontally on the electrode surface for exosomal miRNAs detection. Combined with the advantages of electrochemical biosensors such as ultra-sensitivity, cost-effective, the designed method may offer new perspective for cancer early point-of-care (POC) diagnosis.
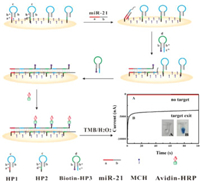
As seen in Fig. 1, two kinds of DNA hairpin probes named HP1 and HP2 are firstly immobilized on the gold electrode (GE) surface through Au-S bond. Both of the probes consist of the same stem and loop sequences but different toehold region specifically selecting the target miR-21 and another hairpin probe with biotin modification termed biotin-HP3, respectively. When miR-21 exists, HP1 is opened to form a HP1- miR-21 duplex, resulting in the loop domain of HP1 exposed. Upon addition of biotin-HP3, it combines with the uncovered loop part of HP1 and makes its own loop region available for hybridization with proximal HP2 on the GE surface. As a result, cascade hybridization chain reaction (HCR) between kinetically trapped HP2 and biotin-HP3 is triggered, resulting in the formation of a DNA "nano-bridge" structure with a large amount of biotin molecule on the GE surface. By adopting affinity reaction between biotin and avidin, the generated "nano-bridge" are linked to massive horseradish peroxidases, which can catalyze the redox reaction of 3, 3′, 5, 5′-tetramethyl benzidine (TMB)-H2O2 efficiently and produce a significant current signal. Theoretically, only one target miR-21 can induce all of the HP2 to assemble into vast DNA "nano-bridge" with enough biotin-HP3, giving rise to a considerable catalytic current signal. Therefore, the target miR-21 can be detected ultra-sensitively by monitoring the current signal. Conversely, in the absence of miR-21, HP1 will not be opened and then activate HCR between HP2 and biotin-HP3 to self-assemble into DNA "nano-bridge", leading to negligible current signal.
|
Download:
|
| Fig. 1. The principle of the electrochemical biosensor. | |
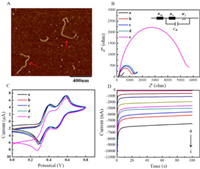
In order to verify the formation of DNA "nano-bridge", atomic force microscope (AFM) imaging was carried out to test the product of HCR reaction of miR-21-HP1-(HP2-HP3)n. As shown in Fig. 2A, a curved DNA long-chain structure with a length of more than 400 nm can be observed in the field of vision (as implied by the red arrow in the figure), indicating the HP1, HP2 and biotin-HP3 can self-assembly into a long DNA "nano-bridge" successfully when the target miR-21 exists.
|
Download:
|
| Fig. 2. (A) AFM imaging of the DNA "nano-bridge". (B) The EIS of various electrodes in 100 mmol/L KCl solution with 5 mmol/L [Fe(CN)6]3−/4− (1:1): (a) bare electrode, (b) HP1/HP2 modified electrode, (c) HP1/HP2 modified electrode with addition of biotin-HP3, (d) HP1/HP2 modified electrode with addition of miR-21, (e) HP1/HP2 modified electrode with addition of biotin-HP3 and miR-21. Insert: The equivalent circuit of EIS. In the circuit, electronic components Raq, Rct, Cdl and W1 represent solution resistance, electron transfer resistance, solution interface capacitance and diffusion impedance, respectively. (C) The CVs of various electrodes in TMB-H2O2 solution: Curves a-e are the same as Fig. 2B. (D) The current-time curves for different concentrations of miR-21 (from a to i): 0 fmol/L, 1 fmol/L, 10 fmol/L, 100 fmol/L, 1 pmol/L, 10 pmol/L, 100 pmol/L, 1 nmol/L and 10 nmol/L. | |
First, electrochemical impedance spectroscopy (EIS) was used to confirm the feasibility of the proposed biosensor. The results of the EIS are fitted with equivalent circuits, and the interface characteristics of the surface modified electrode are reflected by the electronic transfer resistance (Rct) of the equivalent circuit [40, 41]. The negligible Rct (curve a) of the bare GE, as seen in Fig. 2B, exhibits a strong electron transfer capability for process of [Fe(CN)6]3−/4−. On account of the negatively charged phosphate backbones, [Fe(CN)6]3−/4− is far away from the electrode surface, making the Rct of the biosensor constructed with HP1 and HP2 modified electrodes slightly increase (curve b), which in turn proves that HP1 and HP2 are successfully modification of the electrode. The Rct value exhibits only a slight increase when biotin-HP3 is added in the above electrode (curve c), meaning that the cascade hybridization reaction does not occur between the three hairpin probes. Meanwhile, Rct changes little after the target miR-21 was dropped on the HP1/HP2/GE (curve d). However, the value of Rct increases dramatically (curve e) when HP1, HP2, biotin-HP3 and miR-21 coexisted, because only miR-21 can act as an initiator to trigger HCR reaction, leading the formation of the DNA "nano-bridge", and tons of DNA strands are attached to the electrode surface. In short, the EIS showed here demonstrate the feasibility of the sensor.
To further confirm the feasibility of the sensing method of electrochemical biosensor, the cyclic voltammetry (CV) curves of different modified electrodes in TMB-H2O2 and avidin-HRP solution were scanned. As shown in Fig. 2C, we can see two pairs of redox peaks when the bare electrode was immersed into TMB-H2O2 solution (curve a). After surface modification of HP1 and HP2 (curve b), the redox current shows little change. Moreover, the current intensity of HP1/HP2 modified electrode was almost unchanged by adding miR-21 (curve c) or biotin-HP3 (curve d). However, when miR-21 and biotin-HP3 (curve e) were added to HP1/HP2 modified electrode simultaneously, the current signal increased sharply, which ascribes to the initiation ability of miR-21 for DNA "nano-bridges" self-assembly, along with a large number of biotin adhere to the GE surface. All above results are consistent with those of EIS and further suggest that the sensing strategy is feasible.
Experimental parameters were optimized to obtain the optimal performance. The concentration ratio R of HP2 to HP1 (R = CHP2: CHP1) directly affects the length of DNA "nano-bridge" and then the detection sensitivity of the biosensor. Therefore, first of all, R is analyzed experimentally. As shown in Fig. S1A (Supporting information), the catalytic current increases with the increase value of R, reaches its maximum at R = 30, and then decreases with the increase of R value. Therefore, R = 30 was used for subsequent experiments. Subsequently, the effect of incubation time of biotin-HP3 on the length of DNA nanobridge was investigated. As shown in Fig. S1B (Supporting information), the catalytic current reaches its maximum at 60 min. Therefore, 60 min was chosen as the optimal reaction time for biotin-HP3.
Under the optimized experimental conditions, the relationship between the concentration of miR-21 (C) and the catalytic current intensity (I) of the biosensor was investigated by chronoamperometry. As illustrated in Fig. 2D, I increases with the logC from 1 fmol/L to 10 nmol/L. The regression equation (Fig. S2 in Supporting information) is I (nA) = 617.6logC + 478.2 with a correlation coefficient of 0.9919. The LOD is calculated to be 168 amol/L by the 3σ rule, as σ is the standard deviation of the background signal [42]. As expected, the LOD of this method is far less than many recent approaches (Table S2 in Supporting information) [20, 21, 24, 27, 29, 30, 34, 35, 43-51], indicating the excellent sensitivity for exosomal microRNA detection. Compared with the traditional sandwich DNA biosensor, the biosensor proposed here shows better signal efficiency and stability (Fig. S3 in Supporting information). Although some assays based on electrochemistry, exhibited higher sensitivity than our method, those works require either nucleic acid analogues or metal nano-particles, which may need strict control of reaction conditions and high cost. In addition, some assays based on surface enhanced raman scattering (SERS) and surface plasmon resonance (SPR) also show lower LOD than our method. However, due to the lack of uniform and repeatable SERS substrates, the reliable quantification of miRNA using SERS is hindered[44], while the SPR technology is susceptible to the interference of factors such as the composition of samples and environment temperature [52].
In order to evaluate the selectivity of the biosensor to miR-21, the current response values of single base mismatch target (SMT), double bases mismatch target (DMT), three bases mismatch target (TMT), non-complementary target (NCT) and the target miR-21 were investigated under the same conditions. As shown in Fig. 3A, the current intensity induced by miR-21 is significantly higher than that induced by mismatch sequences. Moreover, the sensor can distinguish different types of mismatches and has good selectivity. The proposed approach exhibits excellent selectivity on account of the good discriminative capability of HCR.

|
Download:
|
| Fig. 3. (A) Selectivity of the proposed biosensor: The column diagram of current intensity of the miR-21, SMT, DMT, TMT and NCT, respectively (n = 3). (B) TEM characterization of MCF-7 cell-derived exosomes. Insert: Western-blot analysis of exosomal membrane proteins. | |
In order to clearly demonstrate the reproducibility of the sensing method, 1 pmol/L miR-21 were analyzed by identical and different batches of biosensors for 5 times, respectively. After that, the intra-assay coefficients of variation was 3.74% and inter-assay coefficients of variation was 5.65% (Table S3 in Supporting information). Incubate the used electrode in 90 ℃ water for 5 min, and then rinse it with deionized water several times to revive the sensor. We found that after five cycles of the regeneration process, the signal was only reduced by 8.3% compared to the initial response. The above results show that the biosensor has good stability and regeneration ability.
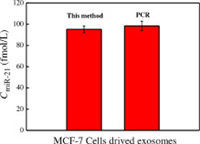
To verify the application of the biosensor in the detection of exosomal miRNAs, we used differential centrifugation to determine the content of exosomal miR-21 isolated from the culture medium of breast cancer cell line MCF-7. The morphology and proteins of the exosomes separated by ultra-centrifugation were characterized. Firstly, the transmission electron microscope (TEM) imaging showed the cup-shaped exosomes with the diameter of ~100 nm in Fig. 3B, which coincides with the previous literatures [53, 54]. Secondly, the exosomal membrane proteins were detected by western blot (WB). Fig. 3B insert shows clear immunoreactive bands corresponding to TSG101 and EpCAM proteins of exosomes, respectively, which was consistent with the parent cells and previous report [24, 41, 55, 56]. In a word, the exsomes can be obtained successfully by ultracentrifugation and be used for exosomal miRNAs detection. Finally, the constructed electrochemical biosensor was used for the quantification of exosomal miR-21 derived from MCF-7. The results were basically consistent with those of PCR (Fig. 4), confirming the practical value of the sensor we designed to detect exosomal miR-21.
|
Download:
|
| Fig. 4. Exosomal miR-21 detection by the proposed electrochemical biosensor and PCR method. | |
All in all, instead of vertical to the electrode surface, the DNA "nano-bridge" constructed here was spread on the surface and be applied in the development of an electrochemical biosensor for amplified detection of exosomal miR-21 for the first time. Due to the numerous "piers", the DNA "nano-bridge" possesses excellent stability, resulting in high reproducibility and reliability of the testing results. Furthermore, this horizontally built DNA nano-structure enables the signal groups on it close to the electrode surface, which increases the efficiency and dependability of signal transmission. At the same time, the tremendous DNA phosphate backbone of DNA "nano-bridge" can hinder the interference from the real clinical samples, leading to low background signal. In addition, cascade signal amplification of HCR makes the electrochemical biosensor to displaying a high degree of sensitivity, and the LOD can be reduced to 168 amol/L. To further improve its performance, further research is needed. For instance, DNAzyme can be used instead of HRP enzyme to reduce testing costs and deduct complicated labeling procedure. And the toehold region of HP1 can become a locked nucleic acid (LNA) modification area to further increase the discriminative ability of the proposed biosensor. In brief, the approach described here lays a foundation for the subsequent construction of sensors that can really be applied to early POCT diagnosis.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThe authors gratefully acknowledge the financial support of Natural Science Foundation of Fujian Province (No. 2020J01545), National Natural Science Foundation of China (No. 21874019), United Fujian Provincial Health and Education Project for Tackling the Key Research, China (No. WKJ2016-2-30), Fujian Science and Technology Innovation Joint Found Project (No. 2019Y9008), Science and Technology Plan Guided Project of Fujian Provincial Science and Technology Department (No. 2020Y0022), Young Top-notch Talent Project of Colleges and Universities of Fujian Province (No. 3002360301).
Supplementary materialsSupplementary material associated with this article can be found, in the online version, at doi: 10.1016/j.cclet.2021.04.056.
| [1] |
R. Kalluri, V.S. LeBleu, Science 367 (2020) eaau6977. DOI:10.1126/science.aau6977 |
| [2] |
J.J. Wang, C. Chen, P.F. Xie, et al., Microbes. Infect. 16 (2014) 283-291. DOI:10.1016/j.micinf.2013.12.001 |
| [3] |
A. Aghebati-Maleki, S. Nami, A. Baghbanzadeh, et al., J. Cell. Physiol. 234 (2019) 21694-21706. DOI:10.1002/jcp.28875 |
| [4] |
M. Groza, A.A. Zimta, A. Irimie, et al., J. Cell. Physiol. 235 (2020) 691-705. DOI:10.1002/jcp.29096 |
| [5] |
H.X. Wang, O. Gires, Cancer Lett. 460 (2019) 54-64. DOI:10.1016/j.canlet.2019.06.012 |
| [6] |
F. Momen-Heravi, B. Saha, K. Kodys, et al., J. Transl. Med. 13 (2015) 261. DOI:10.1186/s12967-015-0623-9 |
| [7] |
L.D. Jiang, Y.W. Gu, Y. Du, J.Y. Liu, Mol. Pharm. 16 (2019) 3333-3349. DOI:10.1021/acs.molpharmaceut.9b00409 |
| [8] |
S. Samanta, S. Rajasingh, N. Drosos, et al., Acta Pharm Sin 39 (2018) 501-513. DOI:10.1038/aps.2017.162 |
| [9] |
K. Jiang, Y. Wu, J. Chen, et al., Chin. Chem. Lett. 32 (2021) 1827-1830. DOI:10.1016/j.cclet.2020.11.031 |
| [10] |
A. Thind, C. Wilson, J. Extracell. Vesicles 5 (2016) 31292. |
| [11] |
M. Salehi, M. Sharifi, J. Cell. Physiol. 233 (2018) 6370-6380. DOI:10.1002/jcp.26481 |
| [12] |
D. Bhagirath, T.L. Yang, N. Bucay, et al., Cancer Res. 78 (2018) 1833-1844. DOI:10.1158/0008-5472.can-17-2069 |
| [13] |
S. Yoshii, Y. Hayashi, H. Iijima, et al., Cancer Sci. 110 (2019) 2396-2407. DOI:10.1111/cas.14084 |
| [14] |
D.G. He, L. Hai, H.Z. Wang, R. Wu, H.W. Li, Analyst 143 (2018) 813-816. DOI:10.1039/C7AN01691C |
| [15] |
B. Kulkarni, P. Kirave, P. Gondaliya, et al., Drug Discov. Today 24 (2019) 2058-2067. DOI:10.1016/j.drudis.2019.06.010 |
| [16] |
Z.C. Zeng, Y.L. Li, Y.J. Pan, et al., Nat. Commun. 9 (2018) 5395. DOI:10.1038/s41467-018-07810-w |
| [17] |
L. Qu, J. Ding, C. Chen, et al., Cancer Cell 29 (2016) 653-668. DOI:10.1016/j.ccell.2016.03.004 |
| [18] |
T. Matsumura, K. Sugimachi, H. Iinuma, et al., Br. J. Cancer 113 (2015) 275-281. DOI:10.1038/bjc.2015.201 |
| [19] |
A.B. Hashkavayi, B.S. Cha, E.S. Lee, S. Kim, K.S. Park, Anal. Chem. 92 (2020) 12733-12740. DOI:10.1021/acs.analchem.0c02745 |
| [20] |
K. Boriachek, M. Umer, M.N. Islam, et al., Analyst 143 (2018) 1662-1669. DOI:10.1039/C7AN01843F |
| [21] |
J. Zhang, L.L. Wang, M.F. Hou, et al., Biosens. Bioelectron. 102 (2018) 33-40. DOI:10.11648/j.ijpp.20180201.17 |
| [22] |
Y.X. Zhao, F. Chen, Q. Li, L.H. Wang, C.H. Fan, Chem. Rev. 115 (2015) 12491-12545. DOI:10.1021/acs.chemrev.5b00428 |
| [23] |
T.D. Canady, N. Li, L.D. Smith, et al., Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 19362-19367. DOI:10.1073/pnas.1904770116 |
| [24] |
Y.K. Xia, L.L. Wang, J. Li, et al., Anal. Chem. 90 (2018) 8969-8976. DOI:10.1021/acs.analchem.8b01143 |
| [25] |
L.Y. Li, L.L. Wang, Q. Xu, et al., ACS Appl. Mater. Interfaces 10 (2018) 6895-6903. DOI:10.1021/acsami.7b17327 |
| [26] |
Q. Hu, J.M. Kong, D.X. Han, L. Niu, X.J. Zhang, ACS Sens. 4 (2019) 235-241. DOI:10.1021/acssensors.8b01357 |
| [27] |
Q.Q. Guo, Y.Q. Yu, H. Zhang, C.X. Cai, Q.M. Shen, Anal. Chem. 92 (2020) 5302-5310. DOI:10.1021/acs.analchem.9b05849 |
| [28] |
L. Liu, H. Lu, R.X. Shi, et al., Anal. Chem. 91 (2019) 13198-13205. DOI:10.1021/acs.analchem.9b03622 |
| [29] |
L.P. Luo, L.L. Wang, L.P. Zeng, et al., Talanta 207 (2020) 120298. DOI:10.1016/j.talanta.2019.120298 |
| [30] |
X.Q. Tang, Y. Wang, L. Zhou, et al., Mikrochim. Acta 187 (2020) 172. DOI:10.1007/s00604-020-4143-9 |
| [31] |
R.R. Huang, N.Y. He, Z.Y. Li, Biosens. Bioelectron. 109 (2018) 27-34. DOI:10.1016/j.bios.2018.02.053 |
| [32] |
L. Tian. J. Qi, O. Oderinde, et al., Biosens. Bioelectron. 110 (2018) 110-117. DOI:10.1016/j.bios.2018.03.045 |
| [33] |
X.L. Zhang, W.M. Li, Y. Zhou, Y.Q. Chai, R. Yuan, Biosens. Bioelectron. 135 (2019) 8-13. |
| [34] |
H.Z. Wang, D.G. He, K.J. Wan, et al., Analyst 145 (2020) 3289-3296. DOI:10.1039/d0an00393j |
| [35] |
R.X. Wang, X.X. Zhao, X.H. Chen, et al., Anal. Chem. 92 (2020) 2176-2185. DOI:10.1021/acs.analchem.9b04814 |
| [36] |
Y.H. Yuan, Y.D. Wu, B.Z. Chi, et al., Biosens. Bioelectron. 97 (2017) 325-331. DOI:10.1016/j.bios.2017.06.022 |
| [37] |
S. Yu, Y.Y. Wang, L.P. Jiang, S. Bi, J.J. Zhu, Anal. Chem. 90 (2018) 4544-4551. DOI:10.1021/acs.analchem.7b04930 |
| [38] |
S. Helmig, K.V. Gothelf, Angew. Chem. Int. Ed. 56 (2017) 13633-13634. DOI:10.1002/anie.201706680 |
| [39] |
J. Chao, J.B. Wang, F. Wang, et al., Nat. Mater. 18 (2019) 273-279. DOI:10.1038/s41563-018-0205-3 |
| [40] |
Z.Q. Ning, Y.J. Zheng, D. Pan, Y.J. Zhang, Y.F. Shen, Biosens. Bioelectron. 150 (2020) 111945. DOI:10.1016/j.bios.2019.111945 |
| [41] |
S. Patra, E. Roy, R. Madhuri, P.K. Sharma, Anal. Chim. Acta 1023 (2018) 271-284. |
| [42] |
Y.H. Wang, D.W. Luo, Y. Fang, et al., Sensor Actuat B: Chem 298 (2019) 126900. DOI:10.1016/j.snb.2019.126900 |
| [43] |
D. He, L. Hai, H. Wang, R. Wu, H.W. Li, Analyst 143 (2018) 813-816. DOI:10.1039/C7AN01691C |
| [44] |
Z. Ramshani, C.G. Zhang, K. Richards, et al., Commun. Biol. 2 (2019) 189. DOI:10.1038/s42003-019-0435-1 |
| [45] |
L. Liu, H. Lu, R.X. Shi, et al., Anal. Chem. 91 (2019) 13198-13205. DOI:10.1021/acs.analchem.9b03622 |
| [46] |
R. Tavallaie, J. McCarroll, M.L. Grand, et al., Nature Nanotech. 13 (2018) 1066-1071. DOI:10.1038/s41565-018-0232-x |
| [47] |
Y.F. Pang, C.G. Wang, L.C. Lu, et al., Biosens. Bioelectron. 130 (2019) 204-213. DOI:10.1016/j.bios.2019.01.039 |
| [48] |
J.U. Lee, W.H. Kim, H.S. Lee, K.H. Park, S.J. Sim, Small 15 (2019) e1804968. DOI:10.1002/smll.201804968 |
| [49] |
G.K. Joshi, S. Deitz-McElyea, T. Liyanage, et al., ACS Nano 9 (2015) 11075-11089. DOI:10.1021/acsnano.5b04527 |
| [50] |
J.X. Zhao, C. Liu, Y.K. Li, et al., J. Am. Chem. Soc. 142 (2020) 4996-5001. DOI:10.1021/jacs.9b13960 |
| [51] |
B. Wang, Z. You, D.H. Ren, Analyst 144 (2019) 2304-2311. DOI:10.1039/c8an02266f |
| [52] |
F. Inci, O. Tokel, S. Wang, et al., ACS Nano 7 (2013) 4733-4745. DOI:10.1021/nn3036232 |
| [53] |
H. Etayash, A.R. McGee, K. Kaur, T. Thundat, Nanoscale 8 (2016) 15137-15141. DOI:10.1039/C6NR03478K |
| [54] |
Y.J. He, F. Deng, S.J... Dai, et al., Biomark. Med. 12 (2018) 177-188. DOI:10.2217/bmm-2017-0305 |
| [55] |
S.A. Melo, H. Sugimoto, J.T. O'Connell, et al., Cancer Cell 26 (2014) 1-15. DOI:10.1016/j.ccr.2014.06.015 |
| [56] |
L. Zhao, R.J. Sun, P. He, X.R. Zhang, Anal. Chem. 91 (2019) 14773-14779. DOI:10.1021/acs.analchem.9b04282 |
 2021, Vol. 32
2021, Vol. 32 

