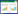扩展功能
文章信息
- 宝钲, 杨茗羽, 张修月, 岳碧松, 范振鑫
- BAO Zheng, YANG Mingyu, ZHANG Xiuyue, YUE Bisong, FAN Zhenxin
- 美洲大蠊microRNA的初步研究
- Preliminary Research of microRNA in Periplaneta americana
- 四川动物, 2019, 38(1): 47-55
- Sichuan Journal of Zoology, 2019, 38(1): 47-55
- 10.11984/j.issn.1000-7083.20180152
-
文章历史
- 收稿日期: 2018-05-10
- 接受日期: 2018-11-14
2. 四川药用动物工程技术研究中心, 四川西昌 615000;
3. 药用美洲大蠊四川省重点实验室, 成都 610031
2. Sichuan Engineering Research Center for Medicinal Animals, Xichang, Sichuan Province 615000, China;
3. Sichuan Key Laboratory for Medicinal American Cockroach, Chengdu 610031, China
microRNA(miRNA)是一类长度18~25 nt的非编码小RNA,广泛存在于动物、植物、微生物中,在转录后水平调控基因表达和翻译(Bartel,2004;Bartel & Chen,2004)。miRNA在生物中的调控方式一般有3种:一种是与靶mRNA的3’UTR完全互补配对,导致mRNA直接被降解(Llave et al., 2002);一种是与靶mRNA的3’UTR不完全互补配对,不影响mRNA的稳定性,只是抑制其翻译(Slack et al., 2000);还有一种是同时具有上述2种方式(Lee & Ambros,2001)。目前大量研究已揭示miRNA在昆虫多种生理活动中均有重要功能,如发育(Cristino et al., 2011;Surridge et al., 2011;Chen et al., 2012)、激素信号传导(Rebijith et al., 2016)、繁殖(Paulo et al., 2017)等。在蜚蠊目Blattodea中,目前仅有德国小蠊Blattella germanica miRNA的相关报道(Cristino et al., 2011)。
美洲大蠊Periplaneta americana属节肢动物门Arthropoda昆虫纲Insecta蜚蠊目,是蜚蠊科Blattidae中体型最大的一种,也是世界上最常见的室内蟑螂之一。美洲大蠊对各类环境适应性强,同时也传播病菌和寄生虫,被认为是世界性卫生害虫(Chen et al., 2015;Salama,2015;Zhang et al., 2016;Li et al., 2017)。但美洲大蠊也是传统的药用昆虫,其提取物具有提高免疫、抗肿瘤、抗炎镇痛、保肝、组织修复等效果(肖小芹等,2007;Wang et al., 2011;Jiang et al., 2012;王鹏飞等,2015;张丹等,2015;吴道勋等,2016)。目前有关美洲大蠊的研究多集中在基因组、繁殖、消化特性和转录组等(Wicher et al., 2006;Nishino et al., 2010;Chen et al., 2013, 2015;Tamaki et al., 2014;Kim et al., 2016;Zhang et al., 2016;晋家正等,2018;Li et al., 2018),美洲大蠊的miRNA及其在基因表达中的调控机制迄今尚未见报道。尤其是雌雄美洲大蠊在形态和寿命上存在明显的差异,雌虫比雄虫体型大,雄虫比雌虫寿命短(陈梦林,2002),这种差异是否与雌雄之间miRNA调控的差异有关?本研究对实验室饲养的雌雄美洲大蠊进行小RNA的高通量测序,去除低质量和受污染的序列后,将所得序列与数据库(NCBI、Rfam)进行比对注释,旨在初步了解美洲大蠊小RNA的组成,包括tRNA、rRNA、snRNA、snoRNA等;将非miRNA的小RNA过滤后,检测已知的miRNA、预测新miRNA并寻找雌雄之间的差异表达miRNA。宝钲等:美洲大蠊microRNA的初步研究
1 材料与方法 1.1 样品准备与小RNA的文库构建和测序美洲大蠊成虫和饲料均由四川好医生攀西药业有限责任公司提供。取饲养于本实验室(温度25 ℃,湿度60%)的雌雄成虫各3只。为避免消化道中细菌对测序结果的干扰,将去除消化道的美洲大蠊搅碎后加入磷酸盐缓冲液离心,加入异丙醇提取总RNA。检测合格的总RNA由北京诺禾致源科技股份有限公司进行建库和高通量测序,测序平台为Illumina HiSeq 2000,测序长度为50 bp。
1.2 数据处理测序完成后,去除低质量和被污染的序列,剩下的序列(干净序列)用于后续分析。
为了防止非miRNA(包括tRNA、rRNA、scRNA、snRNA、snoRNA等)序列影响分析,将得到的干净序列用Blastn与数据库(NCBI、Rfam)中非miRNA进行比较,再利用Repeatmasker找出小RNA中的重复序列,然后注释这些非miRNA并在后续分析中去除。
剩下的未注释序列利用miRDeep2(Friedländer et al., 2011)检测已知的miRNA和预测潜在的新miRNA。因为miRBase 21数据库(ftp://mirbase.org/pub/mirbase/21)中目前还没有美洲大蠊的miRNA数据,但miRNA在物种间具有保守性,所以通过miRBase 21数据库中其他昆虫已知的miRNA对美洲大蠊进行检测。分析所需要的基因组数据来自本实验室测序和组装注释的美洲大蠊基因组(晋家正等,2018)。
1.3 预测靶基因及KEGG通路分析利用miRanda(Enright et al., 2003)和RNAhybrid(Krüger & Rehmsmeier,2006)预测美洲大蠊miRNA的靶基因。miRanda的阈值设定为:分数≥155,自由能ΔG≤-20 kcal/mol。RNAhybrid的阈值设定为:ΔG≤-20 kcal/mol,其余参数为默认值(Enright et al., 2003;Krüger & Rehmsmeier,2006)。为使预测结果更加可信,将2个软件的预测结果取交集进行后续的分析。
通过KEGG通路分析对miRNA的靶基因进行注释。利用KAAS (Moriya et al., 2007)和KEGG mapper software (Kanehisa et al., 2012)对靶基因进行KEGG注释。
1.4 已知miRNA的差异表达将雄性成虫和雌性成虫的miRNA分别进行统计分析以鉴定雌雄之间的差异表达。先将miRNA的表达量归一化,然后再计算P值。
归一化公式:归一化表达量(NE)=实际miRNA数量/clean reads总数×1 000 000。
P值公式:
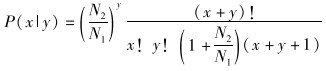
|
式中,N1和x属于相同的组,N1代表clean reads的总数,x代表归一化的表达水平。N2和y属于相同的组,N2代表clean reads的总数,y代表归一化的表达水平(Zhang et al., 2013)。
2 结果 2.1 测序数据、长度分布和其他小RNA的注释通过测序,在雄性成虫和雌性成虫中分别得到17 259 315条和17 249 947条小RNA序列,剔除质量低和被污染的序列后,分别得到12 155 616条和9 847 263条。其中,共有序列占总序列的84.08%,雄性成虫特有的序列占总序列的8.81%,雌性成虫特有的序列占总序列的7.11%(图 1:a)。雌雄成虫中所测得的所有小RNA序列均主要分布于18~23 nt,在22 nt和29 nt处有2个峰值(图 1:b)。在这些序列中,将相同的序列统计为同一种得到唯一序列,最终在雄性成虫和雌性成虫中分别得到2 247 751条和2 051 813条小RNA序列。
|
| 图 1 美洲大蠊雌雄成虫相同序列的小RNA数量和长度分布 Fig. 1 Venn diagram of the same sequence of small RNAs and their length distributions in female and male Periplaneta americana |
| |
通过对小RNA注释,发现许多非miRNA存在,包括rRNA、tRNA、snRNA、snoRNA、scRNA、短小的重复序列、内含子、外显子等(表 1)。为了防止这些序列干扰后续分析,去除注释为非miRNA的序列。最终在雄性成虫和雌性成虫中分别获得11 507 387条和9 207 534条可能为miRNA的序列,这些序列除部分序列没有注释外,绝大多数被注释为miRNA。
| 组别 | 雄性成虫 | 雌性成虫 | |||
| 唯一序列数量/条(占总小RNA的比例) | 全序列数量/条(占总小RNA的比例) | 唯一序列数量/条(占总小RNA的比例) | 全序列数量/条(占总小RNA的比例) | ||
| 总小RNA | 2 247 751(100%) | 12 155 616(100%) | 2 051 813(100%) | 9 847 263(100%) | |
| rRNA | 47 172(2.10%) | 384 358(3.16%) | 57 580(2.81%) | 392 256(3.98%) | |
| tRNA | 29 363(1.31%) | 227 840(1.87%) | 28 868(1.41%) | 218 086(2.21%) | |
| snoRNA | 3 145(0.14%) | 9 626(0.08%) | 2 489(0.12%) | 6 692(0.07%) | |
| snRNA | 3 688(0.16%) | 11 157(0.09%) | 2 796(0.14%) | 9 860(0.10%) | |
| piRNA | 247(0.01%) | 3 866(0.03%) | 183(0.01%) | 2 106(0.02%) | |
| scRNA | 0(0%) | 0(0%) | 1(0%) | 1(0%) | |
| 外显子 | 23(0%) | 80(0%) | 15(0%) | 54(0%) | |
| 内含子 | 1 440(0.06%) | 3 611(0.03%) | 1 958(0.10%) | 5 208(0.05%) | |
| 重复序列 | 3 187(0.14%) | 7 691(0.06%) | 2 401(0.12%) | 5 466(0.06%) | |
| 未注释序列 | 2 159 486(96.07%) | 11 507 387(94.67%) | 1 955 522(95.31%) | 9 207 534(93.50%) | |
在雄性成虫和雌性成虫中分别发现了57种和53种已知的miRNA,去除二者共有的miRNA,共发现59种已知的miRNA。表达量前10的miRNA见表 2,其中,miR-1、miR-10-5p、miR-8-5p、miR-31a等广泛参与了各种组织和细胞的发育、生长和代谢等重要生理过程。
| miRNA名称 | 雄性成虫表达量 | 雌性成虫表达量 |
| miR-1 | 1 421 469 | 840 951 |
| miR-276b-5p | 1 195 999 | 719 123 |
| miR-100 | 616 606 | 280 854 |
| bantam | 254 908 | 106 470 |
| miR-8-5p | 206 294 | 152 100 |
| miR-9a | 203 018 | 92 451 |
| miR-10-5p | 150 950 | 92 034 |
| miR-277-3p | 132 950 | 67 294 |
| miR-9b-5p | 90 860 | 53 344 |
| miR-31a | 69 865 | 26 774 |
同时,在雄性成虫和雌性成虫中预测出152种和94种潜在的新miRNA,去除共有的miRNA,共发现152种新miRNA。这些新miRNA在miRBase 21数据库中尚未报道,可能是美洲大蠊所特有的miRNA。
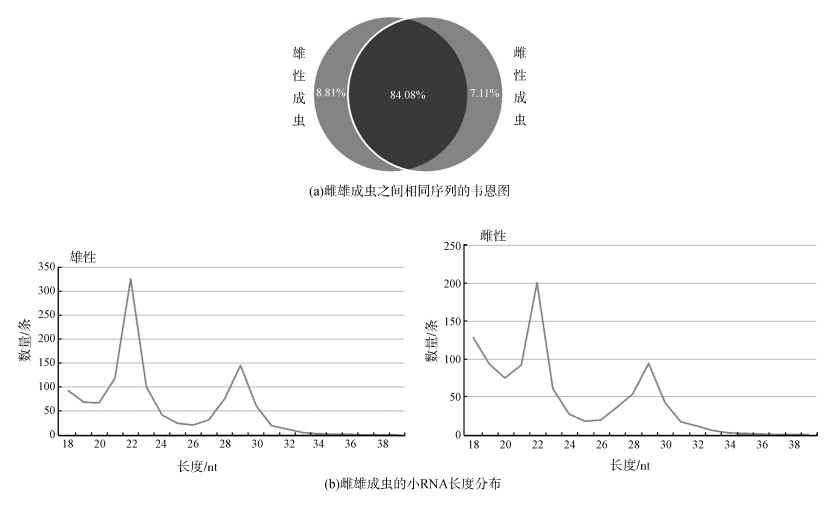
进一步对预测的潜在的新miRNA的靶基因进行预测,将miRanda和RNAhybrid预测结果整合后,共计在雄性成虫和雌性成虫中预测了5 389个和3 823个靶基因,去掉共有的,共5 822个靶基因。KEGG通路分析(图 2)显示了这些靶基因的功能,其中,信号转导通路注释到的靶基因最多,且雄性成虫比雌性成虫多,说明美洲大蠊miRNA调控的靶基因主要是参与信号转导通路的基因。
|
| 图 2 KEGG通路中美洲大蠊雌雄成虫miRNA的靶基因数量 Fig. 2 Numbers of target genes of miRNA in female and male Periplaneta americana annotated by KEGG |
| |
雌雄美洲大蠊成虫差异表达的miRNA的散点图显示仅有1条显著差异表达的miRNA:miR-750,其在雌性成虫中的表达显著高于雄性成虫(图 3)。
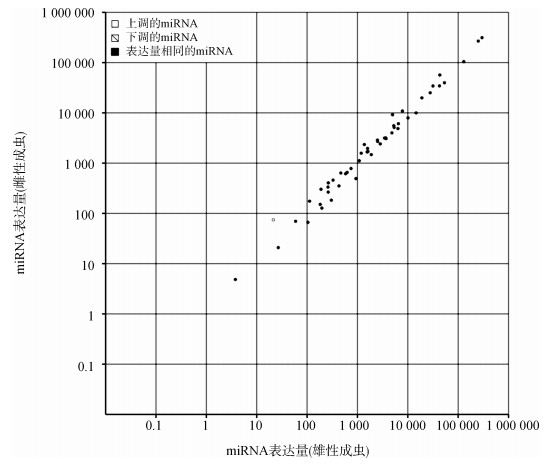
|
| 图 3 美洲大蠊雌雄成虫已知miRNA的差异表达散点图 Fig. 3 The scatter figure of differentially expressed known miRNAs between female and male Periplaneta americana |
| |
miRNA越来越受到研究者的关注,这是由于miRNA被发现在越来越多生物过程中发挥着重要作用,如发育(Reinhart et al., 2000)、细胞分化(Kawasaki & Taira,2003)、细胞增殖(Brennecke et al., 2003)等。很多昆虫的miRNA已被报道(Wei et al., 2009;Jagadeeswaran et al., 2010;Skalsky et al., 2010;Zhang et al., 2012;Huang et al., 2014;He et al., 2016)。本研究是首次对美洲大蠊的miRNA进行报道,在雄性和雌性成虫中分别发现了2 247 751条和2 051 813条miRNA序列。在miRNA序列长度分布上,显示出2个峰值,在21~23 nt的峰代表的是miRNA,在28~30 nt的峰代表的可能是Piwi蛋白互作RNA(piRNA),这是一种与miRNA完全不同的RNA沉默因子(Kawaoka et al., 2011)。这一现象在其他昆虫,包括飞蛾Manduca sexta(Zhang et al., 2012)、桔小实蝇Bactrocera dorsalis(Huang et al., 2014)等都存在相似的结果。与德国小蠊的miRNA (Cristino et al., 2011)相比,美洲大蠊表达量最高的前10个miRNA中,有4个与德国小蠊相同:miR-1、miR-8-5p、miR-10-5p和miR-31a。在德国小蠊中表达、在美洲大蠊中不表达的miRNA有miR-184、miR-276a、miR-279b、miR-375和miR-iab-4as-5p。然而,更多的miRNA在美洲大蠊中表达而未在德国小蠊中发现,如miR-133、miR-277-3p、miR-278、miR-2765-5p、miR-281-5p、miR-316和miR-9b-5p。其中,miR-1在心脏和骨骼肌祖细胞的发育上发挥重要作用(Zhao & Srivastava,2007);miR-10-5p的靶基因Hox基因决定体轴的正确前后样式(Lund,2010);miR-8-5p参与众多生物学过程,包括发育(Belles et al., 2012)、神经肌肉协调(Karres et al., 2007;Rubio et al., 2013)、卵及卵巢形成(Lucas et al., 2015)等;miR-31a可能在蜕皮过程中调控上皮代谢(Yu et al., 2008);miR-276通过调控1个转录共活化基因调控孵化同步性(He et al., 2016);miR-133的其中1个靶基因是结缔组织生长因子(CTGF),其在纤维化过程中发挥重要作用(Duisters et al., 2009);miR-281的靶基因是登革热病毒血清2型(DENV-2),增强在白纹伊蚊Aedes albopictus中DENV-2的病毒复制(Zhou et al., 2014)。本研究结果显示,美洲大蠊的miRNA广泛参与了昆虫组织和细胞的生长、发育、代谢等多种重要的调控机制。此外,美洲大蠊和德国小蠊都存在一些特有的miRNA调控的基因,显示2种蜚蠊目昆虫存在不同的调控机制,这或许是造成二者形态和生理差异的原因之一。
差异表达分析显示,美洲大蠊雌雄成虫miRNA在组成和表达量上几乎没有差异,只有miR-750在雌性成虫中高表达。miR-750在被白斑综合症病毒WSSV感染的斑节对虾Penaeus monodon中表达量显著降低,其预测的靶基因是ATG14基因,但随后的实验证实ATG14基因不是miR-750的靶基因(Kaewkascholkul et al., 2016)。Rebijith等(2016)通过RNAi实验在棕榈蓟马Thrips palmi中证明JH候选受体——过剩气门蛋白是miR-750的靶基因,表明miR-750可能通过调节卵黄蛋白原基因参与免疫、应激反应和激素信号传导。卵黄蛋白原主要存在于非哺乳类性成熟的卵生雌性动物的血淋巴、脂肪体和卵中,所以miR-750在雌性成虫中高表达。总之,对雌雄美洲大蠊进行了小RNA的测序,检测出了57种已知的miRNA,并预测出152种潜在的新miRNA,但这些数据是用软件预测出来的,这些潜在的新miRNA是否具有功能及其相应的靶基因,还需要进一步的验证。这些miRNA的发现为将来美洲大蠊中miRNA的研究提供了基础,其表达量和功能的研究将推进对于美洲大蠊调控网络的理解,并为美洲大蠊的虫害防治和在中药中的应用提供新的视角。
| 陈梦林. 2002. 蟑螂养殖技术[J]. 农村新技术, 10: 21–23. |
| 晋家正, 李午佼, 牟必琴, 等. 2018. 药用美洲大蠊全基因组测序分析[J]. 四川动物, 37(2): 121–126. |
| 王鹏飞, 许润春, 李江维, 等. 2015. 美洲大蠊油脂急性毒性及其拮抗美洲大蠊提取液肝脏保护作用的初步研究[J]. 现代中药研究与实践, 29(6): 34–36. |
| 吴道勋, 邵维莉, 杨贤英, 等. 2016. 美洲大蠊抗肿瘤与免疫调节研究进展[J]. 亚太传统医药, 12(23): 48–51. |
| 肖小芹, 汪世平, 徐绍锐, 等. 2007. 美洲大蠊提取物抗炎、镇痛作用的实验研究[J]. 中国病原生物学杂志, 2(2): 140–143. |
| 张丹, 孙玉红, 李茂, 等. 2015. 美洲大蠊多肽提取物对荷瘤小鼠肿瘤生长及免疫功能的影响[J]. 中国新药杂志, 24(6): 681–686. |
| Bartel DP, Chen CZ. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs[J]. Nature Reviews Genetics, 5(5): 396–400. DOI:10.1038/nrg1328 |
| Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function[J]. Cell, 116(2): 281–297. DOI:10.1016/S0092-8674(04)00045-5 |
| Belles X, Cristino AS, Tanaka ED, et al. 2012. Insect microRNAs: from molecular mechanisms to biological roles[M]. Gilbert LI. Insect molecular biology and biochemistry. New York: Academic Press: 30-56. |
| Brennecke J, Hipfner DR, Stark A, et al. 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila[J]. Cell, 113(1): 25–36. DOI:10.1016/S0092-8674(03)00231-9 |
| Chen Q, Lu L, Hua H, et al. 2012. Characterization and comparative analysis of small RNAs in three small RNA libraries of the brown planthopper (Nilaparvata lugens)[J]. PLoS ONE, 7(3): e32860. DOI:10.1371/journal.pone.0032860 |
| Chen W, Jiang GF, Sun SH, et al. 2013. Identification of differentially expressed genes in American cockroach ovaries and testes by suppression subtractive hybridization and the prediction of its miRNAs[J]. Molecular Genetics and Genomics, 288(11): 627–638. DOI:10.1007/s00438-013-0777-1 |
| Chen W, Liu YX, Jiang GF. 2015. De novo assembly and characterization of the testis transcriptome and development of EST-SSR markers in the cockroach Periplaneta americana[J]. Scientific Reports, 5: 11144. DOI:10.1038/srep11144 |
| Cristino AS, Tanaka ED, Rubio M, et al. 2011. Deep sequencing of organ- and stage-specific microRNAs in the evolutionarily basal insect Blattella germanica (L.)(Dictyoptera, Blattellidae)[J]. PLoS ONE, 6(4): e19350. DOI:10.1371/journal.pone.0019350 |
| Duisters RF, Tijsen AJ, Schroen B, et al. 2009. MiR-133 and miR-30 regulate connective tissue growth factor[J]. Circulation Research, 104(2): 170–178. DOI:10.1161/CIRCRESAHA.108.182535 |
| Enright AJ, John B, Gaul U, et al. 2003. MicroRNA targets in Drosophila[J]. Genome Biology, 5(1): R1. DOI:10.1186/gb-2003-5-1-r1 |
| Friedländer MR, Mackowiak SD, Li N, et al. 2011. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades[J]. Nucleic Acids Research, 40(1): 37–52. |
| He J, Chen Q, Wei Y, et al. 2016. MicroRNA-276 promotes egg-hatching synchrony by up-regulating brm in locusts[J]. Proceedings of the National Academy of Sciences of the United States of America, 113(3): 584–589. DOI:10.1073/pnas.1521098113 |
| Huang Y, Dou W, Liu B, et al. 2014. Deep sequencing of small RNA libraries reveals dynamic expression patterns of microRNAs in multiple developmental stages of Bactrocera dorsalis[J]. Insect Molecular Biology, 23(5): 656–667. DOI:10.1111/imb.2014.23.issue-5 |
| Jagadeeswaran G, Zheng Y, Sumathipala N, et al. 2010. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel-pam microRNAs and microRNA-stars during silkworm development[J]. BMC Genomics, 11(1): 52. DOI:10.1186/1471-2164-11-52 |
| Jiang L, Liu X, Xia CL, et al. 2012. Research advance on chemical constituents and anti-tumor effects of Periplaneta americana L[J]. Medicinal Plant, 3(11): 95–97. |
| Kaewkascholkul N, Somboonviwat K, Asakawa S, et al. 2016. Shrimp miRNAs regulate innate immune response against white spot syndrome virus infection[J]. Developmental & Comparative Immunology, 60: 191–201. |
| Kanehisa M, Goto S, Sato Y, et al. 2012. KEGG for integration and interpretation of large-scale molecular data sets[J]. Nucleic Acids Research, 40(D1): D109–D114. DOI:10.1093/nar/gkr988 |
| Karres JS, Hilgers V, Carrera I, et al. 2007. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila[J]. Cell, 131(1): 136–145. DOI:10.1016/j.cell.2007.09.020 |
| Kawaoka S, Arai Y, Kadota K, et al. 2011. Zygotic amplification of secondary piRNAs during silkworm embryogenesis[J]. RNA, 17(7): 1401–1407. DOI:10.1261/rna.2709411 |
| Kawasaki H, Taira K. 2003. Hes1 is a target of microRNA-23 during retinoic-acid-induced neuronal differentiation of NT2 cells[J]. Nature, 423(6942): 838–842. DOI:10.1038/nature01730 |
| Kim IW, Lee JH, Subramaniyam S, et al. 2016. De novo transcriptome analysis and detection of antimicrobial peptides of the American cockroach Periplaneta americana (Linnaeus)[J]. PLoS ONE, 11(5): e0155304. DOI:10.1371/journal.pone.0155304 |
| Krüger J, Rehmsmeier M. 2006. RNAhybrid: microRNA target prediction easy, fast and flexible[J]. Nucleic Acids Research, 34(suppl_2): W451–W454. |
| Lee RC, Ambros V. 2001. An extensive class of small RNAs in Caenorhabditis elegans[J]. Science, 294(5543): 862–864. DOI:10.1126/science.1065329 |
| Li S, Zhu S, Jia Q, et al. 2018. The genomic and functional landscapes of developmental plasticity in the American cockroach[J]. Nature Communications, 9(1): 1008. DOI:10.1038/s41467-018-03281-1 |
| Li ZQ, He P, Zhang YN, et al. 2017. Molecular and functional characterization of three odorant-binding protein from Periplaneta americana[J]. PLoS ONE, 12(1): e0170072. DOI:10.1371/journal.pone.0170072 |
| Llave C, Xie Z, Kasschau KD, et al. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA[J]. Science, 297(5589): 2053–2056. DOI:10.1126/science.1076311 |
| Lucas KJ, Roy S, Ha J, et al. 2015. MicroRNA-8 targets the Wingless signaling pathway in the female mosquito fat body to regulate reproductive processes[J]. Proceedings of the National Academy of Sciences of the United States of America, 112(5): 1440–1445. DOI:10.1073/pnas.1424408112 |
| Lund AH. 2010. miR-10 in development and cancer[J]. Cell Death & Differentiation, 17(2): 209–214. |
| Moriya Y, Itoh M, Okuda S, et al. 2007. KAAS: an automatic genome annotation and pathway reconstruction server[J]. Nucleic Acids Research, 35(suppl_2): W182–W185. |
| Nishino H, Yoritsune A, Mizunami M. 2010. Postembryonic development of sexually dimorphic glomeruli and related interneurons in the cockroach Periplaneta americana[J]. Neuroscience Letters, 469(1): 60–64. |
| Paulo D, Azeredo-Espin A, Canesin L, et al. 2017. Identification and characterization of microRNAs in the screwworm flies Cochliomyia hominivorax and Cochliomyia macellaria (Diptera: Calliphoridae)[J]. Insect Molecular Biology, 26(1): 46–57. DOI:10.1111/imb.2017.26.issue-1 |
| Rebijith K, Asokan R, Hande HR, et al. 2016. The first report of mirnas from a Thysanopteran insect, Thrips palmi Karny using high-throughput sequencing[J]. PLoS ONE, 11(9): e0163635. DOI:10.1371/journal.pone.0163635 |
| Reinhart BJ, Slack FJ, Basson M, et al. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans[J]. Nature, 403(6772): 901–906. DOI:10.1038/35002607 |
| Rubio M, Montañez R, Perez L, et al. 2013. Regulation of atrophin by both strands of the mir-8 precursor[J]. Insect Biochemistry and Molecular Biology, 43(11): 1009–1014. DOI:10.1016/j.ibmb.2013.08.003 |
| Salama EM. 2015. A novel use for potassium alum as controlling agent against Periplaneta americana (Dictyoptera: Blattidae)[J]. Journal of Economic Entomology, 108(6): 2620–2629. DOI:10.1093/jee/tov239 |
| Skalsky RL, Vanlandingham DL, Scholle F, et al. 2010. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus[J]. BMC Genomics, 11(1): 119. DOI:10.1186/1471-2164-11-119 |
| Slack FJ, Basson M, Liu ZC, et al. 2000. The lin-41 RBCC gene acts in the CC. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor[J]. Molecular Cell, 5(4): 659–669. DOI:10.1016/S1097-2765(00)80245-2 |
| Surridge AK, Lopez-Gomollon S, Moxon S, et al. 2011. Characterisation and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene[J]. BMC Genomics, 12(1): 62. DOI:10.1186/1471-2164-12-62 |
| Tamaki FK, Pimentel AC, Dias AB, et al. 2014. Physiology of digestion and the molecular characterization of the major digestive enzymes from Periplaneta americana[J]. Journal of Insect Physiology, 70: 22–35. DOI:10.1016/j.jinsphys.2014.08.007 |
| Wang XY, He ZC, Song LY, et al. 2011. Chemotherapeutic effects of bioassay-guided extracts of the American cockroach, Periplaneta americana[J]. Integrative Cancer Therapies, 10(3): NP12–NP23. DOI:10.1177/1534735411413467 |
| Wei Y, Chen S, Yang P, et al. 2009. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust[J]. Genome Biology, 10(1): R6. DOI:10.1186/gb-2009-10-1-r6 |
| Wicher D, Söhler S, Gundel M, et al. 2006. Differential receptor activation by cockroach adipokinetic hormones produces differential effects on ion currents, neuronal activity, and locomotion[J]. Journal of Neurophysiology, 95(4): 2314–2325. DOI:10.1152/jn.01007.2005 |
| Yu X, Zhou Q, Li SC, et al. 2008. The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages[J]. PLoS ONE, 3(8): e2997. DOI:10.1371/journal.pone.0002997 |
| Zhang J, Zhang Y, Li J, et al. 2016. Midgut transcriptome of the cockroach Periplaneta americana and its microbiota: digestion, detoxification and oxidative stress response[J]. PLoS ONE, 11(5): e0155254. DOI:10.1371/journal.pone.0155254 |
| Zhang X, Zheng Y, Jagadeeswaran G, et al. 2012. Identification and developmental profiling of conserved and novel microRNAs in Manduca sexta[J]. Insect Biochemistry and Molecular Biology, 42(6): 381–395. DOI:10.1016/j.ibmb.2012.01.006 |
| Zhang XD, Zhang YH, Ling YH, et al. 2013. Characterization and differential expression of microRNAs in the ovaries of pregnant and non-pregnant goats (Capra hircus)[J]. BMC Genomics, 14(1): 1. DOI:10.1186/1471-2164-14-1 |
| Zhao Y, Srivastava D. 2007. A developmental view of microRNA function[J]. Trends in Biochemical Sciences, 32(4): 189–197. DOI:10.1016/j.tibs.2007.02.006 |
| Zhou Y, Liu Y, Yan H, et al. 2014. MiR-281, an abundant midgut-specific miRNA of the vector mosquito Aedes albopictus enhances dengue virus replication[J]. Parasites & Vectors, 7(1): 488. DOI:10.11861s/13071-014-0488-4 |
 2019, Vol. 38
2019, Vol. 38