2. Department of Anatomical and Cellular Pathology, Prince of Wales Hospital, the Chinese University of Hong Kong, Hong Kong 999077, China;
3. Department of Pathology, Huashan Hospital, Fudan University, Shanghai 200040, China;
4. Department of Neurosurgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou 450052, China;
5. Department of Radiotherapy, Huashan Hospital, Fudan University, Shanghai 200040, China
Ping Zhong, Professor 1989.7–1995.6 Huashan Hospital Affiliated Fudan University, General resident
1995.7–2000.6 Huashan Hospital Affiliated Fudan University, Attending physician
1995.7–2006.3 Huashan Hospital Affiliated Fudan University, department administrative secretary
2000.6–2013.11 Huashan Hospital Affiliated Fudan University, Deputy Chief Physician
2006.3–2011.12 Huashan Hospital Affiliated Fudan University, department secretary
2006.3–present Huashan Hospital Affiliated Fudan University, deputy director of department of neurosurgery
2008–present Huashan Hospital Affiliated Fudan University, Master Instructor
2013–present Huashan Hospital Affiliated Fudan University, Chief physician
2014.11–present Member of Chinese Medical Association, Department of Neurosurgery.
Medulloblastoma is a kind of malignant brain tumor with poor prognosis[1-5]. Recent progress in the genomic characterization of medulloblastoma has shown that medulloblastoma is a heterogeneous disease comprised of 4 molecular subgroups: sonic hedgehog (SHH), wingless (WNT), Group 3, and Group 4[4-9]. Signaling pathways or biological programs driving the pathogenesis of Group 3 and Group 4 subgroups have remained largely elusive[10]. Moreover, studies have demonstrated that these two subgroups overlap in both molecular profiling and clinical features[10-13], and Group 3 and Group 4 subgroups together are called Non-SHH/WNT subgroup[14]. There is a controversy in the neurosurgical community that complete resection is prognostically superior to an incomplete resection. Some studies show that there is no prognostic difference between gross total resection and neartotal resection[15]. This is the first large-scale study to investigate the prognostic value of the extent of resection in a subgroup-specific manner in patients from the Chinese population.
2 Methods 2.1 Patient characteristicsSurgical resections took place from January 2002 to December 2013 at the Huashan Hospital (Single centre). Patients were excluded for study inclusion if they had incomplete data about molecular subgroup, extent of resection, EFS, or OS. Specimens of 113 patients with medulloblastoma were stored in accordance with the Ethics Review of Huashan Hospital. Medulloblastomas were diagnosed by two or more pathologists. Complete or incomplete resection was defined based on surgeons' reports and confirmed by postoperative cranial computer tomography (CT). The primary analysis outcome was the effect of extent of resection by molecular subgroup on overall and event-free survival. This study was approved by the Ethics Committee of Huashan Hospital, Fudan University.
2.2 Molecular subgroup determinationFormalin-fixed paraffin embedded (FFPE) tissues from the primary tumor resection were available in all 113 cases. Hematoxylin and eosin (H & E)-stained sections were reviewed by three senior pathologists (YW, HKN, and JX) and tumors were classified according to the 2016 WHO classification of tumors of the central nervous system[16]. We identified Group3 and Group4 subgroups as NON-SHH/WNT subgroups.
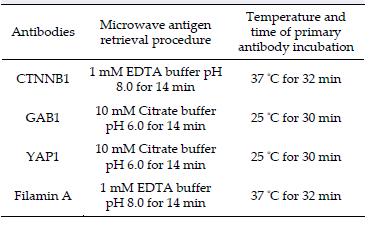
According to a study of medulloblastoma from Fattet et al.[17], the subgroups can be identified by immunohistochemistry. Paraffin sections of 4 μm thickness were examined for immunoreactivity by four antibodies, namely β-catenin (Dako #M3539; 1:30), GAB1 (Abcam #ab27439; 1:100), YAP1 (Santa Cruz #sc-101199; 1:100), and Filamin A (Fitzgerald #10RF113A; 1:400). Tissue slides were dewaxed by xylene and rehydrated in a series of descending grades of alcohol. Antigen retrieval was then done by procedures listed in Table 1. Subsequent steps of CTNNB1 and Filamin A immunostainings were performed on automated Ventana Benchmark XT immunostainer (Ventana Medical Systems, Tucson, AZ, USA). Immunostaining of GAB1 and YAP1 was carried out on Leica BOND-MAX immunostainer (Leica Microsystems Inc., Buffalo Grove, IL, USA). In brief, slides were incubated with the primary antibodies at the temperature and duration listed in Table 1. Endogenous peroxidase activity was blocked by incubation with OptiView Peroxidase Inhibitor for 5 min at room temperature. OptiView HQ Universal Linker was used as the secondary antibody. Signals were detected by 3, 3'-diaminobenzidine tetrahydrochloride (DAB) and counterstained with Mayer's hematoxylin. Positive immunoreactivity was considered according to the criteria stated in studies by Ellison et al. and Min et al.[17, 18].
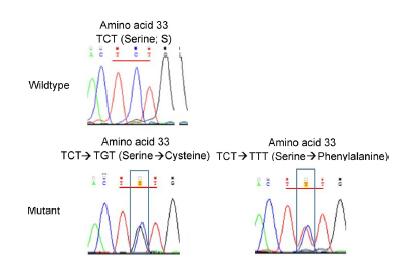
Sequencing was used to confirm the mutation exon 3 of CTNNB1 in FFPE slides. Paraffin sections containing >80% tumor content were dewaxed, washed with ethanol, dried, and scrapped off into microcentrifuge tubes[19]. The tissues were then treated with proteinase K in 10 mM Tris-HCl buffer (pH 8.5) at 55 ℃ for 30 min followed by 98 ℃ for 10 min. The crude cell lysate was centrifuged and the supernatant was taken. PCR amplification was performed in a 20 μL volume containing 1 μL cell lysate, 1 μL of 10 μM forward and reverse primers, and 1 x KAPA2G Robust HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA). Amplification was conducted under the conditions of 95 ℃ for 4 min, followed by 40 cycles of 95 ℃ for 15 s, 64 ℃ for 15 s, and 72 ℃ for 15 s in a PCR machine. PCR products were confirmed by gel electrophoresis and purified by MEGAquickspin TM Total fragment DNA purification kit (iNtRON Biotechnology, Kyungki-Do, Korea). Purified products were sequenced using BigDye Terminator v1.1 Cycle Sequencing Kit with ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
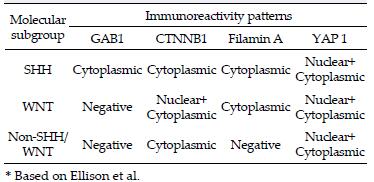
Molecular subgroup determination: immunoreactivity patterns for SHH, WNT, and Non-SHH/WNT subgroups are listed in Table 2. We combined the results of β-catenin immunohistochemistry (IHC) and CTNNB1 exon 3 mutation analysis for determination of WNT medulloblastoma.
Statistical analysis was performed using software IBM SPSS Statistics 19 (IBM Corp., Armonk, NY, USA). Continuous variables between the groups were compared using non-parametric Kruskal-Wallis test. Kaplan-meier survival curve shows event-free survival and overall survival by extent of resection and molecular subgroup. Statistical significance was defined as P < 0.05.
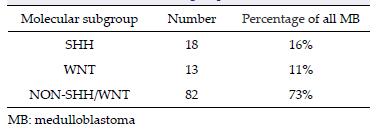
3 Results 3.1 Histological and molecular subgroups of the studyIHC was performed in all 113 cases. CTNNB1 exon 3 mutation analysis was performed in 101 cases (12 cases were unavailable for sequencing). By combining the results of the IHC staining (Figure 1) and CTNNB1 exon 3 mutation data (Figure 2), 18 (16%), 13 (11%), and 82 (73%) of tumors were assigned to SHH, WNT, and Non-SHH/WNT subgroups, respectively[17] (Table 3).
|
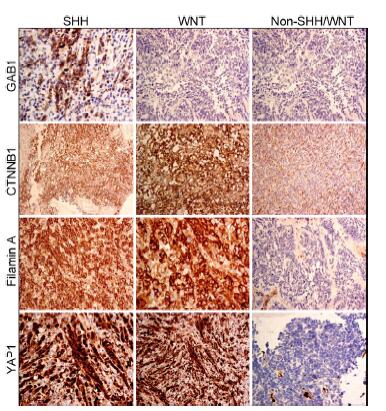
| Figure 1 IHC staining of medulloblastoma FFPE. GAB1, CTNNB1, Filamin A, and YAP1 antibodies were used to determine the different subgroups. IHC: Immunohistochemistry; FFPE: formalinfixed paraffin embedded. |
|
| Figure 2 Sequencing performed to confirm the mutation exon 3 of CTNNB1 in FFPE slides. FFPE: formalin-fixed paraffin embedded. |
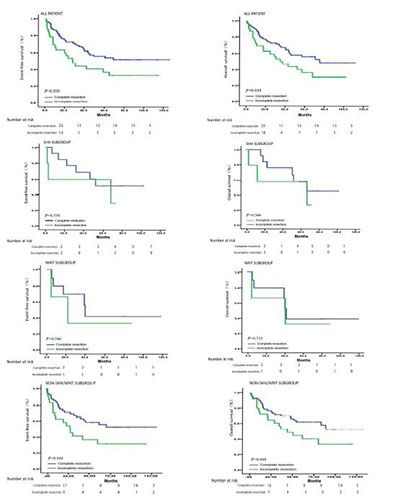
In this study, 81 patients had complete resection (10 patients with WNT tumors, 19 with SHH, 62 with NON-SHH/WNT) and 32 cases had incomplete resection (3 patients with WNT tumors, 5 with SHH, 24 with NON-SHH/WNT). For patients with WNT tumors, incomplete and complete resections were not significantly associated with risk of progression (P = 0.546) and overall survival (P = 0.752). After analysis of patients with SHH tumors, incomplete and complete resections were not significantly associated with risk of progression (P = 0.546) and overall survival (P = 0.566). Incomplete resection was significantly associated with progression for patients with NON-SHH/WNT tumors compared with complete resection (OS: P = 0.048; EFS: P = 0.044) (Figure 3). Therefore, we believe that for the SHH and WNT subgroups of medulloblastoma, the degree of surgical resection does not affect the overall survival and event-free survival of patients, and the impact on the prognosis is limited. For NON-SHH\WNT subgroup of medulloblastoma, the degree of surgical resection has significant effect on the overall survival and event-free survival. We believe that the prognostic benefit of increased extent of resection for patients with medulloblastoma is attenuated after molecular subgroup affiliation is taken into account.
|
| Figure 3 The overall and event-free survival for extent of resection by subgroup. The molecular subgroups are WNT, SHH, and NON-SHH\WNT. Numbers in the parentheses are failure events during that time period. P values are based on the log-rank test across the two strata (Complete resection vs. Incomplete resection). |
In our study, 27 patients were adults, 86 were children, and there were no infants. Because of a lack of infant patients with medulloblastoma, we did not do further study on the effect of age on the molecular groups of patients with medulloblastoma. The rapid progress in the high-throughput genomic analysis technique has increased our knowledge of the molecular mechanism of medulloblastoma. Through the analysis of global gene expression and copy human profiling, medulloblastoma is no longer considered as a single disease. Rather, medulloblastoma refers to heterogeneous diseases comprised at least of four molecular subgroups, which differ by gene expression patterns and abnormalities. Different subgroups have different treatment strategies; for example, the WNT subgroup has the best prognosis after standard surgical removal, postoperative radiotherapy, and chemotherapy. Reports of complete and incomplete resections of medulloblastoma from the Europe[15] demonstrated the different prognosis among the molecular subgroups. Here, we reported a large case series of medulloblastoma based on the Chinese population and investigated the prognostic value for tumor extent of resection among the molecular subgroups.
Some studies[19] classified medulloblastoma into three molecular subgroups (SHH, WNT, Non-SHH/WNT), instead of four subgroups (SHH, WNT, Group 3, and Group 4). There are two main reasons for the adoption of three subgroups: one is that Group 3 and Group 4 medulloblastoma were much less well defined and both showed neuronal/photoreceptor differentiation, and their clinical features were largely similar according to previous studies [10-12, 18-22]. Another reason is the lack of reliable, rapid techniques to differentiate Group 3 and Group 4[18]. Transcriptome profiling analysis remains as the gold standard in molecular subgrouping [21]. Although IHC is a method that can be applied for molecular classification, it is limited by the absence of reliable IHC markers to differentiate Group 3 and Group 4. For instance, IHC positivity for NPR3 and KCNA1 were suggested as the markers for Group 3 and Group 4 medulloblastoma[11], but it was then demonstrated that the specificity and sensitivity of NPR3 and KCNA1 IHC was too low to be used in clinical practice[18, 23]. Thus, in this study, we employed both immunohistochemical analysis of GAB1, YAP1, and Filamin A and sequencing analysis for CTNNB1 mutation to assign the molecular subgroups to 113 medulloblastoma. These antibodies are known to identify the WNT, SHH, and Non-SHH/ WNT subgroups[14], and CTNNB1 mutation analysis is applied to avoid false, positive IHC result[23]. In conclusion, our method has many advantages: fast, low cost, and applicable for classifying the subgroups of medulloblastoma.
Thompson et al.[15] found no significant survival benefit existed for greater extent of resection for patients with WNT and SHH tumors. This is consistent with our findings. According to their study, for patients with group 4 tumors, complete resection conferred a benefit to event-free survival compared with incomplete resection, but with no effect on overall survival. The present study showed that incomplete resection was significantly associated with the progression for patients with NON-SHH/WNT tumors compared with complete resection (OS: P = 0.048; EFS: P = 0.044). A study in Canada[15] identified an event-free survival benefit for complete resection over incomplete resection but no overall survival benefit. The present study found that greater extent of resection (OS: P = 0.038, EFS: P = 0.030) offered event-free survival and overall survival benefit to patients with medulloblastoma. This may be due to our smaller sample size compared with other studies. For patients from the WNT and SHH subgroups, surgical resection of small residual portions is not recommended when the likelihood of postoperative complications is high because there is no definitive benefit to complete resection compared with incomplete resection. For patients in the NON-SHH/WNT subgroup, a secondlook surgery may seem beneficial for survival, but after taking surgical morbidity into account, we remain cautious as to the volume of surgical resection of small residual portions.
In conclusion, the present study is the first to our best knowledge to demonstrate the prognostic value of tumor extent of resection among the molecular subgroups of 113 medulloblastoma diagnosed in China. We consistently found the value of tumor extent of resection among molecular subgroups of Chinese patients with medulloblastoma, and such differences were also reported in American and European studies. The prognostic benefit of increased extent of resection for patients with medulloblastoma is attenuated after the molecular subgroups are taken into consideration. We need further studies to assess the benefit of surgical resection of small residual portions. We believe our first research data on Chinese patients could further enhance the understanding of medulloblastoma. We hope that all newly diagnosed medulloblastoma patients in China should be evaluated for molecular subgroups, and tailor-made treatment should be given based on the molecular classification.
Conflict of interests
All contributing authors have no conflict of interests.
| [1] | Gottardo NG, Hansford JR, McGlade JP, Alvaro F, Ashley DM, Bailey S, Baker DL, Bourdeaut F, Cho YJ, Clay M, Clifford SC, Cohn RJ, Cole CH, Dallas PB, Downie P, Doz F, Ellison DW, Endersby R, Fisher PG, Hassall T, Heath JA, Hii HL, Jones DT, Junckerstorff R, Kellie S, Kool M, Kotecha RS, Lichter P, Laughton SJ, Lee S, McCowage G, Northcott PA, Olson JM, Packer RJ, Pfister SM, Pietsch T, Pizer B, Pomeroy SL, Remke M, Robinson GW, Rutkowski S, Schoep T, Shelat AA, Stewart CF, Sullivan M, Taylor MD, Wainwright B, Walwyn T, Weiss WA, Williamson D, Gajjar A. Medulloblastoma Down Under 2013:A report from the third annual meeting of the International Medulloblastoma Working Group. Acta Neuropathol, 2014, 127(2): 189–201. DOI:10.1007/s00401-013-1213-7 |
| [2] | Dubuc AM, Morrissy AS, Kloosterhof NK, Northcott PA, Yu EP, Shih D, Peacock J, Grajkowska W, van Meter T, Eberhart CG, Pfister S, Marra MA, Weiss WA, Scherer SW, Rutka JT, French PJ, Taylor MD. Subgroup-specific alternative splicing in medulloblastoma. Acta Neuropathol, 2012, 123(4): 485–499. DOI:10.1007/s00401-012-0959-7 |
| [3] | Ramaswamy V, Remke M, Bouffet E, Faria CC, Perreault S, Cho YJ, Shih DJ, Luu B, Dubuc AM, Northcott PA, Schüller U, Gururangan S, McLendon R, Bigner D, Fouladi M, Ligon KL, Pomeroy SL, Dunn S, Triscott J, Jabado N, Fontebasso A, Jones DT, Kool M, Karajannis MA, Gardner SL, Zagzag D, Nunes S, Pimentel J, Mora J, Lipp E, Walter AW, Ryzhova M, Zheludkova O, Kumirova E, Alshami J, Croul SE, Rutka JT, Hawkins C, Tabori U, Codispoti KE, Packer RJ, Pfister SM, Korshunov A, Taylor MD. Recurrence patterns across medulloblastoma subgroups:An integrated clinical and molecular analysis. Lancet Oncol, 2013, 14(12): 1200–1207. DOI:10.1016/S1470-2045(13)70449-2 |
| [4] | Schroeder K, Gururangan S. Molecular variants and mutations in medulloblastoma. Pharmgenomics Pers Med, 2014, 7: 43–51. |
| [5] | Gerber NU, Mynarek M, von Hoff K, Friedrich C, Resch A, Rutkowski S. Recent developments and current concepts in medulloblastoma. Cancer Treat Rev, 2014, 40(3): 356–365. DOI:10.1016/j.ctrv.2013.11.010 |
| [6] | Hooper CM, Hawes SM, Kees UR. Gene expression analyses of the spatio-temporal relationships of human medulloblastoma subgroups during early human neurogenesis. PLoS One, 2014, 9(11): e112909. DOI:10.1371/journal.pone.0112909 |
| [7] | Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gururangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. Novel mutations target distinct subgroups of medulloblastoma. Nature, 2012, 488(7409): 43–48. DOI:10.1038/nature11213 |
| [8] | Paul A Northcott, David JH Shih, John Peacock, Livia Garzia, Sorana Morrissy, Thomas Zichner, Adrian M Stütz, Andrey Korshunov. Juri Reimand, Steven E Schumacher, Rameen Beroukhim, David W Ellison, Christian R Marshall, Anath C Lionel, Stephen Mack, Adrian Dubuc, Yuan Yao, Vijay Ramaswamy, Betty Luu, Adi Rolider, Florence Cavalli, Xin Wang, Marc Remke, Xiaochong Wu, Readman YB Chiu, Andy Chu, Eric Chuah, Richard D Corbett, Gemma R Hoad, Shaun D Jackman, Yisu Li, Allan Lo, Karen L Mungall, Ka Ming Nip, Jenny Q Qian, Anthony GJ Raymond, Nina Thiessen, Richard J Varhol, Inanc Birol, Richard A Moore, Andrew J Mungall, Robert Holt, Daisuke Kawauchi, Martine F Roussel, Marcel Kool, David TW Jones, Hendrick Witt, Africa Fernandez-L, Anna M Kenney, Robert J WechslerReya, Peter Dirks, Tzvi Aviv, Wieslawa A Grajkowska, Marta Perek-Polnik, Christine C Haberler, Olivier Delattre, Stéphanie S Reynaud, François F Doz, Sarah S Pernet-Fattet, Byung-Kyu Cho, Seung-Ki Kim, Kyu-Chang Wang, Wolfram Scheurlen, Charles G Eberhart, Michelle Fèvre-Montange, Anne Jouvet, Ian F Pollack, Xing Fan, Karin M Muraszko, G.Yancey Gillespie, Concezio Di Rocco, Luca Massimi, Erna MC Michiels, Nanne K Kloosterhof, Pim J French, Johan M Kros, James M Olson, Richard G Ellenbogen, Karel Zitterbart, Leos Kren, Reid C Thompson, Michael K Cooper, Boleslaw Lach, Roger E McLendon, Darell D Bigner, Adam Fontebasso, Steffen Albrecht, Nada Jabado, Janet C Lindsey, Simon Bailey, Nalin Gupta, William A Weiss, László Bognár, Almos Klekner, Timothy E Van Meter, Toshihiro Kumabe, Teiji Tominaga, Samer K Elbabaa, Jeffrey R Leonard, Joshua B Rubin, Linda M Liau, Erwin G Van Meir, Maryam Fouladi, Hideo Nakamura, Giuseppe Cinalli, Miklós Garami, Peter Hauser, Ali G Saad, Achille Iolascon, Shin Jung, Carlos G Carlotti, Rajeev Vibhakar, Young Shin Ra, Shenandoah Robinson, Massimo Zollo, Claudia C Faria, Jennifer A Chan, Michael L Levy, Poul HB Sorensen, Matthew Meyerson, Scott L Pomeroy, Yoon-Jae Cho, Gary D Bader, Uri Tabori, Cynthia E Hawkins, Eric Bouffet, Stephen W Scherer, James T Rutka, David Malkin, Steven C Clifford, Steven JM Jones, Jan O Korbel, Stefan M Pfister, Marco A Marra, Michael D Taylor. Subgroupspecific structural variation across 1, 000 medulloblastoma genomes. Nature, 2012, 488(7409): 49–56. DOI:10.1038/nature11327 |
| [9] | Peikai Chen, Yubo Fan, Tsz-kwong Man, YS Hung, Ching C Lau, Stephen TC Wong. A gene signature based method for identifying subtypes and subtype-specific drivers in cancer with an application to medulloblastoma. Bmc Bioinformatics, 2013, 14(18): 1–13. |
| [10] | Li KK, Lau KM, Ng HK. Signaling pathway and molecular subgroups of medulloblastoma. Int J Clin Exp Pathol, 2013, 6(7): 1211–1222. |
| [11] | Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, Pfister S, Taylor MD. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol, 2011, 29(11): 1408–1414. DOI:10.1200/JCO.2009.27.4324 |
| [12] | Wefers AK, Warmuth-Metz M, Pöschl J, von Bueren AO, Monoranu CM, Seelos K, Peraud A, Tonn JC, Koch A, Pietsch T, Herold-Mende C, Mawrin C, Schouten-van Meeteren A, van Vuurden D, von Hoff K, Rutkowski S, Pfister SM, Kool M, Schüller U. Subgroup-specific localization of human medulloblastoma based on pre-operative MRI. Acta Neuropathol, 2014, 127(6): 931–933. DOI:10.1007/s00401-014-1271-5 |
| [13] | Ramaswamy V, Remke M, Bouffet E, Faria CC, Perreault S, Cho YJ, Shih DJ, Luu B, Dubuc AM, Northcott PA, Schüller U, Gururangan S, McLendon R, Bigner D, Fouladi M, Ligon KL, Pomeroy SL, Dunn S, Triscott J, Jabado N, Fontebasso A, Jones DT, Kool M, Karajannis MA, Gardner SL, Zagzag D, Nunes S, Pimentel J, Mora J, Lipp E, Walter AW, Ryzhova M, Zheludkova O, Kumirova E, Alshami J, Croul SE, Rutka JT, Hawkins C, Tabori U, Codispoti KE, Packer RJ, Pfister SM, Korshunov A, Taylor MD. Recurrence patterns across medulloblastoma subgroups:An integrated clinical and molecular analysis. Lancet Oncol, 2013, 14(12): 1200–1207. DOI:10.1016/S1470-2045(13)70449-2 |
| [14] | Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, Taylor RE, Bailey S, Clifford SC, Gilbertson RJ. Medulloblastoma:clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol, 2011, 121(3): 381–396. DOI:10.1007/s00401-011-0800-8 |
| [15] | Thompson E M, Hielscher T, Bouffet E, Michael D Taylor. Prognostic value of medulloblastoma extent of resection after accounting for molecular subgroup:A retrospective integrated clinical and molecular analysis. Lancet Oncology, 2016, 17(4): 484–495. DOI:10.1016/S1470-2045(15)00581-1 |
| [16] | Perreault S, Ramaswamy V, Achrol AS, Chao K, Liu TT, Shih D, Remke M, Schubert S, Bouffet E, Fisher PG, Partap S, Vogel H, Taylor MD, Cho YJ, Yeom KW. MRI surrogates for molecular subgroups of medulloblastoma. AJNR Am J Neuroradiol, 2014, 35(7): 1263–1269. DOI:10.3174/ajnr.A3990 |
| [17] | Fattet S, Haberler C, Legoix P, Varlet P, Lellouch-Tubiana A, Lair S, Manie E, Raquin MA, Bours D, Carpentier S, Barillot E, Grill J, Doz F, Puget S, Janoueix-Lerosey I, Delattre O. Beta-catenin status in paediatric medulloblastomas:Correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. Journal of Pathology, 2009, 218(1): 86–94. DOI:10.1002/path.v218:1 |
| [18] | Min HS, Lee JY, Kim SK, Park SH. Genetic grouping of medulloblastomas by representative markers in pathologic diagnosis. Transl Oncol, 2013, 6(3): 265–272. DOI:10.1593/tlo.12382 |
| [19] | Goschzik T, Zur Mühlen A, Kristiansen G, Haberler C, Stefanits H, Friedrich C, von Hoff K, Rutkowski S, Pfister SM, Pietsch T. Molecular stratification of medulloblastoma:Comparison of histological and genetic methods to detect Wnt activated tumours. Neuropathol Appl Neurobiol, 2015, 41(2): 135–144. DOI:10.1111/nan.12161 |
| [20] | Ramaswamy V, Remke M, Shih D, Wang X, Northcott PA, Faria CC, Raybaud C, Tabori U, Hawkins C, Rutka J, Taylor MD, Bouffet E. Duration of the pre-diagnostic interval in medulloblastoma is subgroup dependent. Pediatric Blood Cancer, 2014, 61(7): 1190–1194. DOI:10.1002/pbc.25002 |
| [21] | Perreault S, Ramaswamy V, Achrol AS, Chao K, Liu TT, Shih D, Remke M, Schubert S, Bouffet E, Fisher PG, Partap S, Vogel H, Taylor MD, Cho YJ, Yeom KW. MRI surrogates for molecular subgroups of medulloblastoma. Ajnr American Journal of Neuroradiology, 2014, 35(7): 1263. DOI:10.3174/ajnr.A3990 |
| [22] | Schroeder K, Gururangan S. Molecular variants and mutations in medulloblastoma. Pharmgenomics Pers Med, 2014, 7: 43–51. |
| [23] | Bien-Willner GA, Lopez-Terrada D, Bhattacharjee MB, Patel KU, Stankiewicz P, Lupski JR, Pfeifer JD, Perry A. Early recurrence in standard-risk medulloblastoma patients with the common idic(17)(p11.2) rearrangement. Neuro Oncol, 14: 831–840. DOI:10.1093/neuonc/nos086 |