文章信息
- 陈发棣, 陈素梅, 宋爱萍, 蒋甲福, 赵楠, 苏江硕
- CHEN Fadi, CHEN Sumei, SONG Aiping, JIANG Jiafu, ZHAO Nan, SU Jiangshuo
- 乙烯调控植物耐涝机制的研究进展
- Progress in mechanisms of ethylene mediated tolerance of plant to waterlogging
- 南京农业大学学报, 2018, 41(2): 203-208
- Journal of Nanjing Agricultural University, 2018, 41(2): 203-208.
- http://dx.doi.org/10.7685/jnau.201709025
-
文章历史
- 收稿日期: 2017-09-14
水分是影响植物生长的重要环境因子之一, 决定着植物形态、生理生化代谢及地理分布, 适量的水分是保证植物生长健壮的先决条件。然而, 台风暴雨、长期阴雨及温室效应等导致江河泛滥, 土壤排水不良及海平面上升, 均使得土壤水分过多, 涝害频发。涝渍导致植物生长受抑, 产量下降, 严重时甚至导致植株死亡, 涝害已成为农业的主要灾害之一[1]。
植物应答涝渍胁迫依次通过信号传导物质诱导合成、代谢适应和形态适应(形成通气组织、不定根等)等过程来完成[2], 包括诱导低氧蛋白、转录因子、乙烯合成、活性氧(reactive oxygen species, ROS)产生与清除等。涝渍胁迫后数小时内即可检测到早期信号——乙烯(ethylene, ETH)的产生[3], 在植物耐涝应答中起着关键的作用[4]。为此, 本文对乙烯介导的植物耐涝机制研究进展进行归纳、综述。
1 涝渍胁迫下植物中乙烯的产生及其合成关键酶调控机制乙烯由一系列酶催化合成, 其中ACC(1-氨基环丙烷-1-羧酸)合成酶(ACS)为乙烯合成限速酶, 受ACS基因编码[5]。大多数植物都存在多个ACS基因, 同一种植物中不同ACS基因在不同发育阶段、器官和外界刺激后的表达具有特异性[6]。涝渍胁迫下, ACS家族特定成员受明显诱导表达。涝害会诱导沼生酸模(Rumex palustris)RpACS1的表达, 酶活性检测也显示受害植株中ACS活性显著高于未受涝害的植株[7]; 酸模(Rumex acetosa)中的RaACS也受涝渍胁迫诱导表达[8]。水稻OsACS1、OsACS2、OsACS3基因在低氧条件下的表达量各异, OsACS1在嫩茎中表达而在根中不表达; OsACS2与OsACS1相反, 仅在根中表达; 而OsACS3主要在黄化的种子中表达[9]; OsACS5在受涝渍胁迫的种子中表达量显著高于通风条件下生长的种子[10]。在拟南芥中, 涝渍胁迫会诱导AtACS5、AtACS7和AtACS8的表达[11]。受到涝渍胁迫后, 植物根中产生的乙烯前体ACC通过木质部运输到茎中, 然后被氧化为乙烯, 从而介导植物的耐涝性[12]。
不同刺激响应因子在转录和转录后水平调控ACS, 从而调控乙烯的合成[5, 13]。不同家族的转录因子能够诱导或抑制ACS表达, 如拟南芥的病菌响应激酶(mitogen-activated protein kinase, MAPK)MPK3和MPK6可以调控WRKY33的活性, 而WRKY33通过结合ACS2/6启动子的W-box元件, 从而促进后者的表达, 以增加乙烯的合成[14]; 还原型谷胱甘肽(GSH)通过促进WRKY33的表达, 也促进ACS2/6的转录[15]。拟南芥ABI4(AP2/ERF转录因子)通过直接结合AtACS4和AtACS8的启动子区, 抑制乙烯合成[16]。研究还发现HY5(ELONGATED HYPOCOTYL 5)可结合拟南芥乙烯响应因子AtERF11启动子的G-box区, 从而促进其表达, 而AtERF11则直接结合AtACS2/5启动子区, 抑制其转录, 从而减少乙烯的合成[17]。AtERF022通过抑制AtACS7表达和乙烯信号路径参与调控体细胞胚发生[18]。苹果MdERF2通过直接结合MdACS1的启动子而抑制其表达, 而MdERF3也能结合MdACS1启动子从而促进其表达, 且MdERF3在转录和翻译水平上被MdERF2抑制[19]; MdERF2还可结合MdACS3a的启动子, 抑制其表达, 从而延迟果实熟化[6]。拟南芥ETO1(ethylene over producer 1)是乙烯合成的负调控因子, 通过蛋白酶体途径促进ACS(ACS4/5/9)降解, 从而抑制乙烯合成[20]; EOL1(ETO1-like)和EOL2也具有类似功能[21]。最近研究发现, AtACS7的降解需要E3连接酶XBAT32[22]和AtACS7非催化反应的N端[23]。
目前, 涝渍胁迫下乙烯合成调控机制的研究还鲜见报道。在涝渍胁迫过程中, 拟南芥NAC(NAM、ATAF1/2和CUC2)转录因子SPEEDY HYPONASTIC GROWTH(SHYG)直接调节ACO5(ACC OXIDASE5)的表达, 而SHYG亦受乙烯诱导表达。涝渍胁迫下, ET-SHYG-ACO5路径在叶柄细胞快速膨大而使叶片迅速上翘的过程中起着重要作用[24]。然而, 涝渍胁迫下植物如何调控乙烯合成限速酶编码基因ACS从而调控乙烯合成的机制还有待进一步研究。
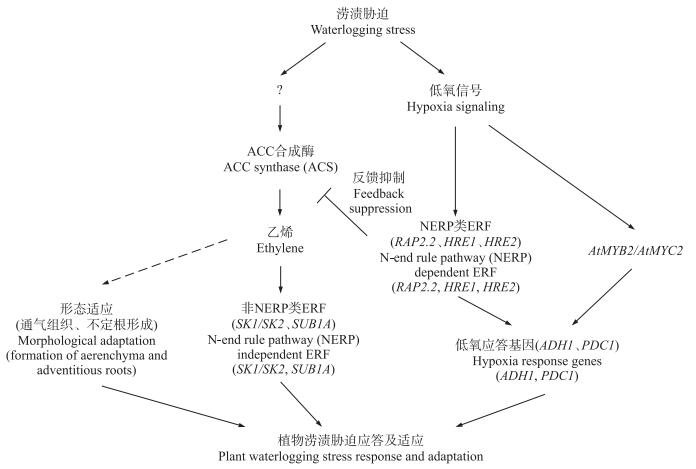
2 乙烯响应因子非NERP类ERF家族成员应答涝渍胁迫的机制已知涝渍胁迫应答中发挥重要作用的转录因子有MYB、bZIP、bHLH和ERF等[4]。ERF类转录因子家族成员均含有1个或2个保守的AP2/ERF DNA结合域[25], 是植物特有的与胁迫应答有关的转录因子超家族[26]。其中, Ⅶ ERF分为N-末端规则代谢途径(N-end rule pathway, NERP)和非NERP两类, 其调控耐涝机制的研究最为深入[27-28]。
涝渍胁迫下, 非NERP类ERF家族成员通过不同策略应答涝渍胁迫。湿生植物水稻通过Ⅶ ERF调控“耐受(quiescence)”和“逃逸(escape)”2种不同的生存策略。“逃逸”策略由Ⅶ ERF家族SNORKEL1/SNORKEL2(SK1/SK2)调控, 涝渍胁迫乙烯信号诱导SK1/SK2表达, 进而激活赤霉素(GA)途径促进水稻节间伸长生长, 从而使水稻“逃出”水淹环境[29](图 1)。“耐受”是深水水稻以减少能量消耗和抑制淹水部位生长而应对涝渍胁迫的策略, “耐受”策略由乙烯诱导Ⅶ ERF家族SUB1A-1表达来完成。SUB1A由SUB1位点编码, 该位点编码SUB1A、SUB1B和SUB1C 3个基因。目前已知SUB1A有SUB1A-1和SUB1A-2这2个等位基因, 耐淹水稻品种均含有SUB1A-1, 而不耐淹品种中则为SUB1A-2基因, 两者的区别在于第186位氨基酸分别为丝氨酸与脯氨酸[30]。耐淹水稻中, SUB1A-1受乙烯诱导表达; 在不耐水淹品种中过量表达SUB1A-1能增加其对淹水胁迫的耐性, 且SUB1C的表达受抑制, 表明SUB1A-1是水稻耐水淹的一个主效基因[31]。一方面, SUB1A-1通过诱导赤霉素信号途径抑制蛋白SLR1和SLRL1(DELLA蛋白)的表达来抑制赤霉素反应, 从而限制淹水部位的伸长生长[32]; 另一方面, SUB1A-1抑制水淹下的乙烯产生, 进而抑制淹水部位伸长生长; 此外, SUB1A-1还抑制SUB1C表达[33], 进而抑制淀粉酶活性, 减少淀粉和糖的分解, 从而使植株存活更长时间[34]。然而, 部分耐涝野生种中没有SUB1A基因, 表明存在着SUB1A以外的调控机制。此外, SUB1A-1蛋白含有N-端基序, 但并不被NERP规则降解。这些都表明植物耐涝分子机制十分复杂, 尚有待进一步研究。
|
图 1 植物耐涝性应答机制 Figure 1 Strategies for plants to cope with waterlogging |
研究表明, 涝渍土壤中氧溶解量只有空气中氧容量的1/320 000[35], 并且土壤微生物和植物根系呼吸会进一步消耗土壤中有限的氧, 导致植物氧化磷酸化依赖的生理活动受到抑制, 从而影响其生长与存活。因此, 低氧是涝渍的主要胁迫因子[36]。涝渍低氧胁迫使得供给ATP的氧化磷酸化有氧呼吸受抑制, 导致依赖于ATP的生理过程受到限制[37], 植物转而以无氧呼吸来缓解能量亏缺的状态[3]。在厌氧条件下, 植物通过乙醇发酵[丙酮酸脱羧酶(PDC)和乙醇脱氢酶(ADH)]、乳酸发酵[乳酸脱氢酶(LDH)]或丙酮酸生成丙氨酸的丙氨酸发酵[丙氨酸转氨酶(AlaAT)]途径提供能量[38]。其中, 乙醇发酵路径为涝渍低氧土壤中植物的主要能量供给路径, ADH1和PDC1为乙醇发酵路径的关键酶, 其发酵水平越高, 植物耐涝性越强[39]。
ADH1受低氧信号诱导。低氧胁迫下乙烯受体突变株ctr中ADH1基因上调表达[40]。ADH1和PDC1基因启动子区均含有低氧响应元件ARE(anaerobic response element)。ARE元件包括GC-motif及GT-motif, 若ARE元件缺失则导致低氧下该类基因不表达。拟南芥AtMYB2通过结合ADH1启动子区GT-motif(5′-GTT-3′)而诱导ADH1的表达[11]。5′-AAACC(G/A)(G/A)-3′为低氧诱导基因保守的GT-motifs, AtMYB2可以识别含有这一保守低氧应答元件的所有基因, 从而调控发酵代谢[41]。在玉米[42]和小麦[43]中均发现MYB转录因子参与了低氧的应答。Atmyc2突变株中ADH1的表达受抑制, 而共同表达AtMYB2与AtMYC2的植株在正常供氧条件下即可检测到ADH1的高表达, 表明AtMYB2对ADH1的诱导表达需要AtMYC2的协同作用[44]。由此可知, 乙醇发酵路径关键基因ADH1的表达受多种转录因子调控。
3.2 NERP类Ⅶ ERF家族成员调控低氧应答的开启与关闭NERP代谢是指具有保守N-端基序的蛋白在甲硫氨酸氨肽酶的作用下去除N-端第1位蛋氨酸(Met), 使第2位Cys暴露后被氧化(Cysox), 随后在精氨酸转移酶作用下形成Arg-Cysox复合体, 从而被泛素连接酶PRT6降解的过程[45]。拟南芥的NERP类Ⅶ ERF包括RAP2.12(AtERF75)、RAP2.2(AtERF74)、RAP2.3(AtERF72)、HRE1(AtERF73)和HRE2(AtERF71)[46-47]。
RAP2.12通过NERP代谢路径改变其亚细胞定位与稳定性, 从而开启或关闭低氧信号应答。Gibbs等[46]发现RAP2.12蛋白(NERP类蛋白)受低氧诱导, 非低氧胁迫下其与膜定位的乙酰辅酶A结合蛋白ACBP1和ACBP2互作定位于膜上; 低氧胁迫下, RAP2.12蛋白与乙酰辅酶A结合蛋白分离, 转而定位于细胞核中, 并与低氧响应基因的调控元件结合, 从而激活PDC1、ADH1等低氧响应基因的表达; 而在正常供氧下或低氧胁迫解除后, RAP2.12通过保守的N-末端规则代谢途径被泛素化降解[47](图 1)。去除RAP2.12保守的N-端基序导致该蛋白在细胞膜与细胞核均有定位, 且不再随氧浓度恢复而降解; 而将该蛋白N-端第2位的Cys替换成Ala, 其定位与去除了N-端基序后的定位相一致; 相比野生型及RAP2.12 N-端修饰的突变型, 转35S::RAP2.12的拟南芥植株表现出较强耐涝性, 表明其N-端基序(NH2-MCGGAI/L)决定了氧浓度依赖的亚细胞定位及蛋白稳定性, 从而在转录和翻译后水平调控植物的低氧应答[46-47]。
NERP类ERF蛋白(RAP2.2、ERF73/HRE1、ERF71/HRE2)调控乙醇发酵基因应答低氧胁迫, 该类蛋白的降解与否并不决定低氧应答的开启与关闭(图 1)。低氧胁迫诱导RAP2.2表达, RAP2.2超表达增加了PDC1和ADH1的表达, 植株低氧存活率高于rap2.2突变体植株; RAP2.2表达受乙烯诱导, 乙烯受体ctr及ein1、ein2、ein3突变体中低氧对RAP2.2的诱导作用下降, 相反, RAP2.2通过抑制乙烯合成相关基因ACS2、ACS7及ACS9表达而反馈抑制乙烯合成, 表明乙烯及乙烯信号参与了RAP2.2的调控[40]。RAP2.2与ATCTA元件结合[48], ADH1启动子区亦含有此元件, 但RAP2.2是否与ADH1启动子区直接结合尚缺乏可靠证据。rap2.2突变体植株中, ADH1和PDC1的表达降低不明显, 推测可能其他ERF协同参与低氧应答, RAP2.2超表达株中ERF73、ERF71、ERF4受诱导表达[40]; hre1/hre2双突变体拟南芥幼苗的低氧存活率比hre1或hre2单突变体及野生型明显降低, 组成型超表达HRE1或HRE2能迅速诱导低氧响应基因(如PDC1、ADH1等)的表达从而提高低氧耐性[40, 49]。因此, 推测ERF73、ERF71、ERF4在RAP2.2的下游起作用, 可能激活ADH1、PDC1、LDH1等基因表达。研究还发现RAP2.2超表达植株中AtMYB2表达下调, 暗示RAP2.2可通过其他转录因子调控ADH1表达[40]。可见, ADH1受乙烯信号诱导表达, 且乙烯信号路径(ERF)与低氧信号路径(AtMYB2)之间存在交互对话。
4 乙烯介导的耐涝形态适应除了前文所述的乙烯介导的叶片向上运动及茎快速生长以外, 通气组织的形成也是植物耐涝适应性反应之一[12]。涝渍胁迫时通气组织在根和叶片中形成, 在植物中形成气体通道[50-51]。植物体的通气组织不仅可以向根系输送氧, 而且也有助于有害气体如CH4、CO2、N2等的排放[52]。大量试验表明, 乙烯是通气组织形成的信号物质, 在涝渍胁迫条件下乙烯大量合成并引发一系列下游级联反应, 进而诱导程序性细胞死亡(PCD)形成通气组织[52-53]。
不定根的形成也是植物涝渍胁迫的形态适应之一。不定根可以替代涝渍损伤的根系, 促进氧向无氧环境的根中渗透。涝渍胁迫1 h后即检测到乙烯积累, 表明乙烯在ROS信号上游起作用。乙烯激发的ROS启动了不定根的生长, 从而提供给表皮一个信号, 发生PCD, 便于侧根发生[54]。菊花涝渍胁迫耐性存在基因型差异, 耐涝的菊花材料快速诱导乙烯信号合成, 从而促进PCD、通气组织形成及不定根发生, 改善低氧状况, 减少乙醇发酵产物对根系的伤害[55-56]。通气组织及不定根生成受乙烯诱导, 目前乙烯信号调控涝渍应答形成通气组织和气生根的具体分子机制尚不清楚。
5 结语与展望乙烯是响应植物涝渍胁迫的早期信号, 在涝渍胁迫适应性中起着十分重要的作用。涝渍胁迫下乙烯通过信号响应因子ERF调控植株生长、低氧胁迫应答以及涝渍形态适应等。以往有关乙烯合成调控的研究多集中在采后衰老、病原侵染引起的乙烯合成等, 而对于涝渍胁迫早期信号——乙烯的合成调控研究尚未见报道; 乙烯通过调控多种转录因子从而调控植物发育、逆境应答等多个生命活动过程, 目前乙烯调控耐涝的信号通路仍局限于ERF家族成员的研究。因此, 关于乙烯调控耐涝机制今后可在以下两方面取得突破:一方面是涝渍胁迫下乙烯合成的调控机制; 另一方面是乙烯调控耐涝性的信号通路, 特别是关键转录因子的挖掘及其调控通路的解析。
| [1] | Herzog M, Striker G G, Colmer T D, et al. Mechanisms of waterlogging tolerance in wheat:a review of root and shoot physiology[J]. Plant, Cell and Environment, 2016, 39(5): 1068-1086. DOI: 10.1111/pce.12676 |
| [2] | Phukan U J, Mishra S, Shukla R K. Waterlogging and submergence stress:affects and acclimation[J]. Critical Reviews in Biotechnology, 2016, 36(5): 956-966. DOI: 10.3109/07388551.2015.1064856 |
| [3] | Christianson J A, Llewellyn D J, Dennis E S, et al. Comparisons of early transcriptome responses to low-oxygen environments in three dicotyledonous plant species[J]. Plant Signaling and Behavior, 2010, 5(8): 1006-1009. DOI: 10.4161/psb.5.8.12231 |
| [4] | Bailey-Serres J, Fukao T, Gibbs D J, et al. Making sense of low oxygen sensing[J]. Trends in Plant Science, 2012, 17(3): 129-138. DOI: 10.1016/j.tplants.2011.12.004 |
| [5] | Booker M A, DeLong A. Producing the ethylene signal:regulation and diversification of ethylene biosynthetic enzymes[J]. Plant Physiology, 2015, 169(1): 42-50. |
| [6] | Li T, Tan D, Liu Z, et al. Apple MdACS6 regulates ethylene biosynthesis during fruit development involving ethylene-responsive factor[J]. Plant and Cell Physiology, 2015, 56(10): 1909. DOI: 10.1093/pcp/pcv111 |
| [7] | Rieu I, Cristescu S M, Harren F J M, et al. RP-ACS1, a flooding-induced 1-aminocyclopropane-1-carboxylate synthase gene of Rumex palustris, is involved in rhythmic ethylene production[J]. Journal of Experimental Botany, 2005, 56(413): 841-849. DOI: 10.1093/jxb/eri078 |
| [8] | Veen H V, Sasidharan R. Two rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms[J]. The Plant Cell, 2013, 25(11): 4691-4707. DOI: 10.1105/tpc.113.119016 |
| [9] | Zarembinski T I, Theologis A. Anaerobiosis and plant growth hormones induce two genes encoding 1-aminocyclopropane-1-carboxylate synthase in rice(Oryza sativa L.)[J]. Molecular Biology of the Cell, 1993, 4(4): 363-373. DOI: 10.1091/mbc.4.4.363 |
| [10] | Zhou Z, Vriezen W, Caeneghem W V, et al. Rapid induction of a novel ACC synthase gene in deepwater rice seedlings upon complete submergence[J]. Euphytica, 2001, 121(2): 137-143. DOI: 10.1023/A:1012059425624 |
| [11] | Lee S C, Mustroph A, Sasidharan R, et al. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia[J]. New Phytologist, 2011, 190(2): 457-471. DOI: 10.1111/j.1469-8137.2010.03590.x |
| [12] | Sasidharan R, Voesenek L A. Ethylene-mediated acclimations to flooding stress[J]. Plant Physiology, 2015, 169(1): 3-12. DOI: 10.1104/pp.15.00387 |
| [13] | Rodrigues M A, Bianchetti R E, Freschi L. Shedding light on ethylene metabolism in higher plants[J]. Frontiers in Plant Science, 2014, 5(5): 665. |
| [14] | Li G, Meng X, Wang R, et al. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis[J]. PLoS Genetics, 2012, 8(6): e1002767. DOI: 10.1371/journal.pgen.1002767 |
| [15] | Riddhi Datta D K, Asma S, Saptarshi H, et al. Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via WRKY33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress[J]. Plant Physiology, 2015, 169(4): 2963-2981. |
| [16] | Dong Z, Yu Y, Li S, et al. Abscisic acid antagonizes ethylene production through the ABI4-mediated transcriptional repression of ACS4 and ACS8 in Arabidopsis[J]. Molecular Plant, 2016, 9(1): 126-135. DOI: 10.1016/j.molp.2015.09.007 |
| [17] | Li Z, Zhang L, Yu Y, et al. The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis[J]. The Plant Journal, 2011, 68(1): 88-99. DOI: 10.1111/j.1365-313X.2011.04670.x |
| [18] | Nowak K, Wójcikowska B, Gaj M D. ERF022 impacts the induction of somatic embryogenesis in Arabidopsis through the ethylene-related pathway[J]. Planta, 2015, 241(4): 967-985. DOI: 10.1007/s00425-014-2225-9 |
| [19] | Li T, Jiang Z, Zhang L, et al. Apple(Malus domestica)MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription[J]. The Plant Journal, 2016, 88(5): 735-748. DOI: 10.1111/tpj.2016.88.issue-5 |
| [20] | Wang K L, Yoshida H, Lurin C, et al. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein[J]. Nature, 2004, 428(6986): 945. DOI: 10.1038/nature02516 |
| [21] | Christians M J, Gingerich D J, Hansen M, et al. The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in arabidopsis by controlling type-2 ACC synthase levels[J]. The Plant Journal, 2009, 57(2): 332-345. DOI: 10.1111/tpj.2009.57.issue-2 |
| [22] | Lyzenga W J, Booth J K, Stone S L. The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7[J]. The Plant Journal, 2012, 71(1): 23-34. DOI: 10.1111/tpj.2012.71.issue-1 |
| [23] | Xiong L, Xiao D, Xu X, et al. The non-catalytic N-terminal domain of ACS7 is involved in the post-translational regulation of this gene in Arabidopsis[J]. Journal of Experimental Botany, 2014, 65(15): 4397-4408. DOI: 10.1093/jxb/eru211 |
| [24] | Rauf M, Arif M, Fisahn J, et al. NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis[J]. The Plant Cell, 2013, 25(12): 4941-4955. DOI: 10.1105/tpc.113.117861 |
| [25] | Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice[J]. Plant Physiology, 2006, 140(2): 411-432. DOI: 10.1104/pp.105.073783 |
| [26] | Berens M L, Berry H M, Mine A, et al. Evolution of hormone signaling networks in plant defense[J]. Annual Review of Phytopathology, 2017, 55: 401-425. DOI: 10.1146/annurev-phyto-080516-035544 |
| [27] | Sasidharan R, Mustroph A. Plant oxygen sensing is mediated by the N-end rule pathway:a milestone in plant anaerobiosis[J]. The Plant Cell, 2011, 23(12): 4173-4183. DOI: 10.1105/tpc.111.093880 |
| [28] | Bailey-Serres J, Lee S C, Brinton E. Waterproofing crops:effective flooding survival strategies[J]. Plant Physiology, 2012, 160(4): 1698-1709. DOI: 10.1104/pp.112.208173 |
| [29] | Hattori Y, Nagai K, Furukawa S, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water[J]. Nature, 2009, 460(7258): 1026-1030. DOI: 10.1038/nature08258 |
| [30] | Niroula R K, Pucciariello C, Ho V T, et al. SUB1A-dependent and-independent mechanisms are involved in the flooding tolerance of wild rice species[J]. The Plant Journal, 2012, 72(2): 282-293. DOI: 10.1111/j.1365-313X.2012.05078.x |
| [31] | Septiningsih E M, Hidayatun N, Sanchez D L, et al. Accelerating the development of new submergence tolerant rice varieties:the case of Ciherang-Sub1 and PSB Rc18-Sub1[J]. Euphytica, 2015, 202(2): 259-268. DOI: 10.1007/s10681-014-1287-x |
| [32] | Fukao T, Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice[J]. Proc Natl Acad Sci USA, 2008, 105(43): 16814-16819. DOI: 10.1073/pnas.0807821105 |
| [33] | Peña-Castro J M, van Zanten M, Lee S C, et al. Expression of rice SUB1A and SUB1C transcription factors in arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism[J]. The Plant Journal, 2011, 67(3): 434-446. DOI: 10.1111/j.1365-313X.2011.04605.x |
| [34] | Jung K H, Seo Y S, Walia H, et al. The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors[J]. Plant Physiology, 2010, 152(3): 1674-1692. DOI: 10.1104/pp.109.152157 |
| [35] | Armstrong W, Drew M. Root growth and metabolism under oxygen deficiency[M]//Waisel Y, Eshel A, Kafkafi U. Plant Roots: the Hidden Half. New York: Marcel Dekker, 2002: 729-761. |
| [36] | Voesenek L, Bailey-Serres J. Flooding tolerance:O2 sensing and survival strategies[J]. Current Opinion in Plant Biology, 2013, 16: 647-653. DOI: 10.1016/j.pbi.2013.06.008 |
| [37] | Bailey-Serres J, Voesenek L. Flooding stress:acclimations and genetic diversity[J]. Annual Review of Plant Biology, 2008, 59: 313-339. DOI: 10.1146/annurev.arplant.59.032607.092752 |
| [38] | Ismond K P, Dolferus R, de Pauw M, et al. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway[J]. Plant Physiology, 2003, 132(3): 1292-1302. DOI: 10.1104/pp.103.022244 |
| [39] | Sairam R, Kumutha D, Ezhilmathi K, et al. Physiology and biochemistry of waterlogging tolerance in plants[J]. Biologia Plantarum, 2008, 52(3): 401-412. DOI: 10.1007/s10535-008-0084-6 |
| [40] | Hinz M, Wilson I W, Yang J, et al. Arabidopsis RAP2.2:an ethylene response transcription factor that is important for hypoxia survival[J]. Plant Physiology, 2010, 153(2): 757-772. DOI: 10.1104/pp.110.155077 |
| [41] | Hoeren F U, Dolferus R, Wu Y, et al. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene(ADH1)by low oxygen[J]. Genetics, 1998, 149(2): 479-490. |
| [42] | Mattana M, Vannini C, Espen L, et al. The rice Mybleu transcription factor increases tolerance to oxygen deprivation in Arabidopsis plants[J]. Physiologia Plantarum, 2007, 131(1): 106-121. DOI: 10.1111/ppl.2007.131.issue-1 |
| [43] | Lee T G, Jang C S, Kim J Y, et al. A Myb transcription factor(TaMyb1)from wheat roots is expressed during hypoxia:roles in response to the oxygen concentration in root environment and abiotic stresses[J]. Physiologia Plantarum, 2007, 129(2): 375-385. |
| [44] | Abe H, Urao T, Ito T, et al. Arabidopsis AtMYC2(bHLH)and ATMYB2(MYB)function as transcriptional activators in abscisic acid signaling[J]. The Plant Cell, 2003, 15(1): 63-78. DOI: 10.1105/tpc.006130 |
| [45] | Varshavsky A. The N-end rule pathway and regulation by proteolysis[J]. Protein Science, 2011, 20(8): 1298-1345. DOI: 10.1002/pro.666 |
| [46] | Gibbs D J, Lee S C, Isa N M, et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants[J]. Nature, 2011, 479(7373): 415-418. DOI: 10.1038/nature10534 |
| [47] | Licausi F, Kosmacz M, Weits D A, et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization[J]. Nature, 2011, 479(7373): 419-422. DOI: 10.1038/nature10536 |
| [48] | Welsch R, Maass D, Voegel T, et al. Transcription factor RAP2. 2 and its interacting partner SINAT2:stable elements in the carotenogenesis of Arabidopsis leaves[J]. Plant Physiology, 2007, 145(3): 1073-1085. DOI: 10.1104/pp.107.104828 |
| [49] | Hess N, Klode M, Anders M, et al. The hypoxia responsive transcription factor genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis are differentially regulated by ethylene[J]. Physiologia Plantarum, 2011, 143(1): 41-49. DOI: 10.1111/ppl.2011.143.issue-1 |
| [50] | Vartapetian B. Plant anaerobic stress as a novel trend in ecological physiology, biochemistry, and molecular biology:2. Further development of the problem[J]. Russian Journal of Plant Physiology, 2006, 53(6): 711-738. DOI: 10.1134/S102144370606001X |
| [51] | Steffens B, Geske T, Sauter M. Aerenchyma formation in the rice stem and its promotion by H2O2[J]. New Phytologist, 2011, 190(2): 369-378. DOI: 10.1111/j.1469-8137.2010.03496.x |
| [52] |
孔妤, 王忠, 顾蕴洁, 等. 乙烯利诱导水稻根内通气组织形成的研究[J].
中国水稻科学, 2009, 23(1): 65-70.
Kong Y, Wang Z, Gu Y J, et al. Induction of ethephon on aerenchyma formation in rice roots[J]. Chinese Journal of Rice Science, 2009, 23(1): 65-70. (in Chinese with English abstract) |
| [53] |
马月花, 郭世荣, 杜南山, 等. 低氧胁迫对黄瓜幼苗生长和形态结构及有关酶活性的影响[J].
南京农业大学学报, 2016, 39(2): 213-219.
Ma Y H, Guo S R, Du N S, et al. Effect of hypoxia stress on growth, morpho-anatomical acclimation and activity of involved enzymes of cucumber seedlings[J]. Journal of Nanjing Agricultural University, 2016, 39(2): 213-219. DOI: 10.7685/jnau.201506001 (in Chinese with English abstract) |
| [54] | Steffens B, Kovalev A, Gorb S N, et al. Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling[J]. The Plant Cell, 2012, 24(8): 3296-3306. DOI: 10.1105/tpc.112.101790 |
| [55] | Yin D, Chen S, Chen F, et al. Morphological and physiological responses of two chrysanthemum cultivars differing in their tolerance to waterlogging[J]. Environmental and Experimental Botany, 2009, 67(1): 87-93. DOI: 10.1016/j.envexpbot.2009.06.006 |
| [56] | Yin D, Chen S, Chen F, et al. Morpho-anatomical and physiological responses of two Dendranthema species to waterlogging[J]. Environmental and Experimental Botany, 2010, 68(2): 122-130. DOI: 10.1016/j.envexpbot.2009.11.008 |




