2. 上海海洋大学 水产与生命学院, 上海 201306
I-SceI是一种由Ⅰ型内含子编码的归位内切酶(homing endonucleases),又称巨核酸酶(meganuclease)。该基因序列是在酿酒酵母(Saccharomyces cerevisiae)线粒体中核糖体大亚基(21S rRNA)上的一个内含子中发现的[1],它由235个氨基酸编码,有特异的核酸内切酶活性,其识别序列长度为18 bp,可酶切核酸特异序列产生一个4 bp的切口[2]。巨核酸酶产生DNA双链缺口后,诱导细胞内的修复系统进行同源重组而实现基因整合[3]。巨核酸酶I-SceI系统因其识别序列短,构建简单且整合效率高而被应用到多种生物的转基因中,包括链霉菌(Streptomyces coelicolor)[4]、破伤风杆菌(Clostridium spp.)[5]、果蝇(Drosophila melanogaster)[6]、冈比亚按蚊(Anopheles gambiae) [7, 8]、小鼠 [9, 10]、牛[11],以及爪蟾(Xenopus)[12, 13]、青鳉(Oryzias latipes)[14]和斑马鱼[15, 16]等水生动物中。上述研究大多是将带有I-SceI识别位点的转基因载体和I-SceI酶的蛋白共注射到胚胎中,耗时长且工作量大。本研究分别构建表达绿色荧光和红色荧光的质粒与体外转录的I-SceI mRNA共注射到斑马鱼的胚胎,观察转荧光蛋白基因斑马鱼的荧光率,评估I-SceI的转基因介导效率。
1 材料与方法 1.1 实验材料实验所用AB系斑马鱼(Danio rerio)由中国科学院神经科学研究所、国家斑马鱼模式动物中心的杜久林研究员惠赠、珠江水产研究所水产生物技术研究室培育,培育与孵化水温为25~28 ℃。编码Tgf 2转座酶的辅助质粒pCS2-gfTP和Tgf 2转座子供体质粒pTgf2-EF1α-eGFP由上海海洋大学农业部淡水水产种质资源重点实验室邹曙明教授惠赠。
1.2 I-SceI转基因供体质粒的构建对pTgf2-EF1α-eGFP 质粒进行Spe Ⅰ和Xho Ⅰ双酶切,切开质粒的左臂,酶切产物经1.5%琼脂糖凝胶电泳并回收纯化骨架质粒片段pTgf2+。合成两条单链长引物[I-SceI-L-F(SpeⅠ)和I-SceI-L-R(Xho Ⅰ)],其两端带有粘性末端。将2条引物按1∶1的比例混合后在72 ℃下孵育30 min,然后取混合液和回收纯化的骨架质粒片段pTgf2+连接转化,即获得目的质粒的左末端。同样的方法连入右末端的两条长引物,最后得到目的质粒pI-SceI-EF1α-eGFP。同时将目的质粒的eGFP质粒替换为RFP即得到另一个目的质粒pI-SceI-EF1α-RFP。
1.3 巨核酸酶I-SceI mRNA 的体外转录根据NCBI上登录的编码I-SceI的编码区序列(GenBank 登录号GU575293.1),在其编码区上游设计加上kozak序列和酶切位点,由上海生工合成。合成序列连接到pCS2+表达质粒上,即得到巨核酸酶I-SceI的辅助质粒pCS2-I-SceI。用限制性内切酶Not I 酶切辅助质粒pCS2-I-SceI,酶切产物经Cycle-Pure kit(OMEGA,USA)纯化,并进行电泳检测。以该回收纯化产物作为模板,采用SP6 mMessage mMachine kit(Ambion,USA)试剂盒进行体外转录合成,体外转录出的巨核酸酶I-SceI的mRNA存于-80 ℃冰箱备用。
| 表1 构建I-SceI转基因供体质粒所用的引物 Tab.1 Primers used for construction of the I-SceI donor plasmid |
显微注射前一晚,将循环水缸中分开饲养的雌雄斑马鱼亲鱼按雌雄比例 2∶1放入产卵盒中暂养,用挡板将雌雄分开。第二天清晨显微注射前抽去挡板,让斑马鱼进行交配、产卵受精。将受精卵吸出后整齐排列到琼脂糖制作的平板凹槽中。以显微操作仪和玻璃注射针对处于1-2细胞期的斑马鱼受精卵进行显微注射,在受精卵的动物极注射。每个胚胎注射供体质粒pI-SceI-EF1α-eGFP(或pI-SceI-EF1α-RFP)和I-SceI酶mRNA混合液2 nL,供体质粒浓度约为 50 ng/μL,I-SceI酶mRNA 100 ng/μL。注射后的受精卵放入装有无菌水的培养皿中,25~28 ℃孵育。每隔4 h挑除死胚胎,加换新水。
1.5 荧光蛋白表达的观察和胚胎荧光率的统计受精卵注射24 h、48 h和3 d后,在激光共聚焦显微镜(Carl-Zeiss,Germany)下观察胚胎荧光蛋白和红色荧光蛋白的表达情况,并统计注射48 h后胚胎的荧光率。
2 结果 2.1 I-SceI转基因供体质粒和辅助质粒通过设计长引物直接退火的方式,将I-SceI的2个序列识别位点连入酶切好的骨架质粒即得到I-SceI转基因供体质粒pI-SceI-EF1α-eGFP和pI-SceI-EF1α-RFP,此供体质粒的结构包含巨核酸酶的2个识别位点,其识别区域共18 bp。在2个识别区域之间是EF1α启动子和目的基因eGFP(或RFP),见图1a。
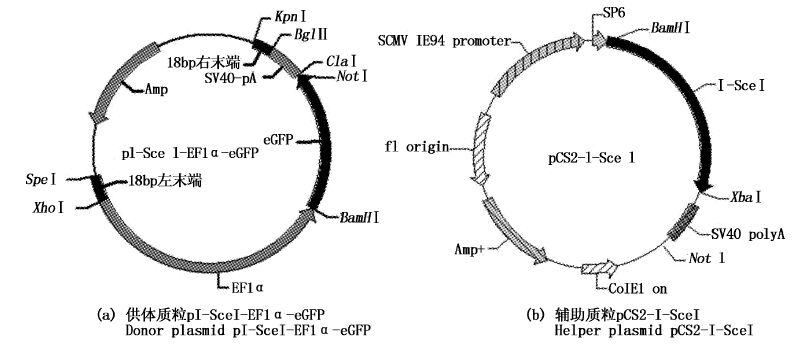
|
图1 巨核酸酶I-SceI转基因供体质粒和辅助质粒示意图 Fig.1 Schematic diagram of I-SceI transgenic donor plasmid and helper plasmid |
辅助质粒pCS2-I-SceI含有巨核酸酶I-SceI的编码区,可在体外转录I-SceI酶的mRNA,注射到胚胎内可翻译成蛋白、介导供体质粒的切割与整合。
2.2 显微注射与胚胎中荧光蛋白检测受精卵进行供体质粒pI-SceI-EF1α-eGFP/RFP与I-SceI mRNA的共注射,注射24 h、48 h和3 d后,在胚胎中可分别观察到绿色荧光蛋白和红色荧光蛋白的表达。2种荧光蛋白的表达均遍布全身,从头部、躯干到尾部均可观察到荧光信号。且随着胚胎的发育,荧光信号逐渐增强,红色荧光尤为明显(图2)。
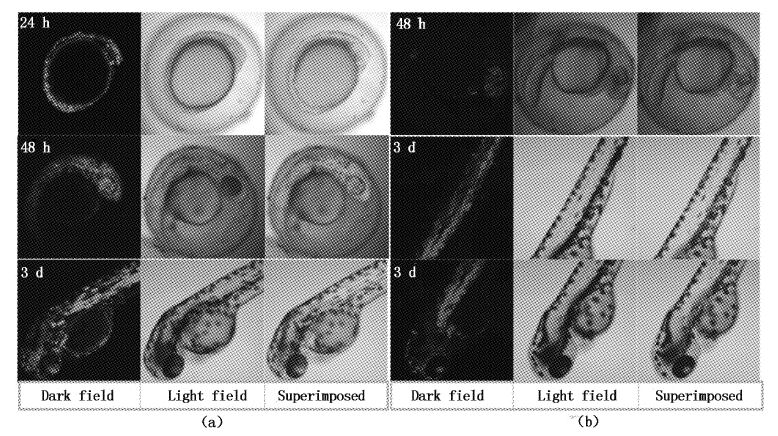
|
图2
激光共聚焦显微镜观察斑马鱼胚胎的荧光
Fig.2
Fluorescent protein detection of zebrafish embryo by confocal laser scanning microscope
(a) 注射供体质粒pI-SceI-EF1α-eGFP和I-SceI的mRNA; (b) 注射供体质粒pI-SceI-EF1α-RFP和I-SceI的mRNA。 (a) Microinjecting of donor plasmid pI-SceI-EF1α-eGFP and I-SceI mRNA; (b) Microinjecting of donor plasmid pI-SceI-EF1α-RFP and I-SceI mRNA. |
在显微注射供体质粒pI-SceI-EF1α--eGFP和I-SceI的mRNA 48 h后,随机抽取3组胚胎,在荧光显微镜下统计其荧光率。每组抽检50~70个胚胎进行统计,其荧光率在70.59%~78.79%之间,3组胚胎的平均荧光率为75.86%(表2)。
| 表2 I-SceI介导的转eGFP基因斑马鱼的荧光率检测 Tab.2 Positive rate in eGFP gene transferred Danio rerio mediated by I-SceI |
早期获得的转基因动物都是通过向刚受精的胚胎中直接注入环状或线性化的质粒DNA,这种方法容易产生嵌合体且后代的可遗传率偏低(普遍低于5%),因此获得转基因动物的成功率很低,需要进行大量的转基因注射与筛选[17]。为了提高转基因动物的基因整合效率和可遗传率,需寻求新的转基因介导方法。近年来转座子和巨核酸酶I-SceI等新的转基因介导方法受到关注[18]。转座子或巨核酸酶I-SceI注射到受精卵后可促使转植基因与宿主基因组更早的发生融合,从而提高转植基因在转基因首代中的表达效率与整合率[19]。巨核酸酶I-SceI在裂解反应产生DNA双链缺口后仍留在识别位点上,减弱了细胞体内连接酶的活性,避免移植基因自连形成多聚体的效应,所以移植基因可大量地在细胞中以线性、短序列状态存在。同时巨核酸酶还可以诱导细胞内的修复系统聚集在DNA双链缺口处进行同源重组而达到基因整合的目的[20],因此巨核酸酶I-SceI可提高转基因效率。此外与转座子系统相比,巨核酸酶I-SceI的识别序列更短,只有18 bp,便于构建[3, 14]。
普通限制性内切酶的识别序列很短,一般3到8个碱基,而巨核酸酶的识别序列在12到40碱基,一般为18个碱基,因此理论上,每七百亿个碱基才会出现一个酶切位点,所以鱼类基因组一般都不会出现巨核酸酶I-SceI的酶切位点,可避免受巨核酸酶I-SceI的酶切。在基因转植时,将巨核酸酶I-SceI以及带有其识别位点的供体质粒一起注射到鱼受精卵中,巨核酸酶I-SceI会切开供体质粒、产生双链缺口,然后再诱导宿主中的修复系统对酶切产生的片段进行同源重组,从而达到整合进鱼类基因组的目的[20]。
自1988年COLLEAUX等开发巨核酸酶I-SceI以来[2],该系统已在不同物种的转基因上应用,但相关研究大多是将带有I-SceI识别位点的转基因载体和I-SceI酶的蛋白共注射到胚胎中来获得转基因动物,耗时长且工作量大。BABARYKA等在应用酵母双杂交系统构建转基因斑马鱼的相关研究中,将体外合成I-SceI的mRNA,与带有I-SceI识别位点与GAL4基因的表达载体,共注射于斑马鱼胚胎,筛选获得GAL4转基因斑马鱼,然后再向转基因斑马鱼子代胚胎中注射带有UAS-GFP的质粒或者UAS-RFP的质粒,即可得到转绿色荧光和红色斑马鱼[16]。本研究直接将I-SceI识别位点加在目标基因eGFP和RFP的表达载体上,通过一次注射即可产生带荧光的斑马鱼,而且对用I-SceI mRNA取代巨核酸酶I-SceI蛋白进行注射的转基因效果的验证也更加直观和简洁。
本研究通过体外转录获得巨核酸酶I-SceImRNA,代替巨核酸酶I-SceI蛋白进行注射,操作更方便、成本更低,也取得了较好的转植效率。I-SceI介导的转基因斑马鱼胚胎在48 h的平均荧光率达到75.86%,与Tgf 2转座子在斑马鱼中的荧光率(76.6%)相近[21]。后期观察胚胎在发育到3~10 d期间,仍有荧光的表达,排除了是游离质粒的瞬时表达[22]。
本研究证实了巨核酸酶I-SceI mRNA可介导斑马鱼中的转基因,并提高基因转植效率。下一步将对首代的转基因斑马鱼进行基因组水平上的检测,确定其整合位点和拷贝数,并进行传代,对其可遗传性进行验证等,以便对此转基因方法做更全面准确的评估。
| [1] | JACQUIER A, DUJON B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene[J]. Cell, 1985, 41(2): 383-394. |
| [2] | COLLEAUX L, D'AURIOL L, GALIBERT F, et al. Recognition and cleavage site of the intron-encoded omega transposase[J]. Proceedings of the National Academy of Sciences of the United States of America, 1988, 85(16): 6022-6026. |
| [3] | PLESSIS A, PERRIN A, HABERT J E, et al. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus[J]. Genetics, 1992, 130(3): 451-460. |
| [4] | SIEGL T, PETZKE L, WELLE E, et al. I-SceI endonuclease: a new tool for DNA repair studies and genetic manipulations in streptomycetes[J]. Applied Microbiology and Biotechnology, 2010, 87(4): 1525-1532. |
| [5] | ZHANG N, SHAO L J, JIANG Y, et al. I-SceI-mediated scarless gene modification via allelic exchange in Clostridium[J]. Journal of Microbiological Methods, 2015, 108: 49-60. |
| [6] | BELLAICHE Y, MOGILA V, PERRIMON N. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila[J]. Genetics, 1999, 152(3): 1037-1044. |
| [7] | WINDBICHLER N, PAPATHANOS P A, CATTERUCCIA F, et al. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos[J]. Nucleic Acids Research, 2007, 35(17): 5922-5933. |
| [8] | WINDBICHLER N, MENICHELLI M, PAPATHANOS P A, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito[J]. Nature, 2011, 473(7346): 212-215. |
| [9] | COHEN-TANNOUDJI M, ROBINE S, CHOULIKA A, et al. I-SceI-induced gene replacement at a natural locus in embryonic stem cells[J]. Molecular and Cellular Biology, 1998, 18(3): 1444-1448. |
| [10] | CHOULIKA A, PERRIN A, DUJON B, et al. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae[J]. Molecular and Cellular Biology, 1995, 15(4): 1968-1973. |
| [11] | BEVACQUA R J, CANEL N G, HIRIART M I, et al. Simple gene transfer technique based on I-SceI meganuclease and cytoplasmic injection in IVF bovine embryos[J]. Theriogenology, 2013, 80(2): 104-113. |
| [12] | PAN F C, CHEN Y L, LOEBER J, et al. I-SceI meganuclease-mediated transgenesis in Xenopus[J]. Developmental Dynamics, 2006, 235(1): 247-252. |
| [13] | NEDELKOVSKA H, ROBERT J. Optimized transgenesis in Xenopus laevis/gilli isogenetic clones for immunological studies[J]. Genesis, 2012, 50(3): 300-306. |
| [14] | THERMES V, GRABHER C, RISTORATORE F, et al. I-SceI meganuclease mediates highly efficient transgenesis in fish[J]. Mechanisms of Development, 2002, 118(1/2): 91-98. |
| [15] | GRABHER C, JOLY J S, WITTBRODT J. Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI[J]. Methods in Cell Biology, 2004, 77: 381-401. |
| [16] | BABARYKA A, KUHN E, KOSTER R W. In vivo synthesis of meganuclease for generating transgenic zebrafish Danio rerio[J]. Journal of Fish Biology, 2009, 74(2): 452-457. |
| [17] | SOROLDONI D, HOGAN B M, OATES A C. Simple and efficient transgenesis with meganuclease constructs in zebrafish[J]. Methods in Molecular Biology, 2009, 546: 117-130. |
| [18] | 邹曙明, 杜雪地, 蒋霞云. 鱼类活性DNA转座子的发掘与应用概况[J]. 上海海洋大学学报, 2012, 21(5): 656-661. ZOU S M, DU X D, JIANG X Y. Overview of fish DNA transposon discovery and into application[J]. Journal of Shanghai Ocean University, 2012, 21(5): 656-661. |
| [19] | 叶星, 田园园, 高风英. 转基因鱼的研究进展与商业化前景[J]. 遗传, 2011, 33(5): 494-503. YE X, TIAN Y Y, GAO F Y. Progress in transgenic fish techniques and application[J]. Hereditas, 2011, 33(5): 494-503. |
| [20] | GRABHER C, WITTBRODT J. Meganuclease and transposon mediated transgenesis in medaka[J]. Genome Biology, 2007, 8(s1): S10. |
| [21] | 吴芳, 叶星, 邹曙明, 等. Tgf2转座系统在转RFP基因斑马鱼上的应用[J]. 中国水产科学, 2014, 21(4): 647-654. WU F, YE X, ZOU S M, et al. Application of the Tgf2 transposon system to RFP transgenic Danio rerio construction[J]. Journal of Fishery Sciences of China, 2014, 21(4): 647-654. |
| [22] | 王瑶, 蒋霞云, 邹曙明. PiggyBac转座元件的构建及其在团头鲂基因组中的转基因效率[J]. 上海海洋大学学报, 2014, 23(2): 161-166. WANG Y, JIANG X Y, ZOU S M. The study on transgenic efficiency of PiggyBac transposon in the genome of Megalobrama amblycephala[J]. Journal of Shanghai Ocean University, 2014, 23(2): 161-166. |
2. College of Fisheries and Life Science, Shanghai Ocean University, Shanghai 201306, China
 2015, Vol. 24
2015, Vol. 24


