文章信息
- 张红生, 程金平, 王健康, 詹成芳
- ZHANG Hongsheng, CHENG Jinping, WANG Jiankang, ZHAN Chengfang
- 水稻种子活力相关基因鉴定及分子调控机制
- Advances in identification of genes related to seed vigor and molecular mechanism in regulation in rice
- 南京农业大学学报, 2019, 42(2): 191-200
- Journal of Nanjing Agricultural University, 2019, 42(2): 191-200.
- http://dx.doi.org/10.7685/jnau.201807015
-
文章历史
- 收稿日期: 2018-07-05
种子活力是由多基因控制的、复杂的数量性状, 种子活力高低与种子形成发育和成熟、种子休眠和萌发、种子贮藏和劣变、幼苗建成及耐逆性等多个过程相关[1-2]。国际种子检验协会(ISTA)将种子活力定义为:种子活力是决定种子或种子批在发芽和出苗期间的活性水平和行为等种子特性的综合表现; 高活力种子在田间表现发芽快、出苗整齐, 对不良环境因子抵抗力强, 幼苗表现健壮[2]。种子活力对播种后幼苗出苗速率和整齐度、幼苗生长和作物产量与品质影响甚大, 所以种子活力一直是种子学的重要研究内容[2-5]。水稻是全球最重要的粮食作物, 近年来, 随着分子生物学技术手段的改进, 尤其是高通量测序技术与各种组学技术的发展, 在水稻种子活力形成、控制种子活力关键基因的鉴定和克隆、高活力种子形成的分子机制与调控等方面取得了很大进展[4-7]。本文对水稻种子活力鉴定方法与评价指标、种子活力形成关键基因鉴定和基因克隆、种子活力形成与调控分子机制等研究进展进行综述, 为进一步利用生物技术手段, 尤其是分子标记辅助选择(marker-assisted selection, MAS)技术改良水稻品种的种子活力, 培育适合我国水稻轻简栽培的新品种。
1 种子活力鉴定方法与评价指标种子活力形成于种子发育的脱水阶段, 终止于种子萌发后的幼苗形成。种子发育是指从受精开始, 形成完整的胚, 之后不断积累贮藏物质, 再历经干燥脱水、休眠, 直到种子完全成熟, 经历早期胚形态发生和后期成熟2个阶段[2, 5, 8]; 经过一定时间休眠后, 在适宜环境条件下, 具有活力的种子快速吸水, 恢复代谢, 生物大分子和细胞器活化修复, 合成大量的mRNA, 调节GA、ABA及IAA等激素含量, 刺激细胞分裂, 胚根尖端突破种皮外伸, 胚芽开始伸长, 当胚根、胚芽伸出种皮并发育到一定程度, 形成具有根、茎、叶的完整幼苗, 完成种子萌发(图 1-A)[8-12]。种子发育与萌发过程中, 伴随着新mRNA和蛋白质合成, 积累淀粉、脂肪和蛋白质等贮藏物质, 保证种子萌发时有充足的能源物质[5, 8, 12](图 1-B)。在田间条件下, 种子萌发常伴随着干旱、低温和盐害等非生物胁迫, 种子活力受到严重影响, 难以形成正常的幼苗, 造成产量下降; 或者进入二次休眠状态以应对恶劣的环境[5, 8](图 1-C)。
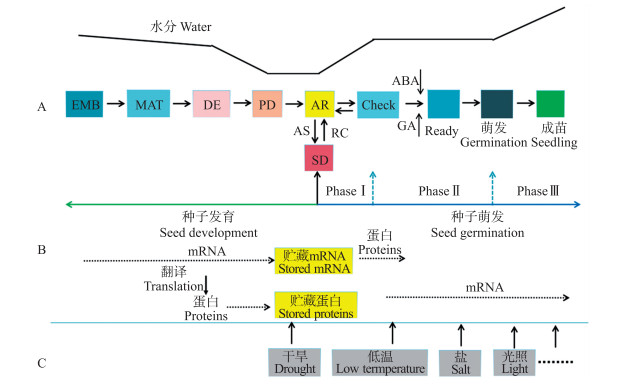
|
图 1 种子活力形成过程中的生化变化及其影响因素[12]
Fig. 1 The biochemical changes during seed vigor formation and influencing factors[12]
A.种子发育、休眠、萌发、成苗4个阶段的简化模型; B.种子发育与萌发过程中mRNA和蛋白质等转录和翻译; C.干旱、低温、盐害及光等影响田间种子活力的非生物胁迫因子。 EMB与MAT分别是指种子胚胎形成与成熟过程; DE与PD分别指干燥脱水与休眠后期种子成熟过程; AR、Check与Ready分别指无休眠具有活力的种子、种子快速吸水后转折点及其萌发准备阶段; AS、SD与RC分别指非生物逆境胁迫、二次休眠与破休眠种子恢复过程; ABA与GA分别为脱落酸与赤霉素。 A.The simplified model of seed development, dormancy, germination and seedling stages; B.The transcription and translation of mRNA and protein during seed development and germination; C.The effect of abiotic stresses on seed vigor in fields, such as drought, low temperature, salt and light. EMB:Embryogenesis; MAT:Maturation; DE:Desiccation; PD:Primary dormancy; AR:("after ripened" seeds) non ̄dormant seeds; Check: Control checkpoint involving ABA and a possible reinduction of the maturation program; Ready:Final step before radicle emergence; AS:Abiotic stress; SD:Secondary dormancy; RC:Recover conditions; ABA:Abscisic acid; GA:Gibberellins. |
水稻种子活力的鉴定通常在种子萌发期和苗期进行。在种子萌发期的鉴定方法有滤纸培养法和模拟田间直播法。滤纸培养法是将一定数量的饱满种子(50或100粒)等间距均匀点播于带有滤纸的培养皿(直径为9cm)中, 加入一定量蒸馏水, 放置在恒温或变温光照培养箱中培育, 定时观察统计水稻种子发芽或成苗状态[13-17]。模拟田间直播法是将水稻干种子直接播于不同深度的湿润土壤中, 在模拟田间或网室可控环境条件下, 观察统计种子萌发、幼苗出土情况[18-19]。该方法的优点是接近大田实际环境, 可以同时观察种子萌发过程中的顶土能力。在水稻幼苗期, 常用的种子活力鉴定方法有水培和土培2种方法。水培鉴定方法是利用96孔的播种板(盘)将种子悬于含有营养液的容器中, 在室温下将其培养至2叶1心时鉴定幼苗生长状态[20-23], 土培鉴定方法是用田间土壤替代营养液[24]。非生物胁迫下水稻种子活力鉴定方法取决于胁迫条件, 如低温胁迫处理, 可以将种子放置于低温环境下培养[25-26], 盐害和模拟干旱胁迫(PEG胁迫)则将营养液替换成相应浓度盐溶液[13, 16, 20, 23]或PEG溶液[20]。
种子活力鉴定方法不同, 衡量水稻种子活力的指标也有多种。目前, 水稻种子活力评价指标主要有种子萌发相关指标、幼苗形成相关指标、种子耐逆性相关指标和种子生理生化相关指标等4类。
1) 种子萌发相关指标, 包括种子萌发率(germination rate, GR/germination percentage, GP)、发芽指数(germination index, GI)、活力指数(vigor index, VI)、成苗率(seedling percentage, SP)、吸胀率(imbibition rate)、胚芽鞘长(coleoptile length, CL)和胚根长(radicle length, RL)等[13-17, 20, 25-27]。
2) 幼苗形成相关指标, 包括芽长(shoot length, SL)、根长(root length, RL)、中胚轴长(mesocotyl length, ML)、芽干质量(shoot dry weight, SDW)、根干质量root dry weight, RDW)、根/芽干质量比(SDW/RDW)、相对苗长(relative shoot length, R-SL)、相对苗干质量(relative seedling dry weight, R-SDW)等[18-19, 21-22, 28-30]。
3) 种子耐逆性相关指标, 包括幼苗成活率(seedling survival percentage, SSP)、幼苗冷害等级(cold- tolerance ranking, CTR)和盐害等级(salt tolerance ranking, STR)等[20, 23, 31]。
4) 种子生理生化相关指标, 包括超氧化物歧化酶(superoxide dismutase, SOD)、过氧化氢酶(catalase, CAT)、谷胱甘肽过氧化物酶(glutathione reductase, GR)活性, 过氧化氢(H2O2)、丙二醛(malondialdehyde, MDA)含量[27], 电导率(electrical conductivity, EC)以及物质动员量(weight of mobilized seed reserve, WMSR)、物质利用率(seed reserve utilization efficiency, SRUE)等[29]。
这4类种子活力相关性指标从不同层面上反映水稻种子活力, 相互之间存在显著的关联性[17, 20-21]。Cheng等[20]以GP与GI为活力指标, 分析发现干旱与盐胁迫下GP (3和10 d)与GI (10 d)显著正相关, 盐胁迫下的GP (3 d)与正常条件下的GP (3 d)与GI (10 d)显著正相关。Xu等[21]对SDW、RDW与MRL (maximum root length)等种子活力指标研究发现, 在幼苗30和40 d时, SDW与RDW均呈显著正相关性, RDW与MRL间也显著正相关。Xie等[17]利用水稻重组自交系群体(RIL)对正常和低温条件种子活力检测, 发现在1、5、8和11号染色体上的5个QTL位点可以用RL、RL等多个种子活力指标共同检测到, 表明用不同指标检测种子活力的结果可以相互比较与验证。
2 水稻种子活力相关QTL鉴定和基因克隆 2.1 基于分子标记技术的QTL鉴定和基因克隆利用重组自交系(recombinant inbred line, RIL)、回交自交系(backcrossinbred line, BIL)、染色体片段置换系(chromosome segment substitution line, CSSL)和近等基因系(nearly isogenic line, NIL)等不同遗传群体, 结合分子标记技术至少鉴定了1600多个与种子萌发、休眠、幼苗形成和种子耐逆性等种子活力相关的数量性状位点(quantitative trait loci, QTL)(http://archive.gramene.org.)。Wang等[14]以粳稻地方品种‘大关稻’与籼稻‘IR28’杂交构建的RIL群体, 以水培处理3、10d种子的GP和GI为指标, 检测到10个种子活力相关QTL位点。Xu等[21]利用珍汕97/明恢63杂交构建的RIL群体, 采用SDW、RDW、RL和SDW/RDW等指标, 在水培后30和40d时, 分别检测到16个和20个幼苗形成相关的QTL位点。Wang等[32]利用‘韭菜青’与‘籼稻IR26’构建的RIL群体, 对早、中、晚3个不同收获时期的水稻种子进行休眠性检测, 鉴定到8个加性QTL位点, 其中2对是上位性连锁位点。迄今, 至少有11个种子活力相关QTL被精细定位(表 1)。Li等[15]利用‘珍汕97’和‘日本晴’构建的CSSL群体, 以种子萌发2d的GR为指标, 在第2、5、6和10号染色体上分别检测到1个种子萌发相关的QTL位点, 并对第2染色体上的主效位点qGR2进行了精细定位。Xie等[17]利用珍汕97/明恢63杂交构成的RIL群体, 以胚芽鞘长(CL)和胚根长(RL)为指标, 鉴定到8个种子活力相关位点, 并对2个主效QTL位点qSV- 1与qSV-5 c进行了精细定位。Li等[25]以粳稻材料‘USSR5’和籼稻品种‘N22’杂交构建的RIL群体为材料, 鉴定到qLTG- 7、qLTG-9和qLTG-12等3个控制种子低温萌发的QTL, 贡献率分别为9.50%、12.12%和7.08%, 进一步利用回交群体将贡献率最大的qLTG- 9精细定位于L9-25D与ID-1两个标记之间。Gu等[33]鉴定到1个源自野生稻的控制种子休眠的QTL位点qSD12, 将其精细定位于75 kb内。Hori等[34]利用CSSL群体与BIL群体分别检测到7个和2个穗发芽相关QTL位点, 2个群体共同定位到的1个位于第3染色体短臂上QTL, 可能与Fujino等[26]报道的低温萌发基因qLTG 3-1是同一位点。Sdr1、Sdr6、Sdr9和Sdr10等4个休眠相关位点[35-37]和1个幼苗形成相关位点qSSL 1 b[38]等也已经被精细定位。
| QTL | 染色体 Chromosome |
区间或连锁标记 CMarker interval |
控制性状 CIdentified traits |
连锁性状 CLinked traits |
参考文献 CReference |
| qSSL1b | 1 | IND1-3与IND1-4 | 幼苗芽长Seedling shoot length | 株高Plant height | [38] |
| qGR2 | 2 | OsMADS29 | 萌发率Germination rate | — | [15] |
| qLTG3-1 | 3 | RM14240-RM14275 | 低温萌发Low-temperature germination | 穗发芽Pre-harvest sprouting | [26, 34] |
| qLTG-9 | 9 | L9-25D与ID-1 | 低温萌发Low-temperature germination | — | [25] |
| qSV-1 | 1 | CAPS5-CAPS7 | 种子活力Seed vigor | — | [17] |
| qSV-5c | 5 | CAPS1-CAPS2 | 种子活力Seed vigor | — | |
| Sdr1 | 3 | C1488 | 种子休眠Seed dormancy | 抽穗期Heading date | [35-36] |
| qSD12 | 12 | RM28645-SD12m50 | 种子休眠Seed dormancy | — | [33] |
| Sdr6 | 1 | RM6902 | 种子休眠Seed dormancy | — | [37] |
| Sdr9 | 6 | RM5963 | 种子休眠Seed dormancy | — | |
| Sdr10 | 6 | RM3207 | 种子休眠Seed dormancy | — |
在QTL精细定位基础上, 利用图位克隆技术至少克隆了8个水稻种子活力关键基因, 分别位于水稻第1、3、6和7号染色体上(图 2), 包括2个种子萌发相关基因qLTG3-1[26]和GD1 [39], 5个种子休眠相关基因OsVP1[40]、Sdr4 [41]、qSD 7-1/qPC7[42]、qSD 1-2/OsGA20ox2[43]和OsPLS1[44], 1个是幼苗形成相关基因OsGA 20ox1[45]。另外, 利用生物信息学技术至少克隆了12个水稻种子活力关键基因, 分别位于水稻第1、2、4、5、6、8与10号染色上(图 2), 包括9个种子萌发相关基因OsFbx352 [46]、ARAG1[47]、OsPIP1;1[48]、OsLOL1 [49]、OsSCP46[50]、OsPP 2C51[51]、OsPYL/RCAR5 [52]与OsPYL/RCAR9[53], 3个种子寿命相关基因OsHSP18.2 [54]、OsPIMT1 [55-56]与OsPIMT2[56]。
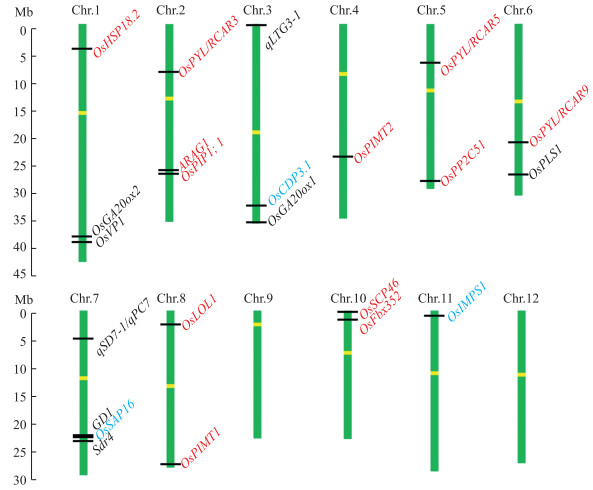
|
图 2 已经克隆的水稻种子活力关键基因 Fig. 2 The cloned genes related to seed-vigor in rice 黑色字体代表图位克隆技术克隆的基因, 包括OsGA 20ox2[43]、OsVP1[40]、qLTG3-1 [26]、OsGA 20ox1[45]、OsPLS1 [44]、qSD 7-1/qPC7[42]、GD1 [39]、Sdr4 [41]; 红色字体代表电子克隆技术克隆的基因, 包括OsHSP18.2 [54]、OsPYL/RCAR3 [53]、ARAG1[47]、OsPIP1; 1 [48]、OsPIMT2 [56]、OsPYL/RCAR5 [53]、OsPP 2C51[51]、OsPYL/RCAR9 [53]、OsLOL1[49]、OsPIMT1[55]、OsSCP46 [50]、OsFbx352 [46]; 蓝色字体代表基于GWAS和组学技术克隆的基因, 包括OsSAP16 [57]、OsCDP3.1 [58]、OsIMPS1[59]; 黄色条框代表染色体上的丝粒。 The black genes were cloned by map-based, including OsGA 20ox2[43], OsVP1[40], qLTG3-1 [26], OsGA 20ox1 [45], OsPLS1 [44], qSD 7-1/qPC7 [42], GD1 [39], Sdr4 [41]; Red genes cloned based on in silica, including OsHSP18.2 [54], OsPYL/RCAR3 [53], ARAG1 [47], OsPIP1;1 [48], OsPIMT2 [56], OsPYL/RCAR5 [53], OsPP 2C51[51], OsPYL/RCAR>9 [53], OsLOL1 [49], OsPIMT1 [55], OsSCP46 [50], OsFbx352 [46]; Blue genes cloned based on GWAS or genomics ways, including OsSAP16 [57], OsCDP3.1 [58] and OsIMPS1 [59]. Yellow boxes represent centromeres on the chromosome. |
近年来, 利用基于连锁不平衡原理的全基因组关联分析(genome-wide association study, GWAS)方法在水稻种子萌发、休眠、幼苗形成与种子耐非生物逆境等关键位点鉴定方面取得了很大进展。Cheng等[20]利用276份籼稻品种, 以水稻种子GP、GI和SSP为指标, 对正常、盐和PEG 3种条件下的种子活力进行分析, 分别检测到12、14和9个与种子活力显著关联的SSR(simple sequence repeat)分子标记。Dang等[28]以SL、RL和SDW为指标, 连续2年对540份水稻种质资源的种子活力进行分析, 检测到27个与水稻种子活力显著关联的位点。Wu等[30]利用270份水稻种质资源, 对种子中胚轴长度进行全基因组关联分析, 检测到13个显著相关位点(-lg P≥8.0), 分别位于1、3、4、5、6与7号染色体。Wang等[31]利用295份水稻资源材料和44 kb的SNP芯片数据, 检测到67个幼苗耐冷相关的QTL位点, 包含56个新位点。
随着高通量测序技术发展, 已经至少定位了14个种子活力相关性状关键基因或候选基因, 分别分布于1、2、3、4、5、7与12号染色体上(表 2)。Lu等[19]以中胚轴长(ML)和芽长(SL)为指标, 采用混合线性模型共鉴定到23个与种子活力相关的位点; 基于基因相对表达分析数据, 发现Os 01g0392100、Os04g0630000、Os01g0904700与Os07g0615000等4个基因极有可能为目的基因。Zhao等[18]利用621不同来源的水稻品种, 在深直播条件下鉴定到13个与中胚轴长(ML)相关的QTL位点; 对2个新的主效位点OsML1和OsML2进行单倍型分析, 推测LOC_Os03g53320和LOC_Os07g24010为目的基因。Shi等[16]以478份水稻资源群体为材料, 结合6 361 920个SNP分子标记, 鉴定到11个与种子活力显著关联位点; 基于单倍型分析数据, 发现位于2号染色体上的耐盐胁迫萌发相关基因OsNRT2.2在亚群间存在显著的变异。Yu等[22]利用295份水稻品种, 对盐胁迫下种子幼苗活力相关性状的全基因组关联分析, 以TDW、R-LW、LW(leaf width)、SL和R-TDW等为指标, 分别检测到13、4、3、2和1个GWAS显著关联的峰值。Wang等[57]利用187份水稻品种在低温条件下共检测到53个显著关联的耐低温萌发QTL, 并利用T-DNA插入突变体验证了候选基因OsSAP16为目的基因, 该基因编码1个含锌指结构域的蛋白, 调控低温环境下的种子萌发(图 2)。Magwa等[60]以350份水稻资源为材料, 在新鲜收获种子(freshly harvested seeds, FHS)和收获后后熟种子(after-ripened seeds, ARS)中分别检测到了16和38个与休眠相关的QTL位点, 包括Sdr 4、GA2OX3、ABI5、EUI1和GH3-2等6个候选基因。Lu等[61]利用453个水稻品种鉴定到了10个休眠相关性位点, 其中1个与玉米胚胎发育基因LOC_Os 03g10110同源。Hsu等[62]通过全基因组关联分析, 结合重组自交系群体连锁分析, 在水稻第1染色体上检测到1个控制胚芽鞘长(CL)的主效QTL位点, 贡献率达27%。
候选基因 Candidate genes |
LOC号 LOC ID |
染色体 Chromosome |
控制性状 Identified traits |
遗传验证 Verification |
基因克隆 Cloned |
参考文献 Reference |
| OsSAP16 | LOC_Os07g38240 | 7 | 低温萌发Low-temperature germination | 是Yes | 否No | [57] |
| OsNRT2.2 | LOC_Os02g02190 | 2 | 耐盐萌发Seed germination under salt stress | 否No | 否No | [16] |
| Os12g0176700 | LOC_Os12g07730 | 12 | 苗期耐盐Salt tolerance at seedling stages | 否No | 否No | [22] |
| Os01g0904700 | LOC_Os01g67770 | 1 | 芽长Shoot length | 否No | 否No | [19] |
| Os07g0615000 | LOC_Os07g42354 | 7 | 芽长Shoot length | 否No | 否No | |
| Os01g0392100 | LOC_Os01g29740 | 1 | 中胚轴长Mesocotyl length | 否No | 否No | |
| Os04g0630000 | LOC_Os04g53770 | 4 | 中胚轴长Mesocotyl length | 否No | 否No | |
| OsML1 | LOC_Os03g53320 | 3 | 中胚轴长Mesocotyl length | 否No | 否No | [18] |
| OsML2 | LOC_Os07g24010 | 7 | 中胚轴长Mesocotyl length | |||
| Sdr4 | LOC_Os07g39700 | 7 | 种子休眠Seed dormancy | 否No | 是Yes | [41, 60] |
| GA2ox3 | LOC_Os01g55240 | 1 | 种子休眠Seed dormancy | 否No | [60] | |
| ABI5 | LOC_Os01g64000 | 1 | 种子休眠Seed dormancy | 否No | ||
| EUI1 | LOC_Os05g40384 | 5 | 种子休眠Seed dormancy | 否No | ||
| GH3-2 | LOC_Os01g55940 | 1 | 种子休眠Seed dormancy | 否No | ||
| Os03g0197300 | LOC_Os03g10110 | 3 | 种子休眠Seed dormancy | 否No | 否No | [61] |
利用转录组(transcriptomics)和蛋白组(proteomics)学技术对不同样本进行大数据分析, 寻找差异表达基因或蛋白, 可以进一步获得与目标性状相关的基因[11-12]。种子活力形成于种子发育脱水阶段, 随着种子发育的成熟, 水分逐渐下降, 种子活力呈上升趋势[27]。长命mRNA在种子萌发早期具有非常重要作用, Sano等[63]利用转录抑制剂放线菌素D(Act D)分别对水稻种子发育10、20、25、30和40 d种子活力进行分析, 发现种子胚在发育10~20 d时种子萌发所需要的长命mRNA就已大量积累, 转录组数据显示至少有529个长命mRNA显著增加。Howell等[64]以水稻品种‘Amaroo’为材料, 对种子萌发1、3、12、24和48 h的胚组织进行转录组和代谢组分析, 发现在萌发3~12 h进程中转录本差异最大。Dametto等[65]以耐低温和低温敏感的2个籼稻姊妹系种子为材料, 在低温(13 ℃)下萌发7 d后进行转录组分析, 获得1 361个显著差异表达基因, 进一步用RT-PCR技术对OsLEA、OsAUX等11个差异表达基因进行了功能验证。
Yang等[11]利用籼稻‘9311’为材料, 通过双向凝胶电泳技术检测到与种子活力相关的显著差异蛋白148个。He等[66-67]以粳稻‘日本晴’为材料, 利用蛋白质双向电泳和液相-质谱蛋白质随机测序方法分析了水稻种子萌发过程中的差异蛋白, 发现差异表达蛋白主要与种子贮藏物质动员、蛋白质前体合成相关。Han等[68]利用无凝胶-磷酸蛋白质组学技术分析了粳稻品种‘日本晴’种子萌发早期的蛋白磷酸化修饰变化, 发现吸胀12 h时, 至少有12种与蛋白质修饰相关的蛋白质大量磷酸化, 说明一些重要的信号传导在吸胀12 h就已经启动种子萌发过程; 其中, 有3个组分涉及到油菜素内酯(brassinosteroid, BR)的信号传导, BR处理可以显著增加种子萌发率; Liu等[69]采用双向凝胶电泳结合质谱分析技术, 分析了ABA和高温处理下水稻品种‘矮培64S’种子吸胀过程中的蛋白质组学信息, 发现7种专一性地与水稻种子萌发相关蛋白, 包括糖基水解酶、淀粉合成酶等; 2016年, Liu等[70]再次通过蛋白组学技术对水稻种子胚开展了研究, 结果从种子萌发过程中共鉴定到了88个显著差异蛋白, 同时发现萌发后期细胞分裂、细胞壁合成和次生代谢被激活, 进而促进幼苗形态建成; Xu等[71]以无休眠的栽培稻品种‘Yannong S’和强休眠的东乡野生稻种子为材料, 在萌发过程中的胚与胚乳中分别鉴定到231种和180种差异表达蛋白; 其中, 胚中的差异表达蛋白主要与氨基酸激活、糖酵解和蛋白质合成相关, 而胚乳中的差异表达蛋白主要与蔗糖裂解、ATP和辅酶CoQ合成以及蛋白质水解等相关; Cheng等[72]利用凝胶双向电泳技术在水稻干种子与吸胀24 h种子中鉴定到10个碳代谢、蛋白合成等相关代谢显著性差异的蛋白; 同时, 发现LOC_Os01g44220、LOC_Os03g07570与LOC_Os06g31070基因参与种子引发(seed priming), 可作为种子引发的生物分子标记; Xu等[58]利用T-DNA插入突变体等转基因材料, 验证了位于3号染色体上cupin家族蛋白基因OsCDP3.1参与水稻种子萌发途径。He等[59]基于蛋白质学信息, 克隆了位于11号染色体上的α-异丙基苹果酸合酶基因OsIMPS1, 并利用T-DNA与CRISPR/Cas9突变体材料, 揭示其调控水稻种子活力作用机制(图 2)。综合蛋白组学结果显示, 水稻种子萌发过程中种子发育、耐脱水和贮藏相关的蛋白总体上显著下调; 而碳代谢、蛋白合成代谢、细胞壁合成代谢、糖酵解相关能量代谢以及GA激素相关代谢的蛋白显著上调[12, 67-70, 72]。
3 水稻种子活力调控的分子机制基于已经克隆的水稻种子活力相关基因研究结果, 水稻种子活力调控可能主要是通过脱落酸(ABA)和赤霉素(GA)2种激素代谢途径协同调控实现的(图 3)。Hobo等[40]报道OsVP1是1个转录因子, 它通过与ABA调控转录的转录因子TRAB1的互作, 间接调控ABA应答元件, 激活诱导ABA控制植物胚的成熟和休眠特性; 该转录因子也可以正向调控种子休眠Sdr4基因, 并依次正调控种子成熟相关蛋白基因OsDOG1-like, 诱导ABA的合成, 抑制萌发相关基因的表达, 实现种子休眠[41]。Gu等[42]报道qSD 7-1/qPC7是1个水稻种子休眠与果皮颜色紧密连锁的基因, 具有“一因多效”的特性, 该基因的表达促进ABA生物合成关键基因的表达, 导致引发休眠的植物激素ABA积累, 调控种子休眠。Song等[46]报道OsFbx352是1个具有F-box结构域的蛋白, 可能参与E3泛素连接酶复合体的形成, 受ABA诱导上调表达, 通过影响ABA合成和分解代谢, 降低ABA含量, 影响种子萌发。Bhatnagar等[51]报道OsPP 2C51是编码蛋白磷酸酶2C的基因, 可直接抑制活性磷酸化的OsbZIP10, 正向调控种子萌发。Li等[50]报道OsSCP46是1个编码丝氨酸羧肽酶基因, 受ABA诱导, 在水稻种子发育过程中高表达, 而突变体scp46籽粒变小, 对ABA的抑制不敏感, 种子萌发变快。Kim等[52]报道OsPYLs是ABA受体家族基因, 其中OsPYL/RCAR3、OsPYL/RCAR5、OsPYL/RCAR9可正向调控ABA响应, 进而影响种子萌发和幼苗生长。Tian等[53]利用过表达转基因植株研究发现OsPYL3和OsPYL9基因具有提高水稻耐旱性和耐冷性特性。
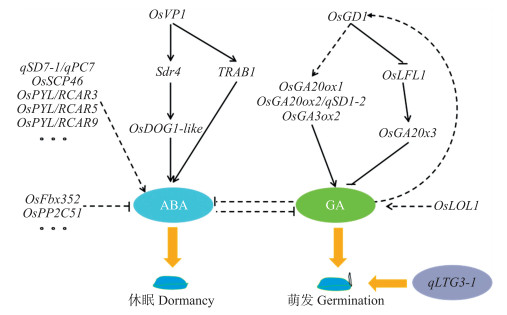
|
图 3 脱落酸(ABA)和赤霉素(GA)调控水稻种子活力的分子机制 Fig. 3 The molecular mechanism of rice seed-vigor regulation with ABA and GA OsVP1正调控Sdr4、OsGOG1-like和TRAB1基因的表达, 促进ABA积累, 引起种子休眠[40-41]; qSD7-1/qPC7[42]、OsSCP46[50]、OsPYL/RCAR3[53]、OsPYL/RCAR5[53]和OsPYL/RCAR9[53]基因正调控ABA, 促进ABA合成, 诱导休眠; OsFbx352[46]和OsPP2C51[51]负调控ABA的合成, 间接促进萌发。OsGD1[39]通过促进OsGA20ox1[45]、OsGA20ox2[43](与qSD1-2为同一基因)和OsGA3ox2合成, 或抑制OsLFL1表达, 抑制GA分解基因OsGA20x3表达, 促进GA合成, 诱导种子萌发; GA也可以通过反馈调节影响OsGD1[39]的表达; 同时, OsLOL1[49]可正调控GA合成。qLTG3-1[26]通过调节组织的细胞液泡化, 引起细胞组织的松弛, 促进种子萌发。 The gene OsVP1 positively regulated expression of Sdr 4, OsGOG1-likeand TRAB1, and further increased accumulation of ABA to induce dormancy of rice seeds[40-41]; The genes qSD 7-1/qPC7 [42], OsSCP46 [50], OsPYL/RCAR3 [53], OsPYL/RCAR5 [53]and OsPYL/RCAR9 [53] positively regulated syntheses of ABA to induce seed dormancy; The genes OsFbx352 [46]and OsPP 2C51 [51]negatively regulated synthesis of ABA indirectly promoting germination of seeds。The genes OsGD1 [39] might up-regulate the expression of OsGA 20ox1 [45], OsGA 20ox2 [43](same as qSD1-2)and OsGA 3ox2, or down-regulate expression of OsLFL1 to inhibite expression of OsGA 20x3, a gene for GA degrading, and increase synthesis of GA to induce germination of rice seeds; The GA might also feedback the expression of OsGD1 [39] and the OsLOL1 [49] up-regulate synthesis of GA to affect the seed germination。The gene qLTG3-1 [26] might regulate vacuolation of tissues covering the embyo and cause tissue weakening to induce seed germination. |
赤霉素(GA)在种子萌发过程中具有重要作用, 种子中GA含量增加, 可促进种子萌发。Guo等[39]利用水稻种子萌发缺陷型突变体gd 1克隆了OsGD1基因, 该基因编码1个B3结构域的转录因子, 通过影响OsLFL1基因表达, 间接调控抑制GA活性的基因OsGA 20x3表达, 或者调控GA合成相关基因OsGA20ox1、OsGA20ox2与OsGA3ox2等的表达, 影响水稻种子萌发。Abe等[45]用图位克隆法克隆的1个水稻幼苗活力相关基因与OsGA 20ox1有关; Ye等[43]克隆的水稻种子休眠相关基因qSD1-2与OsGA20ox2是同一个基因, 与赤霉素的生物合成相关。OsLOL1是1个锌指蛋白, 能与OsbZIP58互作, 激活OsKO2表达, 影响GA生物合成, 进而影响糊粉层PCD过程, 调控水稻种子萌发[49]。
除ABA和GA两大植物激素调控水稻种子活力外, 可能存在其他激素途径共同调控水稻种子活力。Fujino等[26]研究报道水稻低温萌发响应基因qLTG3-1并不参与植物激素的诱导或抑制, 而是通过调节组织的细胞液泡化, 从而引起这些组织的松弛而提高种子萌发; 同时, 该基因存在“一因多效”性, 可能参与种子盐、甘露醇等非生物胁迫。Hori等[34]利用水稻种子穗发芽休眠性状精细定位还发现, 位于3号染色体上的主效QTL位点为该基因的等位基因。此外, Kaur等[54]利用组学鉴定的OsHSP18.2基因, 其功能主要是通过调控活性氧系统(ROS)积累, 影响水稻种子活力、寿命和幼苗建成。
4 展望高活力种子往往具有明显的生长优势和生产潜力, 抵御不良环境条件的能力更强, 对水稻高产、稳产具有十分重要的意义[1-5]。近年来, 随着经济的发展, 农村劳动力日益短缺, 直播稻和机插稻等水稻轻简栽培模式正变得越来越普遍, 具有高活力种子的水稻品种已经成为种业公司发展的核心竞争力。通过构建不同遗传群体和分子遗传图谱, 结合突变体分析和图位克隆技术, 已经获得了一些控制水稻种子活力的关键基因。高通量测序技术结合GWAS分析、转录组与蛋白组学技等术将进一步促进水稻种子活力关键基因的鉴定和克隆。正在兴起的CRISPR/Cas9基因编辑技术具有快捷、成本低等优点, 与GWAS、转录组和蛋白组学技术结合, 为快速鉴定和克隆种子活力关键基因提供了可能。随着越来越多的水稻种子活力基因挖掘与克隆, 有助于阐明水稻种子活力的分子调控机制。通过分子标记辅助选择(MAS)技术、基因编辑等技术培育高活力种子水稻新品种, 满足我国现代种业对高质量种子生产的需求。
| [1] |
孙群, 王建华, 孙宝启. 种子活力的生理和遗传机理研究进展[J]. 中国农业科学, 2007, 40(1): 48-53. Sun Q, Wang J H, Sun B Q. Advances on seed vigor physiological and genetic mechanisms[J]. Scientia Agricultura Sinica, 2007, 40(1): 48-53 (in Chinese with English abstract). DOI:10.3321/j.issn:0578-1752.2007.01.007 |
| [2] |
张红生, 胡晋. 种子学[M]. 2版.北京: 科学出版社, 2015. Zhang H S, Hu J. Seed Science[M]. 2nd ed. Beijing: Science Press, 2015 (in Chinese). |
| [3] |
张安鹏, 钱前, 高振宇. 水稻种子活力的研究进展[J]. 中国水稻科学, 2018, 32(3): 296-303. Zhang A P, Qian Q, Gao Z Y. Research advances on rice seed vigor[J]. Chinese Journal of Rice Science, 2018, 32(3): 296-303 (in Chinese with English abstract). |
| [4] |
Han C, Yang P F. Studies on the molecular mechanisms of seed germination[J]. Proteomics, 2015, 15(10): 1671-1679. DOI:10.1002/pmic.v15.10 |
| [5] |
Bewley J, Bradford K, Hilhorst H, et al. Seeds:Physiology of Development, Germination and Dormancy[M]. New York: Springer, 2013.
|
| [6] |
Wei T, He Z L, Tan X Y, et al. An integrated RNA-Seq and network study reveals a complex regulation process of rice embryo during seed germination[J]. Biochemical and Biophysical Research Communications, 2015, 464(1): 176-181. DOI:10.1016/j.bbrc.2015.06.110 |
| [7] |
李振华, 王建华. 种子活力与萌发的生理与分子机制研究进展[J]. 中国农业科学, 2015, 48(4): 646-660. Li Z H, Wang J H. Advances in research of physiological and molecular mechanism in seed vigor and germination[J]. Scientia Agricultura Sinica, 2015, 48(4): 646-660 (in Chinese with English abstract). |
| [8] |
Bewley J. Seed germination and dormancy[J]. The Plant Cell, 1997, 9: 1055-1066. DOI:10.1105/tpc.9.7.1055 |
| [9] |
Zhu G H, Ye N H, Zhang J H, et al. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis[J]. Plant and Cell Physiology, 2009, 50(3): 644-651. DOI:10.1093/pcp/pcp022 |
| [10] |
Nakashima K, Yamaguchi-Shinozaki K. ABA signaling in stress-response and seed development[J]. Plant Cell Reports, 2013, 32(7): 959-970. DOI:10.1007/s00299-013-1418-1 |
| [11] |
Yang P F, Li X J, Wang X Q, et al. Proteomic analysis of rice(Oryza sativa) seeds during germination[J]. Proteomics, 2007, 7(18): 3358-3368. DOI:10.1002/(ISSN)1615-9861 |
| [12] |
Catusse J, Job C, Job D. Transcriptome-and proteome-wide analyses of seed germination[J]. Comptes Rendus Biologies, 2008, 331(10): 815-822. DOI:10.1016/j.crvi.2008.07.023 |
| [13] |
Wang Z F, Wang J F, Bao Y M, et al. Quantitative trait loci controlling rice seed germination under salt stress[J]. Euphytica, 2011, 178(3): 297-307. DOI:10.1007/s10681-010-0287-8 |
| [14] |
Wang Z F, Wang J F, Bao Y M, et al. Quantitative trait loci analysis for rice seed vigor during the germination stage[J]. J Zhejiang Univ Sci B, 2010, 11(12): 958-964. DOI:10.1631/jzus.B1000238 |
| [15] |
Li M, Sun P L, Zhou H J, et al. Identification of quantitative trait loci associated with germination using chromosome segment substitution lines of rice(Oryza sativa L.)[J]. Theoretical and Applied Genetics, 2011, 123(3): 411-420. DOI:10.1007/s00122-011-1593-9 |
| [16] |
Shi Y Y, Gao L L, Wu Z C, et al. Genome-wide association study of salt tolerance at the seed germination stage in rice[J]. BMC Plant Biology, 2017, 17: 92. DOI:10.1186/s12870-017-1044-0 |
| [17] |
Xie L X, Tan Z W, Zhou Y, et al. Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice[J]. Journal of Integrative Plant Biology, 2014, 56(8): 749-759. DOI:10.1111/jipb.v56.8 |
| [18] |
Zhao Y, Zhao W P, Jiang C H, et al. Genetic architecture and candidate genes for deep-sowing tolerance in rice revealed by non-syn GWAS[J]. Frontiers in Plant Science, 2018, 9: 332. DOI:10.3389/fpls.2018.00332 |
| [19] |
Lu Q, Zhang M C, Niu X J, et al. Uncovering novel loci for mesocotyl elongation and shoot length in indica rice through genome-wide association mapping[J]. Planta, 2016, 243(3): 645-657. DOI:10.1007/s00425-015-2434-x |
| [20] |
Cheng J P, He Y Q, Yang B, et al. Association mapping of seed germination and seedling growth at three conditions in indica rice(Oryza sativa L.)[J]. Euphytica, 2015, 206(1): 103-115. DOI:10.1007/s10681-015-1477-1 |
| [21] |
Xu C G, Li X Q, Xue Y, et al. Comparison of quantitative trait loci controlling seedling characteristics at two seedling stages using rice recombinant inbred lines[J]. Theoretical and Applied Genetics, 2004, 109(3): 640-647. DOI:10.1007/s00122-004-1671-3 |
| [22] |
Yu J, Zao W G, He Q, et al. Genome-wide association study and gene set analysis for understanding candidate genes involved in salt tolerance at the rice seedling stage[J]. Molecular Genetics and Genomics, 2017, 292(6): 1391-1403. DOI:10.1007/s00438-017-1354-9 |
| [23] |
Sabouri H, Rezai A M, Moumeni A, et al. QTLs mapping of physiological traits related to salt tolerance in young rice seedlings[J]. Biologia Plantarum, 2009, 53(4): 657-662. DOI:10.1007/s10535-009-0119-7 |
| [24] |
Guo Z L, Yang W N, Chang Y, et al. Genome-wide association studies of image traits reveal genetic architecture of drought resistance in rice[J]. Molecular Plant, 2018, 11(6): 789-805. DOI:10.1016/j.molp.2018.03.018 |
| [25] |
Li L F, Liu X, Xie K, et al. qLTG-9, a stable quantitative trait locus for low-temperature germination in rice(Oryza sativa L.)[J]. Theoretical and Applied Genetics, 2013, 126(9): 2313-2322. DOI:10.1007/s00122-013-2137-2 |
| [26] |
Fujino K, Sekiguchi H, Matsuda Y, et al. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice[J]. Proc Natl Acad Sci USA, 2008, 105(34): 12623-12628. DOI:10.1073/pnas.0805303105 |
| [27] |
He Y Q, Cheng J P, Li X D, et al. Acquisition of desiccation tolerance during seed development is associated with oxidative processes in rice[J]. Botany, 2016, 94(2): 91-101. DOI:10.1139/cjb-2015-0154 |
| [28] |
Dang X J, Thi T G T, Dong G S, et al. Genetic diversity and association mapping of seed vigor in rice(Oryza sativa L.)[J]. Planta, 2014, 239(6): 1309-1319. DOI:10.1007/s00425-014-2060-z |
| [29] |
Cheng X X, Cheng J P, Huang X, et al. Dynamic quantitative trait loci analysis of seed reserve utilization during three germination stages in rice[J]. PLoS One, 2013, 8(11): e80002. DOI:10.1371/journal.pone.0080002 |
| [30] |
Wu J H, Feng F J, Lian X M, et al. Genome-wide association study(GWAS) of mesocotyl elongation based on re-sequencing approach in rice[J]. BMC Plant Biology, 2015, 15: 218. DOI:10.1186/s12870-015-0608-0 |
| [31] |
Wang D, Liu J L, Li C G, et al. Genome-wide association mapping of cold tolerance genes at the seedling stage in rice[J]. Rice, 2016, 9(1): 61. DOI:10.1186/s12284-016-0133-2 |
| [32] |
Wang L, Cheng J P, Lai Y Y, et al. Identification of QTLs with additive, epistatic and QTL×development interaction effects for seed dormancy in rice[J]. Planta, 2014, 239(2): 411-420. DOI:10.1007/s00425-013-1991-0 |
| [33] |
Gu X Y, Liu T L, Feng J H, et al. The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice[J]. Plant Molecular Biology, 2010, 73(1/2): 97-104. |
| [34] |
Hori K, Sugimoto K, Nonoue Y, et al. Detection of quantitative trait loci controlling pre-harvest sprouting resistance by using backcrossed populations of japonica rice cultivars[J]. Theoretical and Applied Genetics, 2010, 120(8): 1547-1557. DOI:10.1007/s00122-010-1275-z |
| [35] |
Takeuchi Y, Lin S Y, Sasaki T, et al. Fine linkage mapping enables dissection of closely linked quantitative trait loci for seed dormancy and heading in rice[J]. Theoretical and Applied Genetics, 2003, 107(7): 1174-1180. DOI:10.1007/s00122-003-1364-3 |
| [36] |
Lin S Y, Sasaki T, Yano M. Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines[J]. Theoretical and Applied Genetics, 1998, 96(8): 997-1003. DOI:10.1007/s001220050831 |
| [37] |
Marzougui S, Sugimoto K, Yamanouchi U, et al. Mapping and characterization of seed dormancy QTLs using chromosome segment substitution lines in rice[J]. Theoretical and Applied Genetics, 2012, 124(5): 893-902. DOI:10.1007/s00122-011-1753-y |
| [38] |
Zhang A P, Liu C L, Chen G, et al. Genetic analysis for rice seedling vigor and fine mapping of a major QTL qSSL1b for seedling shoot length[J]. Breeding Science, 2017, 67(3): 307-315. DOI:10.1270/jsbbs.16195 |
| [39] |
Guo X L, Hou X M, Fang J, et al. The rice GERMINATION DEFECTIVE1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism[J]. The Plant Journal, 2013, 75(3): 403-416. DOI:10.1111/tpj.2013.75.issue-3 |
| [40] |
Hobo T, Kowyama Y, Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription[J]. Proc Natl Acad Sci USA, 1999, 96(26): 15348-15353. DOI:10.1073/pnas.96.26.15348 |
| [41] |
Sugimoto K, Takeuchi Y, Ebana K, et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice[J]. Proc Natl Acad Sci USA, 2010, 107(13): 5792-5797. DOI:10.1073/pnas.0911965107 |
| [42] |
Gu X Y, Foley M E, Horvath D P, et al. Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice[J]. Genetics, 2011, 189(4): 1515-1524. DOI:10.1534/genetics.111.131169 |
| [43] |
Ye H, Feng J H, Zhang L H, et al. Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice[J]. Plant Physiology, 2015, 169: 2152-2165. |
| [44] |
Yang X, Gong P, Li K Y, et al. A single cytosine deletion in the OsPLS1 gene encoding vacuolar-type H+-ATPase subunit A1 leads to premature leaf senescence and seed dormancy in rice[J]. Journal of Experimental Botany, 2016, 67(9): 2761-2776. DOI:10.1093/jxb/erw109 |
| [45] |
Abe A, Takagi H, Fujibe T, et al. OsGA20ox1, a candidate gene for a major QTL controlling seedling vigor in rice[J]. Theoretical and Applied Genetics, 2012, 125(4): 647-657. DOI:10.1007/s00122-012-1857-z |
| [46] |
Song S, Dai X, Zhang W H. A rice F-box gene, OsFbx352, is involved in glucose-delayed seed germination in rice[J]. Journal of Experimental Botany, 2012, 63(15): 5559-5568. DOI:10.1093/jxb/ers206 |
| [47] |
Zhao L F, Hu Y B, Chong K, et al. ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice[J]. Annals of Botany, 2010, 105(3): 401-409. DOI:10.1093/aob/mcp303 |
| [48] |
Liu C W, Fukumoto T, Matsumoto T, et al. Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination[J]. Plant Physiology and Biochemistry, 2013, 63: 151-158. DOI:10.1016/j.plaphy.2012.11.018 |
| [49] |
Wu J H, Zhu C F, Pang J H, et al. OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination through the modulation of gibberell in biosynthesis in Oryza sativa[J]. The Plant Journal, 2014, 80(6): 1118-1130. DOI:10.1111/tpj.12714 |
| [50] |
Li Z Y, Tang L Q, Qiu J H, et al. Serine carboxypeptidase 46 regulates grain filling and seed germination in rice(Oryza sativa L.)[J]. PLoS One, 2016, 11(7): e0159737. DOI:10.1371/journal.pone.0159737 |
| [51] |
Bhatnagar N, Min M K, Choi E H, et al. The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10[J]. Plant Molecular Biology, 2017, 93(4/5): 389-401. |
| [52] |
Kim H, Hwang H, Hong J W, et al. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth[J]. Journal of Experimental Botany, 2012, 63(2): 1013-1024. DOI:10.1093/jxb/err338 |
| [53] |
Tian X J, Wang Z Y, Li X F, et al. Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice[J]. Rice, 2015, 8(1): 28. DOI:10.1186/s12284-015-0061-6 |
| [54] |
Kaur H, Petla B P, Kamble N U, et al. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress[J]. Frontiers in Plant Science, 2015, 6: e00713. |
| [55] |
Wei Y D, Xu H B, Diao L R, et al. Protein repair l-isoaspartyl methyltransferase 1(PIMT1) in rice improves seed longevity by preserving embryo vigor and viability[J]. Plant Molecular Biology, 2015, 89(4/5): 475-492. |
| [56] |
Petla B P, Kamble N U, Kumar M, et al. Rice proteinl-I soaspartyl methyltransferase isoforms differentially accumulate during seed maturation to restrict deleterious isoAsp and reactive oxygen species accumulation and are implicated in seed vigor and longevity[J]. New Phytologist, 2016, 211(2): 627-645. DOI:10.1111/nph.13923 |
| [57] |
Wang X, Zou B H, Shao Q L, et al. Natural variation reveals that OsSAP16 controls low-temperature germination in rice[J]. Journal of Experimental Botany, 2018, 69(3): 413-421. DOI:10.1093/jxb/erx413 |
| [58] |
Xu E S, Chen M M, He H, et al. Proteomic analysis reveals proteins involved in seed imbibition under salt stress in rice[J]. Frontiers in Plant Science, 2017, 7. |
| [59] |
He Y Q, Cheng J P, He Y, et al. Influence of isopropylmalate synthase OsIPMS1 on seed vigor associated with amino acid and energy metabolism in rice[J]. Plant Biotechnology Journal, 2019, 17(2): 322-337. DOI:10.1111/pbi.12979 |
| [60] |
Magwa R A, Zhao H, Xing Y Z. Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice(Oryza sativa L.)[J]. BMC Genetics, 2016, 17(1): 28. DOI:10.1186/s12863-016-0340-2 |
| [61] |
Lu Q, Niu X J, Zhang M C, et al. Genome-wide association study of seed dormancy and the genomic consequences of improvement footprints in rice(Oryza sativa L.)[J]. Frontiers in Plant Science, 2018, 8: e02213. |
| [62] |
Hsu S K, Tung C W. Genetic mapping of anaerobic germination-associated QTLs controlling coleoptile elongation in rice[J]. Rice, 2015, 8(1): 38. DOI:10.1186/s12284-015-0072-3 |
| [63] |
Sano N, Ono H, Murata K, et al. Accumulation of long-lived mRNAs associated with germination in embryos during seed development of rice[J]. Journal of Experimental Botany, 2015, 66(13): 4035-4046. DOI:10.1093/jxb/erv209 |
| [64] |
Howell K A, Narsai R, Carroll A, et al. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process[J]. Plant Physiology, 2009, 149(2): 961-980. |
| [65] |
Dametto A, Sperotto R A, Adamski J M, et al. Cold tolerance in rice germinating seeds revealed by deep RNAseq analysis of contrasting indica genotypes[J]. Plant Science, 2015, 238: 1-12. DOI:10.1016/j.plantsci.2015.05.009 |
| [66] |
He D L, Han C, Yang P F. Gene expression profile changes in germinating rice[J]. Journal of Integrative Plant Biology, 2011, 53(10): 835-844. DOI:10.1111/jipb.2011.53.issue-10 |
| [67] |
He D L, Han C, Yao J L, et al. Constructing the metabolic and regulatory pathways in germinating rice seeds through proteomic approach[J]. Proteomics, 2011, 11(13): 2693-2713. DOI:10.1002/pmic.v11.13 |
| [68] |
Han C, Yang P F, Sakata K, et al. Quantitative proteomics reveals the role of protein phosphorylation in rice embryos during early stages of germination[J]. Journal of Proteome Research, 2014, 13(3): 1766-1782. DOI:10.1021/pr401295c |
| [69] |
Liu S J, Xu H H, Wang W Q, et al. A proteomic analysis of rice seed germination as affected by high temperature and ABA treatment[J]. Physiologia Plantarum, 2015, 154(1): 142-161. DOI:10.1111/ppl.2015.154.issue-1 |
| [70] |
Liu S J, Xu H H, Wang W Q, et al. Identification of embryo proteins associated with seed germination and seedling establishment in germinating rice seeds[J]. Journal of Plant Physiology, 2016, 196/197: 79-92. DOI:10.1016/j.jplph.2016.02.021 |
| [71] |
Xu H H, Liu S J, Song S H, et al. Proteomics analysis reveals distinct involvement of embryo and endosperm proteins during seed germination in dormant and non-dormant rice seeds[J]. Plant Physiology and Biochemistry, 2016, 103: 219-242. DOI:10.1016/j.plaphy.2016.03.007 |
| [72] |
Cheng J P, Wang L, Zeng P, et al. Identification of genes involved in rice seed priming in the early imbibition stage[J]. Plant Biology, 2017, 19(1): 61-69. DOI:10.1111/plb.2017.19.issue-1 |




