文章信息
- 林占熺, 林冬梅, 刘仲健, 兰思仁
- Zhanxi LIN, Dongmei LIN, Zhongjian LIU, Siren LAN
- 基于形态和基因组证据的禾本科新种——巨菌草
- Cenchrus fungigraminus Z. X. Lin & D. M. Lin & S. R. Lan sp. nov., a new species of Panicoideae (Poaceae): evidence from morphological, nuclear and plastid genome data
- 森林与环境学报,2022, 42(5): 514-520.
- Journal of Forest and Environment,2022, 42(5): 514-520.
- http://dx.doi.org/10.13324/j.cnki.jfcf.2022.05.009
-
文章历史
- 收稿日期: 2022-08-23
- 修回日期: 2022-08-31



Panicoideae is the second-largest subfamily of the grasses with over 3 500 species, mainly distributed in warm temperate and tropical regions[1]. It comprises some important agricultural crops, including sugarcane, maize(or corn), sorghum, and switchgrass. Previous study had combined Pennisetum Rich. and Odontelytrum Hack. into Cenchrus L.[2]. As currently evidence, Cenchrus is a genus of Panicoideae with 107 species[2-3] (https://powo.science.kew.org/). The various species are native to Africa, Asia, Australia, and Latin America, with some of them widely naturalized in Europe and North America, as well as on various oceanic islands. They are annual or perennial grasses. The inflorescence is a very dense, narrow panicle containing fascicles of spikelets interspersed with bristles. There are three kinds of bristle, and some species have all three, while others do not.
During our field work in KwaZulu-Natal of South Africa, we found some populations of Pennisetum which is similar to P. purpureum in 2004, but they are very different in morphological mainly in stem with branches, fascicles sparse. We consider it is a new species and namely "Jujuncao", also given a scientific name, Pennisetum giganteum Z. X. Lin, nov. sp., but was not officially published. Due to Pennisetum was transferred into the Cenchrus, Pennisetum purpureum was changed to Cenchrus purpureus. In addition, the specific epithet "giganteum" of Pennisetum giganteum was used by Pennisetum giganteum A. Rich (1850). Therefore, we formally classify and name the "Jujuncao". Through the project of China Africa Cooperation, some individuals were transplanted to Fuzhou Juncao Germplasm Resources Garden, Fujian Province. After careful observation of the species planting, morphological comparisons with the Cenchrus species, we have described the distinctive features of this species and used nuclear single-copy genes and plastid genome data to evaluate its phylogenetic position, the result strongly supports "Jujuncao" is a new species and sister to C. purpureus of genus Cenchrus.
1 Materials and methods 1.1 Morphological observationsMorphological data of the species described here were based on living plants obtained in wild and further grown in the Fujian Agriculture and Forestry University (FAFU) nursery. Their inflorescence characteristics were observed under a microscope. The type specimen has been deposited in the FAFU herbarium. Photographs of flowering plant were taken by Canon D750.
1.2 Molecular samplingTo explore the phylogenetic position of the new Cenchrus species, we used a total of 17 taxa as ingroups and Panicum pygmaeum, Whiteochloa capillipes and Urochloa reptans as outgroups. The nucleotide sequences used in this study were downloaded from the National Center for Biotechnology Information (NCBI) database. Voucher information and GenBank accession numbers are provided in Table 1. The plastid genome for the newly identified species was deposited in NCBI GenBank database under accession number OP279446.
| Species | Accession | Plastid genome size/bp |
| Ingroups | ||
| Cenchrus alopecuroides | ON206984 | 138 053 |
| C. americanus | KX756179 | 138 172 |
| C. centrasiaticus | MW421597 | 138 294 |
| C. ciliaris | MH286942 | 138 737 |
| C. clandestinus | MW816927 | 138 003 |
| C. compressus | MW816925 | 138 060 |
| C. echinatus | MN078360 | 137 131 |
| C. flaccidus | MW442087 | 138 336 |
| C. longispinus | MN078361 | 137 144 |
| C. polystachios | MW816926 | 136 346 |
| C. purpureus | MF594682 | 138 199 |
| C. fungigraminus | MN000000 | 138 144 |
| Setaria geminata | KU291476 | 139 142 |
| S. italica | MK348609 | 139 261 |
| S. viridis | KT289405 | 138 102 |
| Stenotaphrum secundatum | KY432796 | 138 907 |
| S. subulatum | MN989414 | 139 282 |
| Outgroups | ||
| Whiteochloa capillipes | KU291487 | 139 177 |
| Panicum pygmaeum | MF563373 | 140 996 |
| Urochloa reptans | KU291486 | 140 177 |
Total DNA was extracted from fresh material or from leaves using a modified CTAB procedure and sequenced by the BGISEQ-500 platform[4]. The clean reads were used to assemble the complete plastid genome by the GetOrganelle pipe-line[5], with the plastid genome of C. purpureus(GenBank accession number MF594682) as the reference sequences. The assembled plastid genome was annotated using the Geneious R11.15 (https://www.geneious.com/)[6].The nuclear single-copy genes were clustered using 12 monocots species genome (C. fungigraminus genome data unpublished) by OrthoMCL v.2.0.9 [7].
1.4 Phylogenetic analysisThe data analyses included maximum parsimony (MP), Bayesian inference (BI) and maximum likelihood (ML) methods. The MP analysis was performed in PAUP v.4.0b10[8], the BI analysis was conducted using MrBayes v.3.2.6 [9] and the ML analysis was performed on the CIPRES Science Gateway web server (RAxML-HPC2 on XSEDE 8.1.11)[10].
1.5 Population genetics analysisA total of 22 individuals were used in the analysis, 19 individuals (2 individuals of C. fungigraminus) were collected and sequenced in the study, 3 individuals of C. americanus as outgroup were from NCBI. The Illumina paired-end reads were aligned to the reference genome sequences using BWA[11]. After mapping, SNPs were identified on the basis of the files generated by SAMtools[12]. A neighbor-joining tree was constructed using PHYLIP (http://evolution.genetics.washington.edu/phylip.html) and MEGA5 was used to display the tree.
2 Results 2.1 Morphological analysisThe key characteristics for the newly discovered species and other related members of Cenchrus were compared in Table 2. The new species was similar to C. purpureus. This new species was characterized by plant very tall up to 10 m; stem with branches on nodes; leaves 1.8-4.8 cm wide, fascicles 6-15 per cm, bristles many, dense long hairs. These features distinguished the new species from all other known species of Cenchrus.
| Characters | C. fungigraminus | C. purpureus |
| Stem | Up to 8 m tall, with branches on upper nodes | 2-4 m tall, not branches on nodes |
| Leaf | Up to 5 cm wide | 1-3 cm wide |
| Panicles | Fascicle 6-15 per cm, with 3 spikelets | Fascicle 30-40 per cm, fascicle with 1-3 spikelets |
| Bristles | Dense long hair | Scabrous |
| Spikelets | Spikelets 7-8 mm, pedicellate | Spikelets 5.9-7.0 mm, pedicellate |
| Caryopses | 1.8-2.2 mm | 1.6-1.8 mm |
| Flowering period | May-July | August-October |
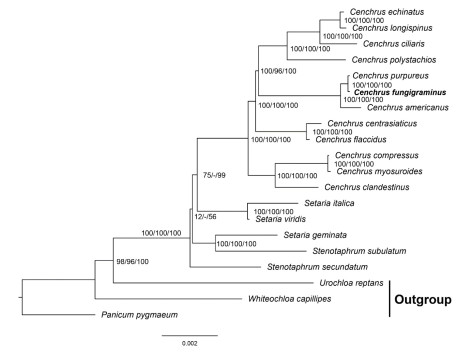
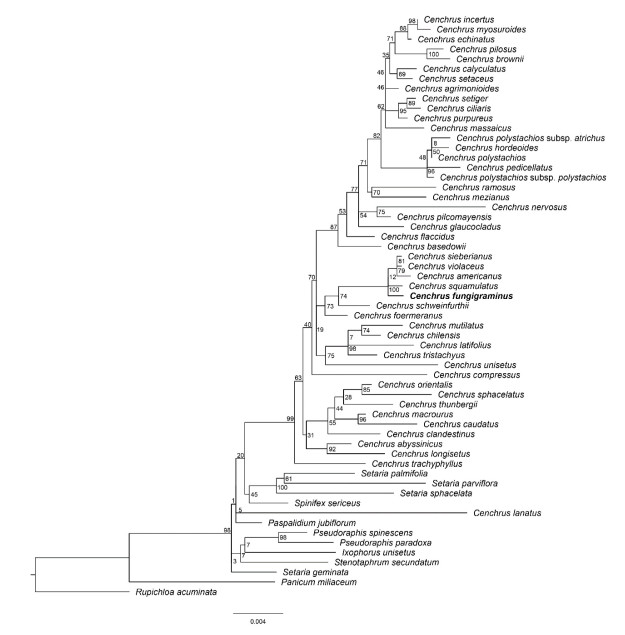
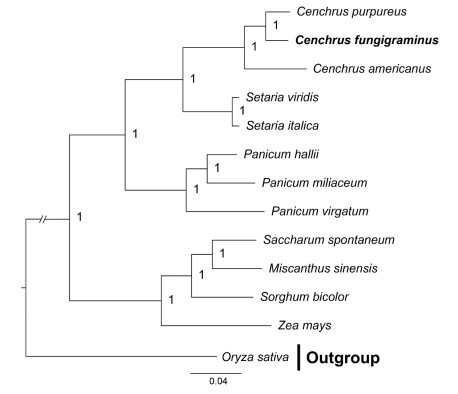
The phylogenetic analysis based on the plastid genome suggested four clades were divided in 17 taxa (Figure 1). All groups with high support values except the second and third clades. The Cenchrus was divided into four clades, C. fungigraminus was located in the third clade, which sister to C. purpureus, and then clustered to C. americanus. The phylogenetic tree based on ndhF and trnL-F showed C. fungigraminus was diverged with C. purpureus (Figure 2). The phylogenetic analysis based on the nuclear single-copy genes also support C. fungigraminus sister to C. purpureus(Figure 3).
|
Note: support values were near the nodes (BPML/ BPMP/PP×100). Note: support values were near the nodes (BPML/ BPMP/PP×100). Figure 1 The phylogenetic analysis of C. fungigraminus and related species based on the plastid genome sequences |
|
Note: support values were near the nodes (ML). Note: support values were near the nodes (ML). Figure 2 The phylogenetic analysis of C. fungigraminus and related species based on the ndhF and trnL-F genes |
|
Note: support values were near the nodes (PP). Note: support values were near the nodes (PP). Figure 3 The phylogenetic analysis of C. fungigraminus and related species based on the nuclear single-copy genes |
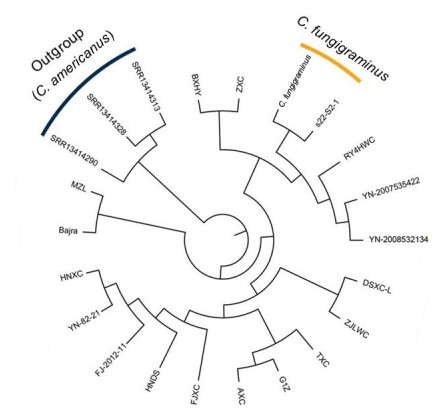
The population analysis of 22 individuals, 19 individuals of C. fungigraminus and C. purpureus, three individuals of C. americanus were considered as outgroups. The phylogenetic tree suggested 19 individuals were clustered into four clades, and the individuals of C. fungigraminus were clustered into one clade (Figure 4).
|
Figure 4 The population structure analysis of C. fungigraminus and related species based on 22 resequencing data |
This new species is thus consistent with the other species of Cenchrus in some morphological characters, and is similar to C. purpureus, but the new species very different by having stem with branches on nodes, leaves larger; fascicles 6-15 per cm, bristles many, yellow, with long hairs; inner bristles length as same as primary bristles spikelets with longer pedicellate (Figure 5).The phylogenetic analysis suggested C. fungigraminus was sister to C. purpureus with full support values (Figure 1-3). The plastid genome size between C. fungigraminus and C. purpureus was similar (Table 1). The population analysis of C. fungigraminus and related species suggested it was an independent clade (Figure 4). Based on morphological and DNA analyses, C. fungigraminus should be a new species.

|
Note: C. fungigraminus: A. plant; B. panicles; C. bristles; D. bristles hair. C. purpureus: E. plant; F. panicles. G. bristles; H. bristles hair. Note: C. fungigraminus: A. plant; B. panicles; C. bristles; D. bristles hair. C. purpureus: E. plant; F. panicles. G. bristles; H. bristles hair. Figure 5 Morphological comparison of C. fungigraminus with C. purpureus |
Cenchrus fungigraminus Z. X. Lin & D. M. Lin & S. R. Lan sp. nov.(巨菌草) (Figure 5)
Type: South Africa, KwaZulu-Natal, elevation 1 071 m, grassy slope, 9 March, 2004, Lin0010 (holotype: FAFU). This new species is similar to C. purpureus Schumacher, but can be distinguished by the characterization of stem with branches on nodes, leaves wider and longer; fascicles 6-15 per cm, bristles many, yellow, with long hairs; bristles unequal, outer shorter, the longest being about four time of the shortest bristles of pedicellate spikelets in each fascicles.
Plants perennial. Culms 5-10 m, erect, pubescent beneath the panicle; stem with branches on upper nodes; nodes glabrous. Sheaths pubescent; ligules 2-3 mm; blades 40-95 cm long, 1.8-4.8 cm wide, flat, pubescent. Panicles terminal, 20-30 cm long, 3.5-4.0 cm wide, fully exerted from the leaf sheaths, erect, greenish-yellow; rachises terete, pubescent. Fascicles 6-15 per cm, fascicle axes 1.5-2.0 mm, with 2-3 spikelets; outer bristles 40-63, 4.5-9.0 mm, greenish-yellow, with long hairs; inner bristles 20-30, 10.0-18.0 mm, yellow, with long hairs. Spikelets 3-4 mm, pedicellate; pedicels of terminal spikelets 0.1-0.2 mm, of other spikelets 1.8-3.0 mm; lower glumes abscent; upper glumes 0.8-3.0 mm, 0-1-veined; lower florets neuter or staminate; lower lemmas 4.0-5.3 mm, 3-5-veined; upper lemmas 4.5-7.0 mm, subcoriaceous, shiny, 5-7-veined, acuminate; anthers 2.7-3.6 mm, penicillate. Caryopses 1.8-2.2 mm.
多年生草本植物。丛生,高5~10 m,直立。花序下被短柔毛;茎上部的节具分枝;节上光滑无毛。叶鞘具短柔毛,叶舌2~3 mm;叶片长40~95 cm,宽1.8~4.8 cm,扁平,具短柔毛。圆锥花序顶生,长20~30 cm,宽3.5~4.0 cm,高出叶鞘,直立,黄绿色;花序轴圆柱状,具短柔毛。每厘米簇生伞房花序6~15个;伞房花序轴1.5~2.0 mm,具2~3小穗;外刚毛40~63根,长4.5~9.0 mm,黄绿色,具长毛;内刚毛20~30根,长10.0~18.0 mm,黄色,具长毛。小穗长3~4 mm,有花梗;顶生小穗花梗长0.1~0.2 mm,其他小穗花梗长1.8~3.0 mm;第一颖缺失;第二颖片0.8~3.0 mm,具0~1脉;第一小花中性式雄性;第一外稃长4.0~5.3 mm,3~5脉;第二外稃4.5~7.0 mm,近革质,有光泽,5~7脉,渐尖;花药2.7~3.6 mm,顶端具细毛。颖果1.8~2.2 mm。
Etymology: Referring to the plant easily to multiply fungi, from the Latin fungi (fungus) and gramin (grass).
Phenology: May-July.
Distribution: South Africa, KwaZulu-Natal.
Habitat: Grassy slope.
| [1] |
SORENG R J, PETERSON P M, ROMASCHENKO K, et al. A worldwide phylogenetic classification of the Poaceae (Gramineae) Ⅱ: an update and a comparison of two 2015 classifications[J]. Journal of Systematics and Evolution, 2017, 55(4): 259-290. DOI:10.1111/jse.12262 |
| [2] |
CHEMISQUY M A, GIUSSANI L M, SCATAGLINI M A, et al. Phylogenetic studies favour the unification of Pennisetum, Cenchrus and Odontelytrum (Poaceae): a combined nuclear, plastid and morphological analysis, and nomenclatural combinations in Cenchrus[J]. Annals of Botany, 2010, 106(1): 107-130. DOI:10.1093/aob/mcq090 |
| [3] |
MARTEL E, PONCET V, LAMY F, et al. Chromosome evolution of Pennisetum species (Poaceae): implications of ITS phylogeny[J]. Plant Systematics and Evolution, 2004, 249(3/4): 139-149. |
| [4] |
DOYLE J J, DOYLE J L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue[J]. Phytochemical Bulletin, 1987, 19: 11-15. |
| [5] |
JIN J J, YU W B, YANG J B, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes[J]. Genome Biology, 2020, 21(1): 1-31. DOI:10.1186/s13059-019-1906-x |
| [6] |
KEARSE M, MOIR R, WILSON A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data[J]. Bioinformatics, 2012, 28(12): 1647-1649. DOI:10.1093/bioinformatics/bts199 |
| [7] |
LI L, STOECKERT C J JR, ROOS D S. OrthoMCL: identification of ortholog groups for eukaryotic genomes[J]. Genome Research, 2003, 13(9): 2178-2189. DOI:10.1101/gr.1224503 |
| [8] |
SWOFFORD D L. PAUP: phylogenetic analysis using parsimony (*and other methods): version 4.0 b10[M]. Sunderland: Sinauer Associates, 2002.
|
| [9] |
RONQUIST F, HUELSENBECK J P. MrBayes 3: bayesian phylogenetic inference under mixed models[J]. Bioinformatics, 2003, 19(12): 1572-1574. DOI:10.1093/bioinformatics/btg180 |
| [10] |
MILLER M A, PFEIFFER W, SCHWARTZ T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees[C]//Proceedings of 2010 Gateway Computing Environments Workshop (GCE). New Orleans: IEEE, 2010: 1-8.
|
| [11] |
LI H, DURBIN R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25(14): 1754-1760. DOI:10.1093/bioinformatics/btp324 |
| [12] |
LI H, HANDSAKER B, WYSOKER A, et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25(16): 2078-2079. |
 2022, Vol. 42
2022, Vol. 42
