文章信息
- 李周坤, 叶现丰, 杨帆, 黄彦, 范加勤, 王辉, 崔中利
- LI Zhoukun, YE Xianfeng, YANG Fan, HUANG Yan, FAN Jiaqin, WANG Hui, CUI Zhongli
- 黏细菌捕食生物学研究进展及其在农业领域的应用潜力
- The predation biology of myxobacteria and its application in agricultural field
- 南京农业大学学报, 2021, 44(2): 208-216
- Journal of Nanjing Agricultural University, 2021, 44(2): 208-216.
- http://dx.doi.org/10.7685/jnau.202010034
-
文章历史
- 收稿日期: 2020-10-29
2. 南京农业大学植物保护学院, 江苏 南京 210095;
3. 南京农业大学作物免疫学重点实验室, 江苏 南京 210095;
4. 中国科学院南京土壤研究所土壤环境与污染修复重点实验室, 江苏 南京 210008
2. College of Plant Protection, Nanjing Agricultural University, Nanjing 210095, China;
3. Key Laboratory of Plant Immunity, Nanjing Agricultural University, Nanjing 210095, China;
4. Key Laboratory of Soil Environment and Pollution Remediation, Institute of Soil Science, Chinese Academy of Sciences, Nanjing 210008, China
植物病害是制约农作物优质高产的重要因素之一, 据统计全球主要农作物的病害损失约占作物总产量的20%~40%, 每年直接经济损失高达数十亿美元[1-2], 其中70%~80%的病害是病原真菌所致[3-4]。至2050年全球粮食产量需要增加70%以满足日益增长的人口需求, 令人担忧的是自2000年以来, 新的真菌类型或者真菌类植物病原菌(fungal-like plant pathogen)呈现逐年增加的趋势[5], 粮食生产安全问题越来越受到关注, 已成为最重要的国际问题之一[6]。
目前, 农业生产上的植物病害主要以化学防治为主, 而过度依赖和滥用化学农药产生了有害生物抗药性、农药残留超标、环境污染等一系列问题, 严重影响我国农业的绿色可持续发展[7]。为了避免过度依赖化学农药的农业病害防治现状, 近年来利用微生物的抗菌、植物免疫调节以及根际或叶际微生物组调控作用, 阻止病原菌入侵植物已成为有效策略[8-10]。生防微生物的抗菌方式具有多样性, 目前, 受到关注比较多的生防微生物主要来自于假单胞菌(Pseudomonas)、芽胞杆菌(Bacillus)、伯克氏菌(Burkholderia)、溶杆菌(Lysobacter)、木霉(Trichoderma)和腐霉(Pythium)等属[11-12], 其中以枯草芽胞杆菌、哈茨木霉和寡雄腐霉等为代表的生防菌已被开发成商业化微生物菌剂, 应用于农业生产的病害控制, 其主要作用机制包括拮抗作用、竞争作用、诱导植物系统抗性等, 且孢子形成特性使其在菌剂长效保存和土壤生存方面具有一定的优势。生防微生物进入环境中受到环境因子的多变性[13]、植物与微生物互作过程中的免疫识别[14]以及植物根际调控[15]等因素的影响, 生防微生物在开放环境中难以定殖且防治效果不稳定, 导致生物菌剂的实际应用受到一定程度的限制。
在自然生态系统中, 存在着大量不同类型的生物体, 这些生物个体之间为争夺养分和空间形成了复杂的生态网络结构。捕食是生物体之间广泛存在的一种相互作用模式, 是构建生态系统群落结构和维持生物多样性的关键过程[16]。目前, 捕食性微生物包括吸血球菌(Vampirococcus)、蛭弧菌(Bdellovibrio)、噬菌弧菌(Bacteriovorax)、Micavibrio、Daptobacter、拟杆菌(Bacteroidetes)和黏细菌(myxobacteria)等[17-18]。其中, 蛭弧菌能够直接入侵细胞周质空间实现对革兰氏阴性细菌的捕食, 被广泛应用于养殖产业中改善水质和治疗水生动物细菌性疾病等方面[19]。此外, 研究人员发现Micavibrio aeruginosavorus可通过黏附到细菌的细胞壁上, 掠夺它们的养分来维持自身的生存和繁殖, 研究成果为治疗多种传染性疾病提供了依据, 从而减缓了微生物耐药性问题[20]。利用微生物的捕食作用能实现对病原微生物的控制, 为农业病害防治提供策略。
黏细菌是一类具有多细胞群体行为特征的革兰氏阴性细菌, 可以捕食包括细菌和真菌在内的多种微生物, 存在于土壤、树皮、朽木、动物粪便、地衣和昆虫等不同类型的环境中[21]。黏细菌具有超大的基因组(大约10 Mb), 能形成类似真菌的不同形态的子实体结构, 被称为“高等原核生物”。该类群大多具有复杂的生活史和生长代谢调控过程以及较强的环境适应能力[22]。然而, 黏细菌作为一类具有捕食特性的新型生防微生物, 其在农业生产过程中植物病害控制方面的应用并未引起太多的关注。本文综述黏细菌对于微生物的捕食策略、抗菌机制、捕食的生态学功能等方面的研究进展, 评估捕食性黏细菌在病害防治方面的应用潜力。同时, 讨论目前黏细菌对于微生物的捕食研究方面所存在的问题及其应对策略, 为黏细菌应用于农业生产过程的病害控制提供理论依据和策略。
1 黏细菌是一类通才型(generalist predator)的微生物捕食者捕食性微生物分布广泛, 在5个门(Proteobacteria、Chloroflexi、Cytophagaceae、Actinobacteria和Nanoarchaeota)中的15个科均发现了捕食性细菌, 包括寄生于硬蜱属的立克次氏体(Rickettsia)等[23]。除细菌外, 真菌中也存在具有捕食能力的类群, 如Arthrobotrys等可以捕食线虫和一些微生物[24]。微生物的捕食作用可能是通才型的(generalist predator), 如黏细菌, 对不同类型的细菌和真菌均具有捕食作用[25-26]; 也可能是专性的(obligate predator), 如蛭弧菌, 通过侵入革兰氏阴性细菌的周质空间进而实现对猎物细胞的分解[27]。然而, 黏细菌和蛭弧菌都可以在固体界面上滑行运动实现捕食, 但是所涉及的分子机制是相互独立的[28]。对于黏细菌的捕食研究开展的较早[29], 其捕食作用与已报道的几种捕食性微生物的作用方式不同。相对于以盘基网柄菌(Dictyostelium discoideum)为代表的细胞吞噬作用、蛭弧菌(Bdellovibrio bacteriovorus)为代表的胞质入侵作用、溶杆菌(Lysobacter)为代表的分泌扩散性抗菌物质以及腐生螺旋体属(Saprospira)为代表的通过黏性物质捕捉猎物(Ixotrophy)等方式[30], 黏细菌利用一种特殊的胞外猎杀机制入侵猎物菌落。
黏细菌的捕食策略类型分为: 1)直接攻击模式(frontal attack)。黏细菌细胞间相互合作形成直接攻击模式, 如黏细菌Myxococcus sp. BS对软腐果胶杆菌等病原细菌的捕食作用[26]。2)狼群围捕攻击模式(wolf pack attack)。黄色黏球菌(Myxococcus xanthus)DZ2对苜蓿中华根瘤菌(Sinorhizobium medicae)AK21的捕食过程中, 由于菌株AK21产生大量的胞外半乳葡聚糖抵御捕食, 因此黏细菌DZ2采取一种先围捕后猎杀的方式实现对猎物的捕食[31]。3)孤立捕食模式(solitary predation)。通常认为黏细菌通过类似群体狩猎的策略捕食猎物[30], 然而研究者也发现黏细菌单个细胞的孤立捕食也能实现对猎物细胞的猎杀[32-34]。基于自然环境中可获取资源的局限性, 多样化的捕食策略有助于黏细菌类群在环境中的适应能力。
2 捕食性黏细菌(predator)与猎物(prey)之间的多重博弈关系目前, 关于黏细菌捕食作用的研究主要集中在细菌捕食方面, 包括黏细菌产生的次级代谢物、裂解酶、外膜囊泡(OMVs)等[30, 35-36], 其中次级代谢物的抗菌活性被认为在黏细菌捕食过程中发挥着重要的作用[37-38], 而外膜囊泡被认为是黏细菌攻击猎物的短距离“运输机”[39]。此外, 黏细菌分泌的蛋白酶或肽酶、溶菌酶等裂解酶可能参与黏细菌的捕食作用[30, 40], 但到目前为止并无直接的证据。在黏细菌捕食大肠杆菌的研究中, 转录组学分析发现上千个基因响应捕食过程, 同时推测猎物细胞壁和蛋白质是黏细菌攻击的首要目标[41]。与对细菌的捕食研究相比, 黏细菌对真菌的捕食研究较少, 仅涉及具有抗真菌活性的几丁质酶、β-1,3-葡聚糖酶等细胞壁裂解酶[42-44]以及抗真菌活性的次级代谢物[45]等。在细菌与真菌互作关系研究中, 伯克霍尔德菌和沙雷氏菌分别进化出三型分泌系统(T3SS)和六型分泌系统(T6SS), 将毒性蛋白直接注入真菌细胞内实现对真菌的猎杀[46-47]。与之不同的是, 黏细菌通过分泌一种新型外膜型β-1,6-葡聚糖酶分解真菌细胞壁中的β-1,6-葡聚糖组分, 进而实现对植物病原真菌的捕食(图 1)[48]。目前具有酶活性的外膜蛋白主要包括膜结合蛋白酶或酯酶等[49-50], 黏细菌来源的外膜型β-1,6-葡聚糖酶是目前已报道的唯一具有糖苷水解酶活性的外膜蛋白, 具有广谱的抗真菌活性, 是黏细菌捕食真菌的关键因子[48]。
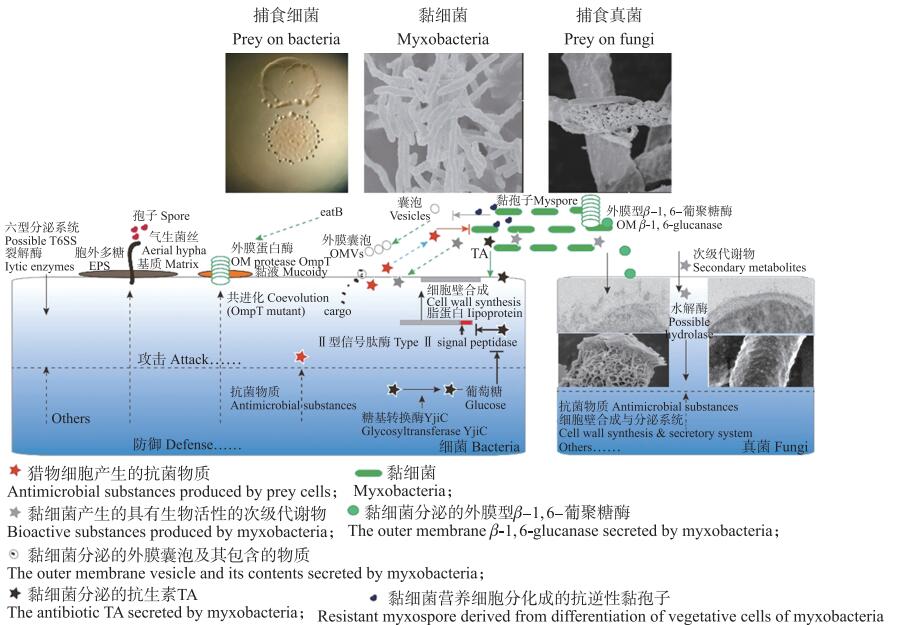
|
图 1 黏细菌与猎物细胞之间的捕食与防御策略 Fig. 1 The attack-defense model between predatory myxobacteria and prey |
捕食性黏细菌通过多种模式攻击猎物以获取营养并建立竞争优势, 而猎物群体面对捕食者的攻击进化出相应的防御策略抵御微生物的捕食, 从而实现种群的自我保护(图 1)。猎物抵御黏细菌捕食的防御策略类型主要分为: 1)产生抑菌物质。黏细菌可以分泌抗生素杀死猎物, 而猎物也可以分泌抗菌物质抑制黏细菌的生长。在黄色黏球菌(M. xanthus)DK1622与天蓝色链霉菌(Streptomyces coelicolor)M45的互作研究中, 链霉菌M45通过气生菌丝和抗菌物质抵御黏细菌的捕食[18], 在黏细菌与芽胞杆菌的互作研究中也有类似的抑制作用报道[51]。此外, 真菌也能够产生具有生物活性的次级代谢物, 如青霉菌产生的抗生素能够抑制炭疽杆菌的生长[52], 具有抑制黏细菌生长的可能。2)形成外围屏障。枯草芽胞杆菌(Bacillus subtilis)NCIB3610和大肠杆菌与黄色黏球菌(M. xanthus)DK1622的互作研究中, 芽胞杆菌和大肠杆菌作为猎物类群通过产生胞外基质和生物膜抵御黏细菌的入侵[53-54]; 根瘤菌利用分泌的胞外半乳葡聚糖保护细胞免受黏细菌的攻击[31]。3)修饰捕食因子结构。黏细菌捕食细菌的一个重要武器是抗生素myxovirescin(TA), 地衣芽胞杆菌通过分泌糖基转移酶(YjiC)对黏细菌分泌的抗生素TA进行葡萄糖糖基化修饰, 减弱其对自身细胞的毒性, 进而逃脱黏细菌的捕食[55](图 1)。4)共进化。地衣芽胞杆菌在黏细菌的捕食压力下进化出对抗生素TA的修饰能力[55]; 在黏细菌与大肠杆菌共进化研究中发现, 大肠杆菌通过增加黏液量降低黏细菌的运动速度, 同时通过突变自身的毒力蛋白-外膜蛋白酶(OmpT)以适应捕食的压力, 而黏细菌则通过突变1个未知的eatB基因从而增强对细菌捕食的适应性[56]。5)改变细胞壁结构组成。黏细菌通过分泌抗菌蛋白β-1,6-葡聚糖酶实现对真菌的猎杀, 然而粗糙脉孢菌等微生物细胞壁不含β-1,6-葡聚糖, 进而避免了黏细菌的捕食[48, 57]。6)其他类型。改变猎物细胞表面成分与细胞形态(如丝状细胞)和增加游动速度等也能够保护猎物逃避捕食[58]。
3 黏细菌捕食行为在土壤菌群生态调控中的功能微生物群落中的捕食涉及原生生物、噬菌体以及具有捕食能力的细菌和真菌等[59]。黏细菌作为微生物食物网结构中的捕食者, 在土壤微生物食物网中代谢活跃, 在土壤生态系统碳循环中起着关键作用[60]。土壤中黏细菌占总细菌群落的比例为0.4%~4.5%, 几乎包含了所有黏细菌科或属, 因此, 黏细菌被认为是土壤细菌群落的重要组成部分[61]。此外, 黏细菌在农田土壤环境中与捕食性细菌存在显著的正相关性, 推测其在农田土壤细菌群落调控方面具有重要的作用[62]。
尽管黏细菌广泛分布, 并且在微生物生态调控中发挥着重要的作用, 但对具体的影响或控制机制的研究并不统一。研究者在开展土壤微生物与植物互作关系研究中发现, 黏细菌Corallococcus sp. EGB能够对植物根际分泌物中麦芽糖和麦芽糖醇具有较强的趋化作用, 使黏细菌向根部定向迁移并定殖。由于黏细菌EGB对包括尖孢镰刀菌在内的多种植物病原真菌和细菌均表现良好的捕食作用[25, 48], 在向根部迁移的过程中, 黏细菌通过捕食作用调控土壤微生物群落结构。其中, Bacillus和Pseudomonas等潜在的病害生防菌以及植物促生菌(PGPR)等丰度上升, 尖孢镰刀菌黄瓜专化型(F. oxysporum f. sp. cucumerinum, FOC)数量明显下降, 从而抑制病害的发生[63]。研究结果为利用捕食性黏细菌调控土壤微生物菌落进而实现植物病害的控制提供了新思路, 同时也暗示着捕食性微生物作为土壤食物网的重要组成部分[64], 在微生物生态系统动态过程调控中起着重要的作用。
4 捕食性黏细菌在植物病害控制方面的应用潜力研究发现来源于不同种属的黏细菌菌株对不同类型的植物病原菌均表现良好的抗菌活性[38, 65-66], 在植物病害生物防治方面表现出潜在的应用价值。盆栽试验中, 黏细菌对病原细菌、真菌和卵菌等造成的植物病害具有良好的生防效果, 表现出较好的土壤定殖能力, 从而保护植物免受病原菌的危害[25, 63, 67-68]。此外, 在水果采后病害控制方面, 黏细菌Corallococcus sp. EGB产生的多种挥发性抗真菌次级代谢物(VOC), 能够有效抑制青霉菌对橘子的侵染[69], 延长水果采后货架期(图 2)。基于黏细菌潜在的生防效果, 研究者进一步开展了田间试验。黏细菌Sorangium cellulosum KYC 3262在辣椒炭疽病的防控试验中连续3年表现出稳定的生防效果, 防控效率与化学杀菌剂相当[70]。黏细菌Corallococcus sp. EGB在连续2年的黄瓜和香蕉枯萎病田间防控试验中也表现出良好的生防效果, 并优于化学药剂处理, 显著提高作物产量[63](图 2)。
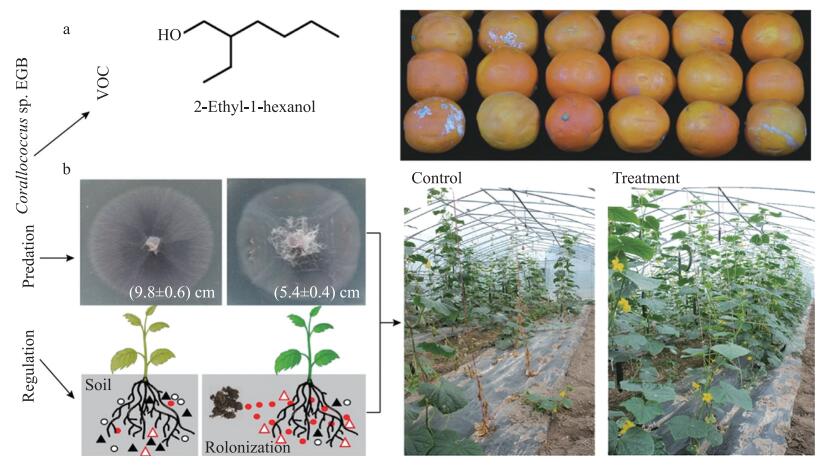
|
图 2 黏细菌Corallococcus sp. EGB在植物真菌病害控制方面的应用性评估 Fig. 2 Application evaluation of of Corallococcus sp. EGB in the biocontrol ofplant fungal disease a. 黏细菌利用挥发性抗菌物质(VOC)抑制气传性植物病原菌的生长[69](VOC: 抑制灰霉病菌对橘子的侵染); b. 黏细菌通过捕食和土壤微生物调控作用控制土传枯萎病害的发生[63](捕食与调控: 抑制黄瓜枯萎病菌对黄瓜的侵染)。 a. Biocidal effects of volatile organic compounds(VOC)produced by the myxobacteria against fungal phytopathogens[69]; b. Predation and microbial community regulation of myxobacteria are involved in the control of soil-borne Fusarium wilt[63]. |
与目前已报道的生防微生物相比, 黏细菌在植物病害控制方面具有显著的特点: 1)能在固体表面滑行运动。黏细菌利用2种不同类型的运动系统(A运动: Adventurous motility; S运动: Social motility)实现在固体界面的滑行运动[71], 降低微生物对土壤水分的要求[72], 而土壤水分含量是生防微生物在土壤中运动的限制因素之一[13]。2)产生丰富的抗菌物质。从黏细菌中已经发现了大量具有抗菌活性的次级代谢产物和酶类[43, 73-74], 是原核生物中仅次于放线菌的第二大次级代谢物来源菌。3)发育形成抗性黏孢子。黏细菌是革兰氏阴性细菌, 但可以分化或诱导分化形成具有抗逆性的孢子[75], 有利于生防菌剂的研发和保存。4)环境适应性强。黏细菌广泛分布于土壤、水体、腐败的树木枯枝落叶、草食类动物的粪便等不同的环境, 具有较强的环境适应力[76-77]。5)能通过多样化的策略捕食真菌和细菌[30, 78]。黏细菌通才型的捕食特性、良好的环境适应性、土壤微生物群落调控能力, 使其被视为是一类新型的生防微生物, 可应用于农业生产过程中植物病害的生防控制。
除了在植物病害生物防治方面, 黏细菌在动物病害控制方面也表现一定的应用潜力。研究人员发现包含不同种属的113株黏细菌对多种动物致病菌均具有高效的捕食作用, 包括肺炎克雷伯菌(Klebsiella pneumoniae)、奇异变形杆菌(Proteus mirabilis)、白色念珠菌(Candida albicans)、肠球菌(Enterococcus)、葡萄球菌(Staphylococcus)等[79]。此外, 黏细菌分泌的具有生物活性的次级代谢物在保护动物健康方面也具有应用潜力, 如S. cellulosum来源的Ambruticin等可以有效抑制多种动物致病菌的生长; 来源于Myxococcus stipitatus的Rhizopodin、S.cellulosum的Epothilone被认为在抗肿瘤方面具有重要的作用[35, 80]。黏细菌通过捕食作用和次级代谢物的抗菌作用使其在致病性微生物引起的动物病害控制方面也具有一定的应用潜力。然而, 目前黏细菌在病害防控中的作用还未引起足够的关注和重视。
5 捕食性黏细菌在植物病害控制方面所面临的问题目前, 捕食性黏细菌通过多种模式对植物病原菌进行捕食或者抑制, 在病害控制方面具有良好的应用潜力, 然而黏细菌捕食生物学研究中还存在一些瓶颈直接制约黏细菌的实际应用。例如: 1)黏细菌捕食行为的复杂性。目前对于黏细菌的捕食相关研究多数集中在行为特征的描述等方面, 已确定的黏细菌捕食因子只有抗生素TA和外膜型β-1,6-葡聚糖水解酶等, 而黏细菌在捕食过程中降解酶与代谢调控、互作关键因子、感知与猎物响应、细胞依赖、群体捕食效率等方面的机制未知, 限制了研究者对黏细菌捕食行为的深入了解。2)黏细菌生长与营养需求的特殊性。黏细菌特殊的生长发育方式导致黏细菌的分离周期长, 且可培养性黏细菌资源有限, 我国只有山东大学、广东省微生物研究所、内蒙古大学、河北大学等在黏细菌菌种资源收集方面建立了良好的基础。同时, 黏细菌生长聚集成团和丰富的胞外多糖等特性使野生型黏细菌的遗传操作难以建立, 具有优良特性的野生型黏细菌的作用机制研究存在瓶颈。此外, 黏细菌生长过程中的自溶特性也导致扩大培养黏细菌受限, 直接限制了黏细菌的实际应用和菌剂的规模化制备。3)研究材料的单一性。目前对于黏细菌的基础研究主要是以黄色黏球菌(M. xanthus)DK1622为材料, 然而黏细菌与植物、微生物共进化过程中, 不同种属之间的特性差异较大, 如黄色黏球菌DK1622分泌的次级代谢物具有良好的抗菌作用, 而黏细菌EGB主要是通过分泌真菌细胞壁裂解酶实现对真菌的抗性作用, 研究材料的单一性直接导致黏细菌捕食研究的进展较为缓慢。
6 研究展望目前, 黏细菌的基础研究主要是以黏细菌为模式生物, 开展发育生物学、种群识别、进化生物学以及生态学等研究, 包括黏细菌子实体的形成、运动性, 多糖的生物合成, 多形态细胞表面受体蛋白TraA(polymorphic cell surface receptor)及其互作蛋白TraB(cohort protein)依赖的外膜融合参与黏细菌细胞识别的作用以及生物多样性等[81-82]。此外, 以黏细菌为种质资源库, 分离筛选一系列具有生物活性的次级代谢物, 黏细菌已成为重要的生物活性物质来源菌[83]。黏细菌早在1941年就被报道具有捕食细菌的能力, 然而黏细菌是如何完成对细菌和真菌的捕食这一关键科学问题至今未知。因此, 解析黏细菌的捕食机制是未来黏细菌研究的重要方向。
黏细菌在自然环境中分布广泛, 具有较高的丰度, 然而已分离培养的黏细菌资源依然较少。广东省微生物研究所科研人员利用病原菌作为被捕食菌构建了直接面向生物防治用途的黏细菌筛选模型[38], 为未来黏细菌分离方法的优化提供了方向。同时, 2010年启动的地球微生物组计划(Earth Microbiome Project), 也为获取不同黏细菌的基因组信息, 构建基因资源库提供了可能。此外, 野生型黏细菌的分子生物学研究体系对于黏细菌研究的深入开展至关重要。研究者前期发现黏细菌细胞分散性、胞外多糖(exopolysaccharide, EPS)、限制-修饰系统(restriction-modification system, R-M system)以及分泌系统等在黏细菌转化过程中起着重要的作用[84]。突破黏细菌胞外多糖的物理屏障, 强化黏细菌生长过程细胞分散性, 建立高效的遗传转化体系对于深入了解黏细菌的抗菌机制具有重要的作用。因此, 为了促进捕食性黏细菌在农业生产过程中的实际应用, 需要深入了解黏细菌捕食的作用机制和生态学功能, 通过基因组学和培养组学等方法获取具有良好抗菌活性的优良菌株; 利用代谢组学、蛋白和转录组学等鉴定参与黏细菌捕食行为的关键因子, 系统解析黏细菌的捕食和代谢调控机制; 结合微生物学、生态学、植物保护等多学科交叉阐明捕食性黏细菌在自然环境中的生态学功能以及与植物、土壤微生物菌群之间的互作关系; 同时, 建立和优化黏细菌规模化培养工艺, 为黏细菌的实际应用提供依据。
| [1] |
Savary S, Willocquet L, Pethybridge S J, et al. The global burden of pathogens and pests on major food crops[J]. Nature Ecology & Evolution, 2019, 3(3): 430-439. |
| [2] |
Savary S, Ficke A, Aubertot J N, et al. Crop losses due to diseases and their implications for global food production losses and food security[J]. Food Security, 2012, 4(4): 519-537. DOI:10.1007/s12571-012-0200-5 |
| [3] |
康振生. 我国植物真菌病害的研究现状及发展策略[J]. 植物保护, 2010, 36(3): 9-12. Kang Z S. Current status and development strategy for research on plant fungal diseases in China[J]. Plant Protection, 2010, 36(3): 9-12 (in Chinese with English abstract). DOI:10.3969/j.issn.0529-1542.2010.03.003 |
| [4] |
Casadevall A. Fungal diseases in the 21st century: the near and far horizons[J]. Pathogens & Immunity, 2018, 3(2): 183-196. |
| [5] |
Fisher M C, Henk D A, Briggs C J, et al. Emerging fungal threats to animal, plant and ecosystem health[J]. Nature, 2012, 484(7393): 186-194. DOI:10.1038/nature10947 |
| [6] |
Keinan A, Clark A G. Recent explosive human population growth has resulted in an excess of rare genetic variants[J]. Science, 2012, 336(6082): 740-743. DOI:10.1126/science.1217283 |
| [7] |
王桂荣, 王源超, 杨光富, 等. 农业病虫害绿色防控基础的前沿科学问题[J]. 中国科学基金, 2020, 34(4): 374-380. Wang G R, Wang Y C, Yang G F, et al. Frontiers in scientific issues of controlling agricultural pests and diseases by environmental-friendly methods[J]. Bulletin of National Natural Science Foundation of China, 2020, 34(4): 374-380 (in Chinese with English abstract). |
| [8] |
Chen T, Nomura K, Wang X L, et al. A plant genetic network for preventing dysbiosis in the phyllosphere[J]. Nature, 2020, 580(7805): 653-657. DOI:10.1038/s41586-020-2185-0 |
| [9] |
Wei Z, Gu Y A, Friman V P, et al. Initial soil microbiome composition and functioning predetermine future plant health[J]. Science Advances, 2019, 5(9): eaaw0759. DOI:10.1126/sciadv.aaw0759 |
| [10] |
Tringe S G. A layered defense against plant pathogens[J]. Science, 2019, 366(6465): 568-569. DOI:10.1126/science.aaz5619 |
| [11] |
Legein M, Smets W, Vandenheuvel D, et al. Modes of action of microbial biocontrol in the phyllosphere[J]. Frontiers in Microbiology, 2020, 11: 1619. DOI:10.3389/fmicb.2020.01619 |
| [12] |
Rahman S F S A, Singh E, Pieterse C M J, et al. Emerging microbial biocontrol strategies for plant pathogens[J]. Plant Science, 2018, 267: 102-111. DOI:10.1016/j.plantsci.2017.11.012 |
| [13] |
Babalola O O. Beneficial bacteria of agricultural importance[J]. Biotechnology Letters, 2010, 32(11): 1559-1570. DOI:10.1007/s10529-010-0347-0 |
| [14] |
Trdá L, Boutrot F, Claverie J, et al. Perception of pathogenic or beneficial bacteria and their evasion of host immunity: pattern recognition receptors in the frontline[J]. Frontiers in Plant Science, 2015, 6: 219. |
| [15] |
Zhalnina K, Louie K B, Hao Z, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly[J]. Nature Microbiology, 2018, 3(4): 470-480. DOI:10.1038/s41564-018-0129-3 |
| [16] |
Erken M, Lutz C, McDougald D. The rise of pathogens: predation as a factor driving the evolution of human pathogens in the environment[J]. Microbial Ecology, 2013, 65(4): 860-868. DOI:10.1007/s00248-013-0189-0 |
| [17] |
Guerrero R, Pedros-Alio C, Esteve I, et al. Predatory prokaryotes: predation and primary consumption evolved in bacteria[J]. Proc Natl Acad Sci USA, 1986, 83(7): 2138-2142. DOI:10.1073/pnas.83.7.2138 |
| [18] |
Pérez J, Moraleda-Muñoz A, Marcos-Torres F J, et al. Bacterial predation: 75 years and counting![J]. Environmental Microbiology, 2016, 18(3): 766-779. DOI:10.1111/1462-2920.13171 |
| [19] |
陈康勇, 钟为铭, 高志鹏. 蛭弧菌在水产养殖中应用研究进展[J]. 水产科学, 2018, 37(2): 283-288. Chen K Y, Zhong W M, Gao Z P. Research progress on utilization of Bdellovibrio in aquaculture[J]. Fisheries Science, 2018, 37(2): 283-288 (in Chinese with English abstract). |
| [20] |
Wang Z, Kadouri D E, Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13[J]. BMC Genomics, 2011, 12: 453. DOI:10.1186/1471-2164-12-453 |
| [21] |
李曙光. 黏细菌的环境分布、季节演替及其相互作用[D]. 济南: 山东大学, 2014. Li S G. Distribution, seasonal succession and intraspecies interactions of myxobacteria[D]. Jinan: Shandong University, 2014(in Chinese with English abstract). |
| [22] |
王春玲, 冯广达, 姚青, 等. 黏细菌基因组学研究进展[J]. 微生物学通报, 2019, 46(9): 2394-2403. Wang C L, Feng G D, Yao Q, et al. Research progress in genomics of myxobacteria[J]. Microbiology China, 2019, 46(9): 2394-2403 (in Chinese with English abstract). |
| [23] |
Jurkevitch E. Predatory behaviors in bacteria: diversity and transitions[J]. Microbe Magazine, 2007, 2(2): 67-73. DOI:10.1128/microbe.2.67.1 |
| [24] |
Barron G L. Predatory fungi, wood decay, and the carbon cycle[J]. Biodiversity, 2003, 4(1): 3-9. DOI:10.1080/14888386.2003.9712621 |
| [25] |
Li Z K, Ye X F, Chen P L, et al. Antifungal potential of Corallococcus sp. strain EGB against plant pathogenic fungi[J]. Biological Control, 2017, 110: 10-17. DOI:10.1016/j.biocontrol.2017.04.001 |
| [26] |
Li Z K, Wang T, Luo X, et al. Biocontrol potential of Myxococcus sp. strain BS against bacterial soft rot of Calla lily caused by Pectobacterium carotovorum[J]. Biological Control, 2018, 126: 36-44. DOI:10.1016/j.biocontrol.2018.07.004 |
| [27] |
Davidov Y, Huchon D, Koval S F, et al. A new alpha-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory[J]. Environmental Microbiology, 2006, 8(12): 2179-2188. DOI:10.1111/j.1462-2920.2006.01101.x |
| [28] |
Zhang Y, Guzzo M, Ducret A, et al. A dynamic response regulator protein modulates G-protein-dependent polarity in the bacterium Myxococcus xanthus[J]. PLoS Genetics, 2012, 8(8): e1002872. DOI:10.1371/journal.pgen.1002872 |
| [29] |
Beebe J M. Studies on the myxobacteria: Ⅰ, distribution in Iowa soils and description of a new species; Ⅱ, Myxobacteria as bacterial parasites; Ⅲ, the morphology and cytology of Myxococcus xanthus[D]. Iowa: Iowa State University, 1941.
|
| [30] |
Berleman J E, Kirby J R. Deciphering the hunting strategy of a bacterial wolfpack[J]. FEMS Microbiology Reviews, 2009, 33(5): 942-957. DOI:10.1111/j.1574-6976.2009.00185.x |
| [31] |
Pérez J, Jiménez-Zurdo J I, Martínez-Abarca F, et al. Rhizobial galactoglucan determines the predatory pattern of Myxococcus xanthus and protects Sinorhizobium meliloti from predation[J]. Environmental Microbiology, 2014, 16(7): 2341-2350. DOI:10.1111/1462-2920.12477 |
| [32] |
Zhang W C, Wang Y, Lu H N, et al. Dynamics of solitary predation by Myxococcus xanthus on Escherichia coli observed at the single-cell level[J]. Applied and Environmental Microbiology, 2019, 86(3): e02286-19. DOI:10.1128/AEM.02286-19 |
| [33] |
McBride M J, Zusman D R. Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli[J]. FEMS Microbiology Letters, 1996, 137(2/3): 227-231. |
| [34] |
Shilo M. Lysis of blue-green algae by myxobacter[J]. Journal of Bacteriology, 1970, 104(1): 453-461. DOI:10.1128/JB.104.1.453-461.1970 |
| [35] |
Kaur R, Kumari A, Kaur R, et al. Myxobacteria: producers of enormous bioactive secondary metabolites[J]. International Journal of Research in Pharmaceutical Sciences, 2018, 9(1): 309-313. |
| [36] |
Evans A G L, Davey H M, Cookson A, et al. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo[J]. Microbiology, 2012, 158(11): 2742-2752. DOI:10.1099/mic.0.060343-0 |
| [37] |
Xiao Y, Wei X M, Ebright R, et al. Antibiotic production by myxobacteria plays a role in predation[J]. Journal of Bacteriology, 2011, 193(18): 4626-4633. DOI:10.1128/JB.05052-11 |
| [38] |
代京莎, 李安章, 朱红惠. 黏细菌在植物病害生物防治中的作用[J]. 生物技术进展, 2016, 6(4): 229-234. Dai J S, Li A Z, Zhu H H. The function of myxobacteria in biological control of plant disease[J]. Current Biotechnology, 2016, 6(4): 229-234 (in Chinese with English abstract). DOI:10.3969/j.issn.2095-2341.2016.04.01 |
| [39] |
Keane R, Berleman J. The predatory life cycle of Myxococcus xanthus[J]. Microbiology, 2016, 162(1): 1-11. DOI:10.1099/mic.0.000208 |
| [40] |
Ensign J, Wolfe R. Characterization of a small proteolytic enzyme which lyses bacterial cell walls[J]. Journal of bacteriology, 1966, 91(2): 524-534. DOI:10.1128/JB.91.2.524-534.1966 |
| [41] |
Livingstone P G, Millard A D, Swain M T, et al. Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding[J]. Microbial Genomics, 2018, 4(2): e000152. |
| [42] |
Hocking D, Cook F D. Myxobacteria exert partial control of damping-off and root disease in container-grown tree seedlings[J]. Canadian Journal of Microbiology, 1972, 18(10): 1557-1560. DOI:10.1139/m72-237 |
| [43] |
Li Z K, Xia C Y, Wang Y X, et al. Identification of an endo-chitinase from Corallococcus sp. EGB and evaluation of its antifungal properties[J]. International Journal of Biological Macromolecules, 2019, 132: 1235-1243. DOI:10.1016/j.ijbiomac.2019.04.056 |
| [44] |
Zhou J, Chen J H, Li Z K, et al. Enzymatic properties of a multi-specific β-(1,3)-glucanase from Corallococcus sp. EGB and its potential antifungal applications[J]. Protein Expression and Purification, 2019, 164: 105481. DOI:10.1016/j.pep.2019.105481 |
| [45] |
Kunze B, Steinmetz H, Höfle G, et al. Cruentaren, a new antifungal salicylate-type macrolide from Byssovorax cruenta (Myxobacteria) with inhibitory effect on mitochondrial ATPase activity[J]. The Journal of Antibiotics, 2006, 59(10): 664-668. DOI:10.1038/ja.2006.89 |
| [46] |
Swain D M, Yadav S K, Tyagi I, et al. A prophage tail-like protein is deployed by Burkholderia bacteria to feed on fungi[J]. Nature Communications, 2017, 8(1): 1-9. DOI:10.1038/s41467-016-0009-6 |
| [47] |
Trunk K, Peltier J, Liu Y C, et al. The type Ⅵ secretion system deploys antifungal effectors against microbial competitors[J]. Nature Microbiology, 2018, 3(8): 920-931. DOI:10.1038/s41564-018-0191-x |
| [48] |
Li Z K, Ye X F, Liu M X, et al. A novel outer membrane β-1,6-glucanase is deployed in the predation of fungi by myxobacteria[J]. The ISME Journal, 2019, 13(9): 2223-2235. DOI:10.1038/s41396-019-0424-x |
| [49] |
Rutten L, Mannie J P B A, Stead C M, et al. Active-site architecture and catalytic mechanism of the lipid A deacylase LpxR of Salmonella typhimurium[J]. Proc Natl Acad Sci USA, 2009, 106(6): 1960-196. DOI:10.1073/pnas.0813064106 |
| [50] |
Fairman J W, Noinaj N, Buchanan S K. The structural biology of β-barrel membrane proteins: a summary of recent reports[J]. Current Opinion in Structural Biology, 2011, 21(4): 523-531. DOI:10.1016/j.sbi.2011.05.005 |
| [51] |
Müller S, Strack S N, Hoefler B C, et al. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus[J]. Applied and Environmental Microbiology, 2014, 80(18): 5603-5610. DOI:10.1128/AEM.01621-14 |
| [52] |
Bills G F, Gloer J B. Biologically active secondary metabolites from the fungi[J]. Microbiology Spectrum, 2016, 4(6): 1-32. |
| [53] |
Müller S, Strack S N, Ryan S E, et al. Predation by Myxococcus xanthus induces Bacillus subtilis to form spore-filled megastructures[J]. Applied and Environmental Microbiology, 2015, 81(1): 203-210. DOI:10.1128/AEM.02448-14 |
| [54] |
Depas W H, Syed A K, Sifuentes M, et al. Biofilm formation protects Escherichia coli against killing by Caenorhabditis elegans and Myxococcus xanthus[J]. Applied and Environmental Microbiology, 2014, 80(22): 7079-7087. DOI:10.1128/AEM.02464-14 |
| [55] |
Wang C, Liu X, Zhang P, et al. Bacillus licheniformis escapes from Myxococcus xanthus predation by deactivating myxovirescin A through enzymatic glucosylation[J]. Environmental Microbiology, 2019, 21(12): 4755-4772. DOI:10.1111/1462-2920.14817 |
| [56] |
Nair R R, Vasse M, Wielgoss S, et al. Bacterial predator-prey coevolution accelerates genome evolution and selects on virulence-associated prey defences[J]. Nature Communications, 2019, 10(1): 1-10. DOI:10.1038/s41467-018-07882-8 |
| [57] |
Maddi A, Dettman A, Fu C, et al. WSC-1 and HAM-7 are MAK-1 MAP kinase pathway sensors required for cell wall integrity and hyphal fusion in Neurospora crassa[J]. PLoS One, 2012, 7(8): e42374. DOI:10.1371/journal.pone.0042374 |
| [58] |
Jousset A. Ecological and evolutive implications of bacterial defences against predators[J]. Environmental Microbiology, 2012, 14(8): 1830-1843. DOI:10.1111/j.1462-2920.2011.02627.x |
| [59] |
Thakur M P, Geisen S. Trophic regulations of the soil microbiome[J]. Trends in Microbiology, 2019, 27(9): 771-780. DOI:10.1016/j.tim.2019.04.008 |
| [60] |
Lueders T, Kindler R, Miltner A, et al. Identification of bacterial micropredators distinctively active in a soil microbial food web[J]. Applied and Environmental Microbiology, 2006, 72(8): 5342-5348. DOI:10.1128/AEM.00400-06 |
| [61] |
Zhou X W, Li S G, Li W, et al. Myxobacterial community is a predominant and highly diverse bacterial group in soil niches[J]. Environmental Microbiology Reports, 2014, 6(1): 45-56. DOI:10.1111/1758-2229.12107 |
| [62] |
Wang W H, Luo X, Ye X F, et al. Predatory Myxococcales are widely distributed in and closely correlated with the bacterial community structure of agricultural land[J]. Applied Soil Ecology, 2020, 146: 103365. DOI:10.1016/j.apsoil.2019.103365 |
| [63] |
Ye X F, Li Z K, Luo X, et al. A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community[J]. Microbiome, 2020, 8(1): 49. DOI:10.1186/s40168-020-00824-x |
| [64] |
Mendes-Soares H, Velicer G J. Decomposing predation: testing for parameters that correlate with predatory performance by a social bacterium[J]. Microbial Ecology, 2013, 65(2): 415-423. DOI:10.1007/s00248-012-0135-6 |
| [65] |
任兴波, 张子良, 赵璞钰, 等. 马铃薯晚疫病菌拮抗黏细菌YR-35的分离鉴定及其代谢产物稳定性[J]. 中国生物防治学报, 2016, 32(3): 379-387. Ren X B, Zhang Z L, Zhao P Y, et al. Isolation and identification of the strain YR-35 resistant to phytophthora infestans and its metabolites[J]. Chinese Journal of Biological Control, 2016, 32(3): 379-387 (in Chinese with English abstract). |
| [66] |
李百元, 谢小林, 张鲜娇, 等. 不同被捕食细菌对新疆盐碱地黏细菌分离的影响[J]. 微生物学报, 2013, 53(4): 379-389. Li B Y, Xie X L, Zhang X J, et al. Influence of different prey strains on isolation of myxobacteria in saline-alkaline soils of Xinjiang[J]. Acta Microbiologica Sinica, 2013, 53(4): 379-389 (in Chinese with English abstract). |
| [67] |
Kim S T, Yun S C. Biocontrol with Myxococcus sp. KYC 1126 against anthracnose in hot pepper[J]. The Plant Pathology Journal, 2011, 27(2): 156-163. DOI:10.5423/PPJ.2011.27.2.156 |
| [68] |
Dahm M, Brzezińska A J, Wrótniak-Drzewiecka W, et al. Myxobacteria as a potential biocontrol agent effective against pathogenic fungi of economically important forest trees[J]. Dendrobiology, 2015, 74: 13-24. DOI:10.12657/denbio.074.002 |
| [69] |
Ye X F, Chen Y, Ma S Y, et al. Biocidal effects of volatile organic compounds produced by the myxobacterium Corrallococcus sp. EGB against fungal phytopathogens[J]. Food Microbiology, 2020, 91: 103502. DOI:10.1016/j.fm.2020.103502 |
| [70] |
Yun S C. Selection and a 3-year field trial of Sorangium cellulosum KYC 3262 against anthracnose in hot pepper[J]. The Plant Pathology Journal, 2014, 30(3): 279-287. DOI:10.5423/PPJ.OA.01.2014.0002 |
| [71] |
Nan B, Zusman D R. Uncovering the mystery of gliding motility in the myxobacteria[J]. Annual Review of Genetics, 2011, 45: 21-39. DOI:10.1146/annurev-genet-110410-132547 |
| [72] |
Spormann A M. Gliding motility in bacteria: insights from studies of Myxococcus xanthus[J]. Microbiology and Molecular Biology Reviews, 1999, 63(3): 621-641. DOI:10.1128/MMBR.63.3.621-641.1999 |
| [73] |
刘新利, 李越中. 黏细菌次级代谢产物及其在农业上的应用价值[J]. 中国农业科技导报, 2007, 9(3): 44-51. Liu X L, Li Y Z. Myxobacterial secondary metabolites and their potential applications in agriculture[J]. Journal of Agricultural Science and Technology, 2007, 9(3): 44-51 (in Chinese with English abstract). DOI:10.3969/j.issn.1008-0864.2007.03.009 |
| [74] |
Kaur R, Singh S, Kaur R, et al. Myxococcus xanthus: a source of antimicrobials and natural bio-control agent[J]. The Pharma Innovation Journal, 2017, 6(11): 260-262. |
| [75] |
Dworkin M. Recent advances in the social and developmental biology of the myxobacteria[J]. Microbiology Review, 1996, 60(1): 70-102. DOI:10.1128/MR.60.1.70-102.1996 |
| [76] |
Dawid W. Biology and global distribution of myxobacteria in soils[J]. FEMS Microbiology Reviews, 2000, 24(4): 403-427. DOI:10.1111/j.1574-6976.2000.tb00548.x |
| [77] |
李曙光, 周秀文, 吴志红, 等. 黏细菌的种群生态及其生存策略[J]. 微生物学通报, 2013, 40(1): 172-179. Li S G, Zhou X W, Wu Z H, et al. Population ecology and survival strategy of myxobacteria[J]. Microbiology China, 2013, 40(1): 172-179 (in Chinese with English abstract). |
| [78] |
Muñoz-Dorado J, Marcos-Torres F, Garcia-Bravo E, et al. Myxobacteria: moving, killing, feeding, and surviving together[J]. Frontiers in Microbiology, 2016, 781. |
| [79] |
Livingstone P G, Morphew R M, Whitworth D E. Myxobacteria are able to prey broadly upon clinically-relevant pathogens, exhibiting a prey range which cannot be explained by phylogeny[J]. Frontiers in Microbiology, 2017, 8: 1593. DOI:10.3389/fmicb.2017.01593 |
| [80] |
刘新利, 李越中. 黏细菌资源与埃博霉素研发[J]. 生物产业技术, 2011(2): 26-32. Liu X L, Li Y Z. Myxobacteria resources and development of epothilone[J]. Biotechnology & Business, 2011(2): 26-32 (in Chinese). |
| [81] |
Cao P, Wall D. Direct visualization of a molecular handshake that governs kin recognition and tissue formation in myxobacteria[J]. Nature Communications, 2019, 10(1): 3073. DOI:10.1038/s41467-019-11108-w |
| [82] |
Yu Y T N, Yuan X, Velicer G J. Adaptive evolution of an sRNA that controls Myxococcus development[J]. Science, 2010, 328(5981): 993. DOI:10.1126/science.1187200 |
| [83] |
Reichenbach H. Myxobacteria, producers of novel bioactive substances[J]. Journal of Industrial Microbiology and Biotechnology, 2001, 27(3): 149-156. DOI:10.1038/sj.jim.7000025 |
| [84] |
Wang J, Hu W, Lux R, et al. Natural transformation of Myxococcus xanthus[J]. Journal of Bacteriology, 2011, 193(9): 2122-2132. DOI:10.1128/JB.00041-11 |




