文章信息
- 王可歆 , 张继婉 , 陈实 . 2016
- WANG Kexin, ZHANG Jiwan, CHEN Shi . 2016
- C9orf 72突变致病的分子机制
- Molecular Pathological Mechanisms Related with C9orf72 Mutation
- 武汉大学学报(理学版), 2016, 62(4): 313-319
- Journal of Wuhan University(Natural Science Edition), 2016, 62(4): 313-319
- http://dx.doi.org/10.14188/j.1671-8836.2016.04.002
-
文章历史
- 收稿日期:2015-03-11

肌萎缩侧索硬化症(amyotrophic lateral sclerosis,ALS)是一种神经退行性疾病.患者的上运动神经元和下运动神经元逐渐发生程序性死亡.神经元不可逆的退行会造成肌肉的萎缩,从而导致患者的运动能力逐渐丧失,最终患者通常死于呼吸系统衰退造成的窒息[1].额颞叶痴呆(frontotemporal dementia,FTD)同样是一种神经退行性疾病,受到影响的细胞主要位于前额叶和前颞叶.FTD发病时间较早,是早发性痴呆的第二大群体[2, 3].FTD患者会在发病后逐渐显现出行为改变、语言表达障碍和执行力受阻等方面的临床表现[3].
C9orf72表达基因的突变是上述两种疾病最主要的致病机制.大约34.2%的ALS患者和25.9%的FTD患者都携带了这种基因突变[4].这两类患者的C9orf72表达基因中插入了大约700到1 600个重复的GGGGCC重复序列[5, 6].如图 1所示,这段重复序列插在C9orf72的内含子里,这个内含子位于C9orf72的两个不翻译的可选外显子1a和1b之间.当RNA转录选择外显子1a时,这段序列位于前体RNA的内含子中,当RNA转录选择外显子1b时,这段序列位于启动子区域[6].
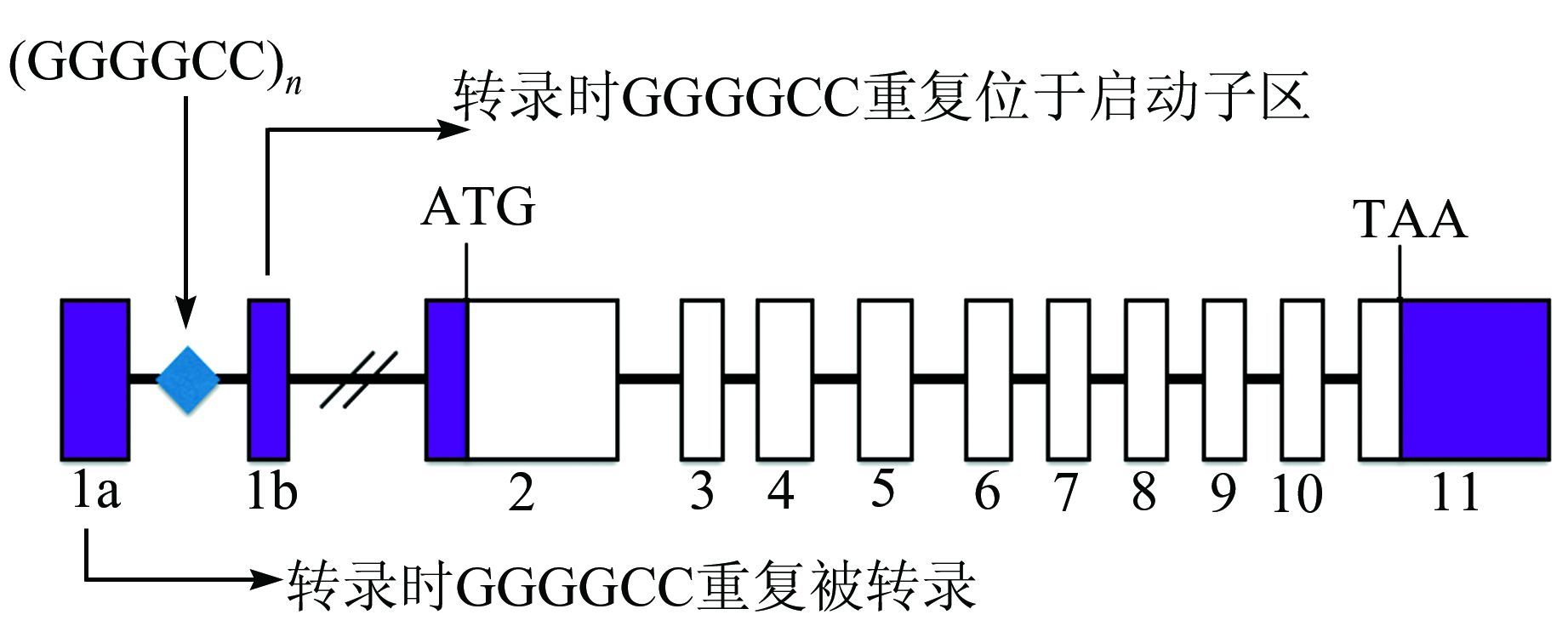
|
| 图 1 突变插入位置示意图 Figure 1 Sketch map of GGGGCC insertion site |
在C9orf72突变的基因型被鉴定出来以后,大量的临床调查发现这种突变是目前鉴定出的ALS和FTD的最主要致病机制,同时发现携带这种基因型的很多患者都出现了ALS和FTD联发的现象.同时,C9orf72的基因突变也被发现与其他的神经退行性疾病如亨廷顿舞蹈症、阿尔兹海默症等有关[7, 8].因为携带这种突变的患者数目众多,而且这种突变造成的疾病谱非常广泛,C9orf72突变的致病机制逐渐成为了研究热点.
由于重复片段插入在C9orf72的非编码区,所以C9orf72蛋白并没有发生氨基酸序列上的改变.由此,如图 2所示,可以简单地将C9orf72突变的致病机制简单分为3类:C9orf72蛋白的功能缺失、GGGGCC长重复RNA的毒性和GGGGCC重复序列翻译产物的神经毒性.
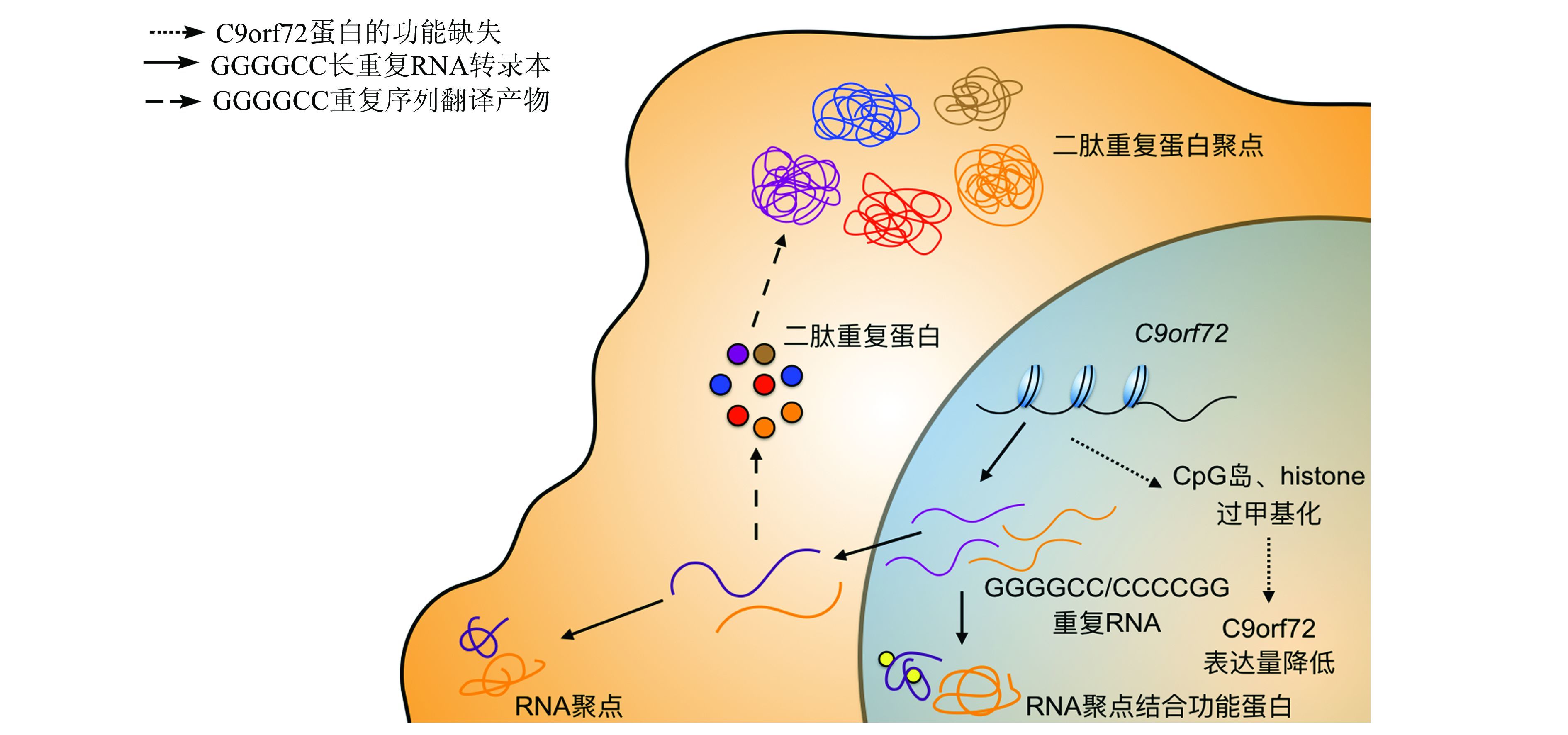
|
| 图 2 C9orf72突变致病的3个致病机制 Figure 2 Sketch map of three pathological pathways |
Belzil等[9]发现携带C9orf72基因突变的人类组织样本的该基因中插入了长GGGGCC片段;Xi等[10]发现,组蛋白H3和H4的3甲基残基可以与之结合,使得插入位点附近的CpG岛甲基化水平的上升.这都可能导致C9orf72转录水平降低.很多研究也表明,C9orf72基因的转录水平在病人组织和诱导多能神经细胞(iPSN)中有显著降低[6, 9, 11~14].另外,一些未携带C9orf722基因突变的散发型ALS和FTD患者样本中也存在C9orf72的表达量降低的现象[15].因此,C9orf72基因表达量的降低被认为是一些神经退行性疾病的致病机制.
1.2 C9orf72的生物学功能生物信息学预测发现C9orf72蛋白含有一个DENN(after differentially expressed in neoplastic versus normal cells)结构域,这预示C9orf72可能是Rab蛋白的GDP (Guanosine diphosphate)-GTP(Guanosine-5′-triphosphate)交换因子[16, 17]. Rab蛋白广泛地参与细胞内的膜运输,它具有两种构象,结合GTP的有活性状态和结合GDP的无活性状态[18].因此,C9orf72可能通过DENN结构调控Rab蛋白构象,影响细胞的内体运输.研究表明,在小鼠神经胶质瘤细胞株Neuro2A、人神经胶质瘤细胞株SH-SY5Y、原代皮层神经元和脊髓运动神经元细胞中,Rab1、5、7、11和C9orf72可以直接相互作用,帮助C9orf72进行胞内囊泡运输和胞外释放,而敲C9orf72会导致胞内运输效率降低.此外,C9orf72也存在于溶酶体中,表明C9orf72可以在蛋白酶体途径和自噬途径中发挥作用[19].
1.3 C9orf72缺失的动物模型人们构建了多种C9orf72基因缺失的动物模型用于研究C9orf72的致病机制.在成年小鼠中枢神经系统中,利用反义寡核苷酸(ASO)将小鼠的C9orf72转录本降低到原水平的30%~40%,小鼠未表现运动强度和运动协调性方面的病理表型;同时,在C9orf72转录本降低小鼠的脊柱、海马、皮质中并未发现C9orf72突变携带者组织、细胞中常出现的TDP-43或是ubiquitin、p62等的蛋白聚集物.这表明,与人类相比,成年小鼠的中枢神经系统对C9orf72缺失有高度耐受性[20].
然而,C9orf72缺失的斑马鱼和线虫出现了严重的神经损伤.在斑马鱼中敲减C9orf72的同源基因,斑马鱼的细胞没有明显的形态学变化,个体却出现了运动方面的细胞病变和行为病变.在敲减C9orf72斑马鱼中高表达人源的C9orf72基因,可以缓解上述病变表型[15].缺失C9orf72同源蛋白的线虫随着年龄的增长,逐渐表现出运动障碍.另外,缺失C9orf72的同源蛋白会提高线虫对于渗透压刺激的敏感性[21].ALS致病基因FUS和TDP-43也参与了调节线虫对渗透压刺激的反应[22, 23],说明与ALS其他致病基因有功能上的联系.
尽管C9orf72在不同物种中具有高度同源性,但不同动物模型之间存在表型差异,可能是由于C9orf72表达水平差异造成的,也可能与敲减C9orf72所处的个体发育阶段有关.
1.4 C9orf72在疾病进程中的作用目前并未证实C9orf72缺失直接导致神经退行性疾病.在患者iPSN细胞中,有针对性地降解含有GGGGCC长重复序列RNA的ASO,可以有效缓解病理表型.但C9orf72进一步降低,不会造成新的或是更严重的病理表型[11].同时,一些临床数据也表明C9orf72功能缺失模型并不成立:在389个ALS患者中的测序表明,C9orf72编码区中未发现其它致病突变,说明ALS发病与C9orf72功能结构域的缺失或突变并无直接联系[24].一般基因功能缺失导致的疾病中,纯合突变的患者表型要明显严重于杂合突变的患者,而携带纯合C9orf72突变患者的临床病理指标并未更加严重[13],因此,C9orf72的缺失可能不是发病的最直接原因.
2 GGGGCC长重复RNA的毒性在人类神经母细胞瘤中过量表达的GGGGCC重复,存在神经毒性可诱导细胞凋亡,且重复序列越长神经毒性越大[25].特异性降解GGGGCC重复RNA的ASO可以在细胞模型中有效缓解患者iPSN的病理表型[11, 20, 26].因此,含GGGGCC重复的RNA具有的神经毒性是C9orf72突变致病的最重要机制[11, 20, 26].
2.1 高重复RNA序列的特殊结构当C9orf72选择外显子1a进行转录时,插入在内含子中的GGGGCC重复序列被转录,导致RNA代谢障碍,如1)RNA聚集:过剩的GGGGCC转录本在转录之后形成稳定的G-四联体结构[27,28]聚集;GGGGCC重复的C9orf72转录本在细胞内形成RNA聚点[6];C9orf72的正义链和反义链上的重复序列被转录形成特异性的RNA聚点[13, 14, 20, 29~31];2)RNA转录终止:重复序列GGGGCC的转录本与单链DNA形成R-环,从而阻止转录的继续进行[32].
2.2 高重复RNA序列结合的重要蛋白CUG/CCUG重复的RNA可大量结合MBNL1蛋白,致使MBNL1蛋白无法行使正常功能,导致强直性肌营养不良[33, 34].因此,与GGGGCC重复RNA结合的蛋白鉴定一直是C9orf72致病机制研究的重点.这类G-四联体结构能够与很多蛋白结合,影响其行使正常功能.
2.2.1 hnRNP家族hnRNP家族在RNA代谢中起重要作用,如:hnRNPA1与前体RNA的代谢相关,并可以和另一个ALS相关蛋白TDP-43相互作用[35],hnRNPA1的阮病毒样结构域突变可以造成多系统蛋白病和ALS[36];hnRNPA3是RNA运输因子,在神经细胞中大量表达[37];hnRNPH能够与多聚鸟嘌呤序列(poly-A)结合,调整RNA剪切中的外显子跳读[38].
G-四联体结构可与hnRNPA1、hnRNPA3、hnRNPH蛋白结合,影响其生物学功能.在携带C9orf72突变患者的iPSN、小脑颗粒细胞中发现,hnRNPA1蛋白和G-四联体有明显的共定位[26, 39].而在体外RNA结合实验中,hnRNPA1却没有特异性地结合G-四联体结构GGGGCC片段.在携带C9orf72突变的患者的海马组织中,hnRNPA3可形成p62阳性/TDP-43阴性的特异性细胞聚集物[40].hnRNPA3通常定位在细胞核中,少量定位在细胞质中[41].携带C9orf72突变患者细胞中,hnRNPA3更多地定位在细胞质[40].但在患者组织和iPSN中,hnRNPA3并未与RNA聚点共定位[25, 26].hnRNPH在患者的小脑组织中可以与30%~70%的RNA聚点共定位[25, 39],在携带C9orf72突变的患者的运动神经元中,也可以与19%的RNA聚点共定位[39].另外,在体外RNA结合实验中,hnRNPH也可与GGGGCCx72相互作用[25].
2.2.2 其他RNA代谢相关蛋白ADARB2是ADAR家族的一个成员.ADAR家族是一类腺苷脱氨酶,可以介导RNA翻译后A-to-I的替换.RNA的A-to-I修饰可以通过替换单碱基完成蛋白质重要氨基酸残基的替换,从而改变该蛋白的活性状态,如:A-to-I修饰造成的GluR2受体中Q-R氨基酸的替换,而Q-R替换的失常会导致严重的神经性病变.散发型ALS患者中也有一些ADAR家族蛋白控制的病理反应.ADARB2可以通过与RNA的竞争性结合调整ADAR家族的功能[42~44].携带C9orf72突变的患者的iPSN中,ADARB2与聚集的G-四联体结构GGGGCCx6.5异常结合.在健康人的iPSN中,敲减ADARB2可导致谷氨酸过敏感,这是C9orf72突变患者iPSN特异的病理特性,说明ADARB2功能缺失是RNA毒性的重要表现[11].
SRSF1和SRSF2属于精氨酸/丝氨酸丰富的剪切蛋白家族.SRSF蛋白广泛参与RNA代谢,如:前体mRNA剪切、RNA运输、RNA降解、RNA翻译等[45, 46].在SH-SY5Y细胞中超量表达GGGGCCx72RNA,会导致SRSF1、SRSF2与RNA聚点的共定位[25],另外,在携带C9orf72突变患者小脑颗粒细胞中,SRSF2可以与33%的RNA聚点共定位;患者的运动神经元中可以与3%的RNA聚点共定位[39].但在携带C9orf72突变的患者的小脑组织中SRSF1和SRSF2只与少量RNA聚点共定位[25].
2.2.3 其他细胞核内蛋白Pur-α蛋白与RNA代谢相关[47].利用GGGGCCx10RNA作为探针,发现在小鼠脊柱裂解液中Pur-α蛋白能与GGGGCC重复RNA特异性结合[47].在携带C9orf72突变的患者小脑中,或过表达GGGGCCx30RNA的果蝇眼组织中,Pur-α蛋白可以形成含有ubiquitin蛋白的细胞聚集物[47].在携带C9orf72突变的患者小脑组织和iPSN中,Pur-α的聚集物与RNA聚点是共定位的[25, 26].但在一些携带C9orf72突变的ALS/FTD患者的iPSN中并未发现Pur-α的聚集[11].
NCL蛋白是一个核仁的磷蛋白,与核糖体的装配与成熟密切相关,对成熟神经细胞的长期维护有重要作用[48].在正常状态下,NCL通常聚集定位在细胞的核仁部位,而在患者细胞中,则通常定位在远离核仁的位置.与RNA结合的实验发现,NCL蛋白可与G-四联体GGGGCCx4RNA特异结合,且携带C9orf72突变的ALS/FTD患者也表现出NCL蛋白造成的病理特征[32].
ALYREF蛋白是细胞核的分子伴侣[39].在患者小脑颗粒细胞中,ALYREF蛋白与26%的RNA聚点共定位,在运动神经元中与29%的RNA聚点共定位[39].
3 GGGGCC重复序列翻译产物的神经毒性尽管GGGGCC重复序列插入在内含子中,这段序列仍可以被翻译:1)由于GGGGCC高度重复序列具有复杂的二级结构,翻译起始不依赖于起始密码子ATG;2)基因的正义链和反义链都可作为模板,从任一个碱基起始翻译,形成poly-GA、poly-GR、poly-GP、poly-PR和poly-PA五种二肽重复蛋白[30,31,49,50].这些异常蛋白可以在携带C9orf72突变患者大脑中形成重要病理标志p62阳性/TDP-43阴性聚集物[30, 31, 49~51].二肽重复蛋白的聚集具有细胞特异性:在携带C9orf72突变患者灰质神经元细胞中出现聚集,但在白质、血管内皮细胞核平滑肌细胞中则未出现聚集[50].
3.1 过表达二肽重复蛋白的毒性携带C9orf72突变患者的组织中,内源性的二肽重复蛋白并没有导致过度的细胞凋亡或坏死倾向[52].而过表达二肽重复蛋白,则表现出细胞毒性[53~55].在果蝇中过表达二肽重复蛋白,尤其是poly-GR和poly-PR,相比GGGGCC重复RNA表现出更强烈的细胞毒性[54].
3.2 二肽重复蛋白毒性与转录本重复长度的关系二肽重复蛋白的毒性与转录本重复的长度密切相关.首先,转录本重复的长度会影响蛋白的翻译水平,如:Poly-GA在重复45次时即可被翻译导致细胞毒性,而poly-GP则需要重复超过145次时才可被翻译[51].其次,翻译蛋白的长度极大地影响二肽重复蛋白的功能和代谢,如:过表达30个重复的二肽重复蛋白时,二肽重复蛋白未发生胞内聚集,但过表达100个重复的poly-GA、poly-GR和poly-PR蛋白则在细胞中发生明显聚集[53, 55].
不同长度的二肽重复蛋白降解途径也有所不同.含30个重复的二肽重复蛋白的降解不依赖于蛋白酶体途径和自噬途径,而含100个重复的二肽重复蛋白的降解却高度依赖于上述途径[53, 55].
3.3 二肽重复蛋白毒性的分子机理1) 影响B23蛋白的定位.B23是一个重要的细胞核蛋白,其定位的改变会造成核仁压力[55].30个重复的poly-GR和poly-PR蛋白定位在核仁上,可导致B23蛋白定位的改变[55].过表达30个重复的poly-GA、poly-GP和poly-PA导致正常细胞中应激颗粒(stress granule)的形成,表明细胞的RNA代谢受到了影响;而过表达30个重复的poly-GR和poly-PR会抑制应激颗粒的形成[55].
2) 影响蛋白酶体复合物的功能.100个重复的poly-PR可以提高蛋白酶体复合物的降解速率,而100个重复的poly-GA、poly-GR和poly-GP会抑制其降解速率[53].
3) 影响TDP-43的表达量.二肽重复单位可以破坏细胞中TDP-43表达的稳态,过表达100个重复的poly-GA、poly-GP和poly-GR可以上调TDP-43的表达,而过表达100个重复的poly-GR则会使TDP-43的表达量降低[53].在携带C9orf72突变的患者额叶皮层、初级运动皮层、齿状回神经细胞和海马CA4区域的神经细胞中,ALS致病蛋白TDP-43与一些二肽重复蛋白聚集物异常聚集[56].
4 结 论本文总结了C9orf72突变致病的3种机制:C9orf72蛋白功能缺失、RNA转录本的细胞毒性和二肽重复蛋白的细胞毒性的分子致病机制.3种致病机制并不是独立作用的,不同的致病机制可能会作用到相同的通路,造成叠加效应.
目前在GGGGCC重复RNA蛋白方面的研究结果没有表现出一致性和可重复性,这可能是由于:1) 组织和细胞的特异性;2) 实验中假阳性结果的干扰;3) 长重复序列的基因组层面的体细胞不稳定性;4) 长重复序列在转录时可能被提前终止;5) 荧光原位杂交等实验的缺陷,使得无法研究特定重复次数的RNA的致病机制.长重复序列在DNA复制过程中往往会发生基因重组,阻碍其全长的体外克隆[25, 26, 49],因此,很难得到理想的实验模型[54].鉴于此,构建合理的长重复RNA疾病模型仍然是目前研究C9orf72突变致病机制的重点.
| [1] | SWINNEN B, ROBBERECHT W. The phenotypic variability of amyotrophic lateral sclerosis[J]. Nature Reviews Neurology, 2014, 10 (11) : 661 –670. |
| [2] | NEARY D, SNOWDEN J, MANN D. Frontotemporal dementia[J]. The Lancet Neurology, 2005, 4 (11) : 771 –780. |
| [3] | SEELAAR H, ROHRER J D, PIJNENBURG Y A L, et al. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: A review[J]. Journal of Neurology, Neurosurgery, and Psychiatry, 2011, 82 (5) : 476 –486. |
| [4] | VAN BLITTERSWIJK M, DEJESUS-HERNANDEZ M, RADEMAKERS R. How do C9orf72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: Can we learn from other noncoding repeat expansion disorders?[J]. Current Opinion in Neurology, 2012, 25 (6) : 689 –700. |
| [5] | RENTON A E, MAJOUNIE E, WAITE A, et al. A hexanucleotide repeat expansion in C9orf72 is the cause of chromosome 9p21-linked ALS-FTD[J]. Neuron, 2011, 72 (2) : 257 –268. |
| [6] | DEJESUS-HERNANDEZ M, MACKENZIE I R, BOEVE B F, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9orf72 causes chromosome 9p-linked FTD and ALS[J]. Neuron, 2011, 72 (2) : 245 –256. |
| [7] | KOHLI M A, JOHN-WILLIAMS K, RAJBHANDARY R, et al. Repeat expansions in the C9orf72 gene contribute to Alzheimer’s disease in Caucasians[J]. Neurobiology of Aging, 2013, 34 (5) : 1519.e5 –1519.e12. |
| [8] | KOUTSIS G, KARADIMA G, KARTANOU C, et al. C9orf72 hexanucleotide repeat expansions are a frequent cause of Huntington disease phenocopies in the Greek population[J]. Neurobiology of Aging, 2015, 36 (1) : 547.e13 –547.e16. |
| [9] | BELZIL V V, BAUER P O, PRUDENCIO M, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood[J]. Acta Neuropathologica, 2013, 126 (6) : 895 –905. |
| [10] | XI Z, ZINMAN L, MORENO D, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion[J]. American Journal of Human Genetics, 2013, 92 (6) : 981 –989. |
| [11] | DONNELLY C J, ZHANG P-W, PHAM J T, et al. RNA toxicity from the ALS/FTD C9orf72 expansion is mitigated by antisense intervention[J]. Neuron, 2013, 80 (2) : 415 –428. |
| [12] | ALMEIDA S, GASCON E, TRAN H, et al. Modeling key pathological features of frontotemporal dementia with C9orf72 repeat expansion in iPSC-derived human neurons[J]. Acta Neuropathologica, 2013, 126 (3) : 385 –399. |
| [13] | FRATTA P, POULTER M, LASHLEY T, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia[J]. Acta Neuropathologica, 2013, 126 (3) : 401 –409. |
| [14] | GIJSELINCK I, VAN LANGENHOVE T, VAN DER ZEE J, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study[J]. The Lancet Neurology, 2012, 11 (1) : 54 –65. |
| [15] | CIURA S, LATTANTE S, LE BER I, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis[J]. Annals of Neurology, 2013, 74 (2) : 180 –187. |
| [16] | ZHANG D, IYER L M, HE F, et al. Discovery of novel DENN proteins: Implications for the evolution of eukaryotic intracellular membrane structures and human disease[J]. Frontiers in Genetics, 2012, 3 : 283 . |
| [17] | LEVINE T P, DANIELS R D, GATTA A T, et al. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs[J]. Bioinformatics, 2013, 29 (4) : 499 –503. |
| [18] | CHERFILS J, ZEGHOUF M. Regulation of small GTPases by GEFs, GAPs, and GDIs[J]. Physiological Reviews, 2013, 93 (1) : 269 –309. |
| [19] | FARG M A, SUNDARAMOORTHY V, SULTANA J M, et al. C9orf72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking[J]. Human Molecular Genetics, 2014, 23 (13) : 3579 –3595. |
| [20] | LAGIER-TOURENNE C, BAUGHN M, RIGO F, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110 (47) : 4530 –4539. |
| [21] | THERRIEN M, ROULEAU G A, DION P A, et al. Deletion of C9orf72 results in motor neuron degeneration and stress sensitivity in C[J]. Plos One, 2013, 8 (12) : e83450 . |
| [22] | VACCARO A, TAUFFENBERGER A, ASH P E A, et al. TDP-1/TDP-43 regulates stress signaling and age-dependent proteotoxicity in Caenorhabditis elegans[J]. Plos Genetics, 2012, 8 (7) : e1002806 . |
| [23] | SAMA R R K, WARD C L, KAUSHANSKY L J, et al. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress[J]. Journal of Cellular Physiology, 2013, 228 (11) : 2222 –2231. |
| [24] | HARMS M B, CADY J, ZAIDMAN C, et al. Lack of C9orf72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis[J]. Neurobiology of Aging, 2013, 34 (9) : 2234.e13 –2234.e19. |
| [25] | LEE Y B, CHEN H J, PERES J N, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic[J]. Cell Reports, 2013, 5 (5) : 1178 –1186. |
| [26] | SAREEN D, O’ROURKE J G, MEERA P, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9orf72 repeat expansion[J]. Science Translational Medicine, 2013, 5 (208) : 208ra149 . |
| [27] | REDDY K, ZAMIRI B, STANLEY S Y R, et al. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures[J]. The Journal of Biological Chemistry, 2013, 288 (14) : 9860 –9866. |
| [28] | FRATTA P, MIZIELINSKA S, NICOLL A J, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes[J]. Scientific Reports, 2012, 2 : 1016 . |
| [29] | MIZIELINSKA S, LASHLEY T, NORONA F E, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci[J]. Acta Neuropathologica, 2013, 126 (6) : 845 –857. |
| [30] | ZU T, LIU Y, BAEZ-CORONEL M, et al. RAN proteins and RNA foci from antisense transcripts in C9orf72 ALS and frontotemporal dementia[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110 (51) : 4968 –4977. |
| [31] | GENDRON T F, BIENIEK K F, ZHANG Y-J, et al. Antisense transcripts of the expanded C9orf72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS[J]. Acta Neuropathologica, 2013, 126 (6) : 829 –844. |
| [32] | HAEUSLER A R, DONNELLY C J, PERIZ G, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease[J]. Nature, 2014, 507 (7491) : 195 –200. |
| [33] | KANADIA R N, URBINATI C R, CRUSSELLE V J, et al. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3[J]. Gene Expression Patterns: GEP, 2003, 3 (4) : 459 –462. |
| [34] | MILLER J W, URBINATI C R, TENG-UMNUAY P, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy[J]. The EMBO Journal, 2000, 19 (17) : 4439 –4448. |
| [35] | BURATTI E, BRINDISI A, GIOMBI M, et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing[J]. The Journal of Biological Chemistry, 2005, 280 (45) : 37572 –37584. |
| [36] | KIM H J, KIM N C, WANG Y D, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS[J]. Nature, 2013, 495 (7442) : 467 –473. |
| [37] | MA A S W, MORAN-JONES K, SHAN J, et al. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein[J]. The Journal of Biological Chemistry, 2002, 277 (20) : 18010 –18020. |
| [38] | XIAO X, WANG Z, JANG M, et al. Splice site strength-dependent activity and genetic buffering by poly-G runs[J]. Nature Structural and Molecular Biology, 2009, 16 (10) : 1094 –1100. |
| [39] | COOPER-KNOCK J, WALSH M J, HIGGINBOTTOM A, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions[J]. Brain, 2014, 137 (7) : 2040 –2051. |
| [40] | MORI K, LAMMICH S, MACKENZIE I R A, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations[J]. Acta Neuropathologica, 2013, 125 (3) : 413 –423. |
| [41] | PAPADOPOULOU C, BOUKAKIS G, GANOU V, et al. Expression profile and interactions of hnRNP A3 within hnRNP/mRNP complexes in mammals[J]. Archives of Biochemistry and Biophysics, 2012, 523 (2) : 151 –160. |
| [42] | CHEN C X, CHO D S, WANG Q, et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains[J]. RNA, 2000, 6 (5) : 755 –767. |
| [43] | HIDEYAMA T, YAMASHITA T, SUZUKI T, et al. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2[J]. The Journal of Neuroscience, 2010, 30 (36) : 11917 –11925. |
| [44] | HIDEYAMA T, YAMASHITA T, AIZAWA H, et al. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons[J]. Neurobiology of Disease, 2012, 45 (3) : 1121 –1128. |
| [45] | TWYFFELS L, GUEYDAN C, KRUYS V. Shuttling SR proteins: more than splicing factors[J]. The FEBS journal, 2011, 278 (18) : 3246 –3255. |
| [46] | SHEPARD P J, HERTEL K J. The SR protein family[J]. Genome Biology, 2009, 10 (10) : 242 . |
| [47] | XU Z, POIDEVIN M, LI X, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110 (19) : 7778 –7783. |
| [48] | HETMAN M, PIETRZAK M. Emerging roles of the neuronal nucleolus[J]. Trends in Neurosciences, 2012, 35 (5) : 305 –314. |
| [49] | MORI K, ARZBERGER T, GR SSER F A, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins[J]. Acta Neuropathologica, 2013, 126 (6) : 881 –893. |
| [50] | ASH P E A, BIENIEK K F, GENDRON T F, et al. Unconventional translation of C9orf72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS[J]. Neuron, 2013, 77 (4) : 639 –646. |
| [51] | MORI K, WENG S-M, ARZBERGER T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS[J]. Science, 2013, 339 (6125) : 1335 –1338. |
| [52] | MANN D M A, ROLLINSON S, ROBINSON A, et al. Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9orf72[J]. Acta Neuropathologica Communications, 2013, 1 (1) : 68 . |
| [53] | YAMAKAWA M, ITO D, HONDA T, et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS[J]. Human Molecular Genetics, 2014, 24 (6) : 1630 –1645. |
| [54] | MIZIELINSKA S, GR NKE S, NICCOLI T, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins[J]. Science, 2014, 345 (6201) : 1192 –1194. |
| [55] | TAO Z, WANG H, XIA Q, et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity[J]. Human Molecular Genetics, 2015, 24 (9) : 2426 –2441. |
| [56] | MACKENZIE I R, ARZBERGER T, KREMMER E, et al. Dipeptide repeat protein pathology in C9orf72 mutation cases: clinico-pathological correlations[J]. Acta Neuropathologica, 2013, 126 (6) : 859 –879. |
 2016, Vol. 62
2016, Vol. 62





