2. 上海海洋大学 港航生态安全研究中心, 上海 201306
压载水系指在船舶航行过程中,控制船舶纵倾、横倾、吃水、稳定或应力而在船上加装的水及其悬浮物[1]。压载舱内沉积物是指船内压载水的沉淀物质[1]。当压载舱装载水后,经过长时间航行,大量的沉积物聚集在压载舱底部,其中包括细菌、浮游动植物死亡个体、淤泥、沙子及有机颗粒等[2]。沉积物与压载水是两个不相同的介质,相互独立,却又相互影响。压载水及其沉积物中携带的生物随船舶在全球海域转运,随着压载水和沉积物的排载,可能在排载海域造成外来生物入侵[3-5]。由于压载舱是密封的舱体,压载舱内处于黑暗和气体交换隔绝状态,导致压载水中的浮游植物和浮游动物的生存受到影响,但微生物对这种环境具有良好的适应能力,因而压载水中微生物具有丰富的生物多样性,这些压载水微生物中可能含有一些致病的种类,例如霍乱弧菌和伤寒[6]、沙门氏菌、铜绿假单胞菌和肺炎克雷伯氏等[7]。这些病原微生物能进入并存活于某些水生生物如牡蛎、蚌以及其他贝类中,直接或间接危及人类健康[8],因而有关压载水微生物的研究备受关注,曾有许多有关压载水及沉积物中微生物检测和多样性研究方面的报道[9-10],但尚未有针对同一船舶压载舱内压载水和沉积物中微生物多样性的比较研究,本文通过高通量测序技术分析了同一压载舱内压载水和沉积物中的微生物多样性,比较连续环境两种介质条件下微生物群落特征的差异和相关性,为压载水及沉积物排放的管理及压载水和沉积物微生物入侵的防控提供理论依据。
1 材料与方法2016年5月在洋山深水港一艘入境船舶压载舱中采集沉积物及压载水样品,用取水器将压载水收集到灭菌蓝口瓶中,用兵工铲采集压载舱底层沉积物表层10 cm以上约为1 500 g样品,迅速放入灭菌塑封袋内。于两小时内将样品低温送回实验室。
可培养细菌计数方法:对压载水样品进行10倍系列稀释,涂布于Zobell 2216培养基平板;取沉积物样品10.0 g放入装有90 mL水的锥形瓶中,180 r/min震荡30 min,静置后取其上清液进行10倍梯度稀释,涂布于培养基平板,25 ℃培养7 d,选取平均菌落数在30~300之间的平板进行计数。所有步骤均按无菌操作进行。
细菌总DNA提取方法:对于沉积物样品,用灭菌药匙挑取样品中心500 mg沉积物,置于2 mL灭菌离心管中;对于压载水样品,取1 L样品进行滤膜过滤,使用3S DNA分离提取试剂盒(上海博彩生物技术有限公司)对细菌进行DNA提取。DNA浓度和纯度利用NanoDrop2000进行检测。
采用引物515F(5′-GTGCCAGCMGCCGCGG-3′)和907(5′-CCGTCAATTCMTTTRAGTTT-3′)对V4和V5可变区进行PCR扩增[11]。PCR反应体系:dd H2O 9.6 μL,d NTP (2.5 mmol/L) 2.0 μL,5 ×Fast Pfu Buffer 4.0 μL,Fast Pfu Polymerase 0.4 μL,10 ng DNA模板,BSA 0.2 μL模板(20 ng/μL) 2.00 μL,引物F(5 μmol / L) 0.8 μL,引物R (5 μmol/L) 0.8 μL。PCR扩增程序为:95 ℃ 3 min; 95 ℃ 30 s,55 ℃ 30 s,72 ℃ 45 s,27个循环; 72 ℃延伸10 min。扩增后的条带进行后续实验。
2%琼脂糖凝胶回收PCR产物,AxyPrep DNA Gel Extraction Kit进行纯化,Tris-HCl洗脱,QuantiFluor-ST定量检测。以Illumina MiSeq平台标准操作规程将纯化后的扩增片段构建PE250的文库。
使用UPARSE软件根据97%的相似度对序列进行OTU聚类。经过序列过滤Qiime和去除嵌合体序列最终得到优质序列,用于后续分析。利用RDP classifier对每条序列进行物种分类注释,比对Silva数据库。根据压载水及沉积物样品结果,通过使用Mothur软件计算群落多样性指数,包括Ace指数[12]、Shannon指数[13]、Chao指数[12]等。
2 结果 2.1 样品序列数特点通过细菌的16S rDNA基因V4-V5区测序,压载水与沉积物样品共产生67 255个序列,平均长度395 bp。所有序列按97%的相似度进行OTU分类,共聚成736个OTU。
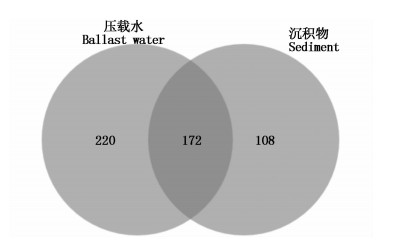
2.2 压载舱中细菌丰度分析压载舱内沉积物样品中可培养细菌总数要高于压载水样品中可培养细菌总数,压载水中可培养细菌总数(total bacterial count, TBC)为9.75×103 CFU/mL,船舶压载舱中沉积物可培养细菌总数为2.725×106 CFU/mL(表 1)。沉积物中可培养细菌总数比压载水中可培养细菌总数高三个数量级。压载水样品的OTU数量为551,船舶沉积物的OTU数量为359(表 1)。压载水样品相比沉积物样品呈现OTU数量较多,但可培养细菌总数较少的特征。结果显示,压载水样品和沉积物样品具有172个相同的物种(图 1)。

|
表 1 压载水与沉积物细菌丰度及多样性指数比较 Tab.1 Comparison of bacterial abundance and diversity index of ballast water and sediment |
|
图 1 Venn图(种水平)
Fig. 1 Venn diagram
|
压载水细菌多样性高于沉积物细菌多样性。压载水中细菌丰度较高、种类较多,物种较为丰富,多样性程度较高,而相比沉积物样品,沉积物样品中细菌种类较为单一,多样性程度较低,物种数量分布不均匀,但优势种的地位更为突出。对于Shannon指数,压载水样品为4.006 6,沉积物样品为1.825,压载水样品Shannon指数是沉积物样品的两倍多(表 1)。
2.4 种类组成和优势类群统计压载水中细菌样品序列信息,包括24门,44纲,108目,167科,283属,392种。分别是变形菌门(Proteobacteria 70.20%)、拟杆菌门(Bacteroidetes 13.38%)、放线菌门(Actinobacteria 10.33%)、Marinimicrobia_SAR406_clade 2.81%、绿弯菌门(Chloroflexi 0.79%)、酸杆菌门(Acidobacteria 0.71%)、单胞菌门(Gemmatimonadetes 0.32%)、蓝藻门(Cyanobacteria 0.17%)、疣微菌门(Verrucomicrobia 0.13%),还有0.63%的OTU序列合并小于0.1%。
统计压载舱中沉积物样品序列信息:包括24门,43纲,93目,138科,218属,280种。分别为变形菌门(Proteobacteria 95.45%)、厚壁菌门(Firmicutes 2.23%)、拟杆菌门(Bacteroidetes 0.92%)、单胞菌门(Gemmatimonadetes 0.32%)、放线菌门(Actinobacteria 0.27%)、浮霉菌门(Planctomycetes 0.25%)、绿弯菌门(Chloroflexi0.12%)、酸杆菌门(Acidobacteria 0.12%),还有0.31%的OTU序列合并小于0.1%。
在门水平上,船舶压载水的优势类群为变形菌(Proteobacteria 70.20%)、拟杆菌(Bacteroidetes 13.38%)、放线菌(Actinobacteria 10.33%)、Marinimicrobia_SAR406_clade 2.81%。压载舱中沉积物的优势类群为变形菌(Proteobacteria 95.45%)、厚壁菌(Firmicutes 2.23%)。压载水与沉积物中变形菌门都是最多的,但沉积物中变形菌门所占的比例更大,占95.45%。其中厚壁菌门在沉积物中含量较高,但在压载水中总含量小于0.1%。在压载水中拟杆菌Bacteroidetes和放线菌Actinobacteria是优势菌种,但在沉积物中拟杆菌和放线菌的含量分别仅占0.92%和0.27%。
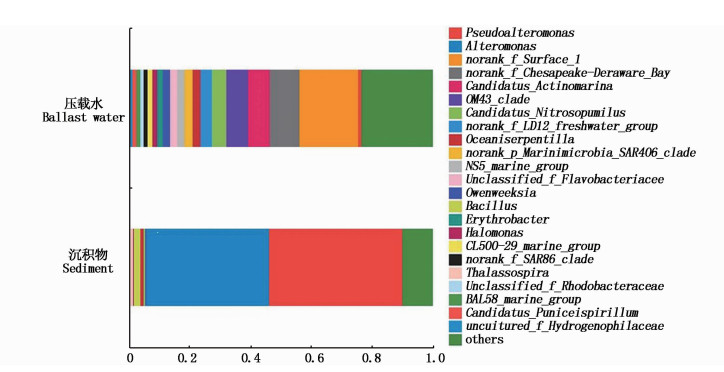
在属水平上,沉积物的优势类群为假交替单胞菌属(Pseudoalteromonas)和交替单胞菌属(Alteromonas),压载水的优势类群为norank_f_Surface_1(图 2)。在沉积物中假交替单胞菌属和交替单胞菌的含量高达44.4%和40.9%,但在压载水中含量都小于2%。在压载水中norank_f_Surface_1高达20.2%,但在沉积物中含量却小于0.1%。
|
图 2 细菌(属)类群组成柱状图
Fig. 2 Bacteria (genus) group composition histogram
|
在同一压载舱中,沉积物中可培细菌总数高于压载水中可培细菌总数。可培养细菌总数是评价生态环境的重要参数。曾有报道,船舶压载水中的可培细菌总数在1.2×105到1.7×106 cells/mL之间[14],船舶压载水沉积物细菌总数在3.20×108到1.90×1011 copies/g之间[11],虽然不是同一压载舱中的压载水和沉积物间的细菌数量比较,但仍显示沉积物中的数量高。在压载舱沉积物细菌中优势类群为γ变形菌纲(Gammaprote obacteria 90.5%),由于γ变形菌纲是革兰氏阴性厌氧杆菌,这类细菌很适宜存活在压载舱这一黑暗、密闭、溶解氧较低的特殊环境中。压载水中的悬浮物质以及死亡生物会沉淀到沉积物中,这些沉积物中丰富的有机和无机营养物质将会被生物体所利用,尤其是微生物[15-16]。相比于压载水,沉积物中含有丰富的营养物质,为细菌提供了充足的营养和良好的环境。不同类型的微生物生活在压载舱的沉积物中,因为沉积物为细菌提供复合营养素和这一特殊的固体基质[17]。BAOYI等比较不同船舶的压载水与沉积物中细菌总数,发现沉积物的细菌总数要比压载水中细菌总数高3~5个数量级。更有学者研究发现压载舱中的沉积物相比海洋中的沉积物具有更高的丰富度[17-20]。本研究显示同一压载舱内沉积物的可培细菌数量要高于压载水中可培细菌数量。
在压载水和沉积物中变形菌门都是优势类群。变形菌门是沉积物及海水环境中丰度较高的细菌,并且占据较大的比例[21-24]。CARR等也发现变形菌门(Proteobacteria)是压载舱沉积物中细菌丰度最高的种类[25];许飞等对南海南沙海区沉积物的16S rDNA序列进行分析,表明变形菌门是明显的优势类群[26];FENG等对长江口及东海沿岸海域沉积物及海水进行研究,结果表明变形菌门是该海域环境中最丰富的种群[27];KERFAHI等在2014年研究发现γ-变形菌纲(Gammaproteobacteria)通常是海洋沉积物中含量较多的纲,一般占海洋微生物的50%以上[28]。在本次研究中γ-变形菌纲也是变形菌门中的重要组成,这个结果与其他海洋沉积物中细菌群落结构组成相一致。
在压载水及沉积物中均发现黄单胞菌科(Xanthomonadaceae)、分枝杆菌属(Mycobacterium)、假单胞菌(Pseudomonas)等致病性微生物,病原性细菌的入侵、传播和爆发会直接或间接危及人类健康。在船舶压载水排放的主要港口及沿海地区,容易造成致病菌的传播和导致动物和人类患病[29-30],因此应加强到港船舶压载水及沉积物的排放监管,对他们的科学管理亦不容忽视。
| [1] |
IMO. International convention for the control and management of ships' ballast water and sediments[S]. London: International Maritime Organization, 2004.
|
| [2] |
MAGLIĆ L, ZEC D, FRANČIĆ V. Ballast water sediment elemental analysis[J]. Marine Pollution Bulletin, 2016, 103(1/2): 93-100. |
| [3] |
PIMENTEL D, LACH L, ZUNIGA R, et al. Environmental and economic costs of nonindigenous species in the united states[J]. BioScience, 2000, 50(1): 53-65. DOI:10.1641/0006-3568(2000)050[0053:EAECON]2.3.CO;2 |
| [4] |
WU H, CHEN C, WANG Q, et al. The biological content of ballast water in China:A review[J]. Aquaculture & Fisheries, 2017, 2(6): 241-246. |
| [5] |
刘艳, 吴惠仙, 薛俊增. 海洋外来物种入侵生态学研究[J]. 生物安全学报, 2013, 22(1): 8-16. LIU Y, WU H X, XUE J Z. The ecology of invasions by marine exotic species[J]. Journal of Biosafety, 2013, 22(1): 8-16. |
| [6] |
胡岚岚. 船舶压载水生态风险评估[D]. 大连: 大连海事大学, 2008. HU L L. Ecological risk assessment of ballast water[D]. Dalian: Dalian Maritime University, 2008. http://cdmd.cnki.com.cn/article/cdmd-10151-2008209856.htm |
| [7] |
IMO. 1st International workshop on guidelines and standards for ballast water sampling[R]. GloBallast Monograph Series No. 9. London: IMO, 2003.
|
| [8] |
ENDRESEN Ø, BEHRENS H L, BRYNESTAD S, et al. Challenges in global ballast water management[J]. Marine Pollution Bulletin, 2004, 48(7/8): 615-623. |
| [9] |
赵丽青, 贾俊涛, 姜英辉, 等. 山东主要港口海域及入境船舶压舱水中病原菌调查结果的多维尺度分析[J]. 中国国境卫生检疫杂志, 2013, 36(5): 334-337. ZHAO L Q, JIA J T, JIANG Y H, et al. MDS analysis on the pathogenic bacterias in Shandong mian Harbor water and entry ship ballast water[J]. Chinese Frontier Health and Quarantine, 2013, 36(5): 334-337. |
| [10] |
LÜ B Y, CUI Y X, TIAN W, et al. Composition and influencing factors of bacterial communities in ballast tank sediments:implications for ballast water and sediment management[J]. Marine Environmental Research, 2017, 132: 14-22. DOI:10.1016/j.marenvres.2017.10.005 |
| [11] |
XIONG J B, LIU Y Q, LIN X G, et al. Geographic distance and PH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau[J]. Environmental Microbiology, 2012, 14(9): 2457-2466. DOI:10.1111/emi.2012.14.issue-9 |
| [12] |
PITTA D W, PINCHAK W E, DOWD S E, et al. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets[J]. Microbial Ecology, 2010, 59(3): 511-522. DOI:10.1007/s00248-009-9609-6 |
| [13] |
SHANNON C E, WEAVER W. The mathematical theory of communication[M]. Urbana: University of Illinois Press, 1971: 3-55.
|
| [14] |
BRINKMEYER R. Diversity of bacteria in ships ballast water as revealed by next generation DNA sequencing[J]. Marine Pollution Bulletin, 2016, 107(1): 277-285. DOI:10.1016/j.marpolbul.2016.03.058 |
| [15] |
WANG L P, LIU L S, ZHENG B H, et al. Analysis of the bacterial community in the two typical intertidal sediments of Bohai Bay, China by pyrosequencing[J]. Marine Pollution Bulletin, 2013, 72(14): 181-187. |
| [16] |
DRAKE L A, DOBLIN M A, DOBBS F C. Potential microbial bioinvasions via ships' ballast water, sediment, and biofilm[J]. Marine Pollution Bulletin, 2007, 55(7/9): 333-341. |
| [17] |
WANG Y, SHENG H F, HE Y, et al. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags[J]. Applied and Environmental Microbiology, 2012, 78(23): 8264-8271. DOI:10.1128/AEM.01821-12 |
| [18] |
ZHENG B H, WANG L P, LIU L S. Bacterial community structure and its regulating factors in the intertidal sediment along the Liaodong Bay of Bohai Sea, China[J]. Microbiological Research, 2014, 169(7/8): 585-592. |
| [19] |
ZHU D C, TANABE S H, YANG C, et al. Bacterial community composition of South China Sea sediments through pyrosequencing-based analysis of 16S rRNA genes[J]. PLoS One, 2013, 8(10): e78501. DOI:10.1371/journal.pone.0078501 |
| [20] |
ZINGER L, AMARAL-ZETTLER L A, FUHRMAN J A, et al. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems[J]. PLoS One, 2011, 6(9): e24570. DOI:10.1371/journal.pone.0024570 |
| [21] |
BOWMAN J P, MCCUAIG R D. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment[J]. Applied and Environmental Microbiology, 2003, 69(5): 2463-2483. DOI:10.1128/AEM.69.5.2463-2483.2003 |
| [22] |
RAVENSCHLAG K, SAHM K, AMANN R. Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard)[J]. Applied and Environmental Microbiology, 2001, 67(1): 387-395. DOI:10.1128/AEM.67.1.387-395.2001 |
| [23] |
FUHRMAN J A, MCCALLUM K, DAVIS A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans[J]. Applied and Environmental Microbiology, 1993, 59(5): 1294-1302. |
| [24] |
ACINAS S G, ANTóN J, RODRíGUEZ-VALERA F. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA[J]. Applied and Environmental Microbiology, 1999, 65(2): 514-522. |
| [25] |
CARR S A, VOGEL S W, DUNBAR R B, et al. Bacterial abundance and composition in marine sediments beneath the Ross Ice Shelf, Antarctica[J]. Geobiology, 2013, 11(4): 377-395. DOI:10.1111/gbi.2013.11.issue-4 |
| [26] |
许飞, 戴欣, 陈月琴, 等. 南沙海区沉积物中细菌和古细菌16S rDNA多样性的研究[J]. 海洋与湖沼, 2004, 35(1): 89-94. XU F, DAI X, CHEN Y Q, et al. Phylogenetic diversity of bacteria and archaea in the Nansha marine sediments, as determined by 16S rDNA analysis[J]. Oceanologia et Limnologia Sinica, 2004, 35(1): 89-94. |
| [27] |
FENG B W, LI X R, WANG J H, et al. Bacterial diversity of water and sediment in the Changjiang estuary and coastal area of the East China Sea[J]. FEMS Microbiology Ecology, 2009, 70(2): 236-248. DOI:10.1111/fem.2009.70.issue-2 |
| [28] |
KERFAHI D, HALL-SPENCER J M, TRIPATHI B M, et al. Shallow water marine sediment bacterial community shifts along a natural CO2 gradient in the Mediterranean Sea off Vulcano, Italy[J]. Microbial Ecology, 2014, 67(4): 819-828. DOI:10.1007/s00248-014-0368-7 |
| [29] |
ALTUG G, GURUN S, CARDAK M, et al. The occurrence of pathogenic bacteria in some ships' ballast water incoming from various marine regions to the Sea of Marmara, Turkey[J]. Marine Environmental Research, 2012, 81: 35-42. DOI:10.1016/j.marenvres.2012.08.005 |
| [30] |
KIM Y, AW T G, TEAL T K, et al. Metagenomic investigation of viral communities in ballast water[J]. Environmental Science & Technology, 2015, 49(14): 8396-8407. |
2. Centre for Research on the Ecological Security of Ports and Shipping, Shanghai 201306, China
 2018,
Vol. 27
2018,
Vol. 27


