文章信息
- 毕研飞, 徐兵划, 郭静, 张永兵, 伊鸿平, 钱春桃, 陈劲枫. 2015.
- BI Yanfei, XU Binghua, GUO Jing, ZHANG Yongbing, YI Hongping, QIAN Chuntao, CHEN Jinfeng. 2015.
- 分子标记辅助甜瓜抗蔓枯病基因聚合及‘白皮脆’品种改良
- Pyramiding disease resistance genes by marker-assisted selection in melon(Cucumis melo L.)and'Baipicui’breed improvement
- 南京农业大学学报, 38(3): 375-380
- Journal of Nanjing Agricultural University, 38(3): 375-380.
- http://dx.doi.org/10.7685/j.issn.1000-2030.2015.03.004
-
文章历史
- 收稿日期:2014-08-20
2. 新疆农业科学院哈密瓜研究中心, 新疆 乌鲁木齐 830091
2. Center of Hami Melon, Xinjiang Academy of Agricultural Sciences, Urumqi 830091, China
甜瓜(Cucumis melo L.)果实香甜,口感优良,深受广大消费者的喜爱。然而,蔓枯病(gummy stem blight)的发生已成为限制甜瓜优质高产的主要因素之一[1, 2, 3, 4]。目前,甜瓜生产上防治蔓枯病的常用方法主要是化学防治,但其效率低而且易引起环境污染。因此,选育甜瓜抗病品种(品系)具有重要的经济价值和生态价值。
国际上已经鉴定并报道了甜瓜蔓枯病抗源PI140471,其抗性基因为Gsb-1 [5],抗源PI420145是由本课题组Joseph等[6]筛选得到的,并命名为Gsb-6 。本课题组前期通过对抗病基因的研究,开发了分别与抗蔓枯病基因Gsb-1和Gsb-6连锁的SSR标记CMCT505和SCAR标记SGSB1800。刘龙洲等[7]以甜瓜JGD-3为抗源鉴定蔓枯病抗性属于多基因控制的数量遗传性状,并发现了与基因位点紧密连锁的3个分子标记CMTCN66、E3S7-1和TJ27。这些分子标记的开发为甜瓜抗蔓枯病育种奠定了良好的基础。
目前抗蔓枯病的甜瓜品种仅携带单个抗病基因(Gsb-1,Gsb-2,Gsb-3,Gsb-4,Gsb-5或Gsb-6),但随着栽培环境的变化,品种的抗性逐渐降低甚至丢失[8, 9, 10]。国内外研究表明,不同抗性基因的聚合有助于提高作物的抗性和抗谱[11, 12, 13],因此,选育高抗聚合基因材料是提高甜瓜蔓枯病抗性的有效途径之一。本文结合苗期人工接种鉴定、分子标记选择和田间抗性统计,初步探究了不同基因聚合后的抗性差异,并选育获得了高抗蔓枯病且商品性优异的改良‘白皮脆’品种(系),为甜瓜优质、抗病和高产育种提供新的遗传资源。 1 材料与方法 1.1 甜瓜抗源材料和接种菌株
甜瓜单基因抗源材料PI140471(含有抗病基因Gsb-1 )和PI420145(含有抗病基因Gsb-6 ),其中PI140471由美国康乃尔大学Molly John教授提供,PI420145是由本课题组的Joseph等筛选得到。聚合基因材料471-145由PI140471与PI420145杂交获得。优质、感病品种‘白皮脆’由新疆农业科学院提供。
蔓枯病菌为本实验室分离纯化并保存的A和A1型菌株,菌株的分生孢子在PDA培养基上经25℃黑暗培养7 d后,再进行4 d间歇紫外灯(12 h紫外/12 h黑暗)处理产生,在显微镜下利用血球计数板将分生孢子悬浮液分别配成5×105、5×107和5×109 mL-1备用。 1.2 苗期接种鉴定
参照Zhang等[9]的方法在苗期接种(3~4片真叶),用微型喷雾器喷洒孢子悬浮液,喷到植株叶片开始滴水为止。接种后用塑料拱棚保湿,相对湿度90%以上,3 d后揭开小拱棚,于7和10 d调查统计病情。叶片侵染分级标准为:0级,无可见侵染;1级,老叶上边缘坏死或斑点<10 mm,新叶无病;2级,老叶同上,新叶边缘坏死;3级,所有叶均有感染,叶坏死面积<25%;4级,25%≤叶坏死面积≤50%;5级,叶坏死面积>50%。根据平均病级(RI)确定蔓枯病抗性级别,高抗(HR):RI<1.0;抗(R):1.0≤RI<2.0;中抗(MR):2.0≤RI<3.0;感(S):3.0≤RI<4.0;高感(HS):RI≥4.0。平均病级计算公式:RI=∑(级值×株数)/总株数。 1.3 DNA提取与分子标记检测
甜瓜DNA的提取参照CTAB法。用于检测抗病基因Gsb-1和Gsb-6的分子标记为本课题组设计,由上海英潍捷基贸易有限公司合成(表 1),扩增产物大小分别为190和1 800 bp。PCR总反应体积为20 μL:ddH2O 12.1 μL,10×PCR Buffer 2.0 μL,2 mmol · L-1 dNTP 1.5 μL,25 mmol · L-1MgCL2 1.2 μL,10 μmol · L-1 3′和5′引物各1.0 μL,5 U · μL-1Taq聚合酶0.2 μL,30 ng · μL-1模板DNA 1.0 μL。扩增反应条件为:94℃ 5 min;94℃ 30 s,55℃ 30 s,72℃ 80 s,35个循环;72℃ 5 min,保持4℃。抗病基因Gsb-1的PCR产物在90 g · L-1聚丙烯酰胺凝胶上电泳,采用改良的银染方法检测。抗病基因Gsb-6的PCR产物用10 g · L-1琼脂糖凝胶电泳检测。
| 基因Gene | 引物Primer | 引物序列Primer sequence | 连锁距离/cMDistance |
| Gsb-1 | CMCT505 | F:5′-GACAGTAATCACCTCATCAAC-3′ R:5′-GGGAATGTAAATTGGATATG-3′ | 5.2 |
| Gsb-6 | SGSB1800 | F:5′-GTTGCGTTCTCTGCTTGGA-3′ R:5′-AGGTAATTGAGGTGTCGTCTTA-3′ | 2.0 |
通过单一抗源PI140471(Gsb-1 )与PI420145(Gsb-6 )杂交,并结合苗期蔓枯病菌梯度接种鉴定与分子标记(CMCT505、SGSB1800)进行筛选,获得聚合2个抗蔓枯病基因的聚合抗源471-145。以表现高抗的聚合抗源471-145为父本,品质优异的感病品种‘白皮脆’为母本进行杂交获得F1,再以‘白皮脆’为轮回亲本进行连续回交3代自交2代,结合苗期接种鉴定与分子标记辅助选择筛选表现高抗且含有2个抗病基因的各世代进行品种改良。 1.5 田间抗性观察与果实品质测定
2014年春季对回交世代BC3F3进行田间蔓枯病抗性观察与果实品质测定。材料种植于南京农业大学江浦实验基地塑料大棚内(甜瓜重茬地,蔓枯病发严重),株距60 cm,行距120 cm,采用吊蔓栽培的方式,单蔓整枝,施肥、排灌、除草等按常规管理。果实品质测定以‘白皮脆’和聚合抗源471-145的果实性状为对照,选择9个果实进行品质测定。单果质量为9个果实的平均值;果实脆度测定采用马庆华等[14]的方法,用英国Stable Micro Systems公司生产的TAXT plus质构仪,采用P/2n针状探头(直径2 mm),测前速度2 mm · s-1,贯入速度1 mm · s-1,测后速度5 mm · s-1,最小感知力为10 g;果肉颜色利用CR-400型色差仪测定;将果实打成汁后,利用糖度计测果实可溶性固形物含量;采用分光光度计法测定维生素C含量。 1.6 数据分析
采用SPSS 19.0软件对数据进行统计分析。 2 结果与分析 2.1 聚合材料抗性基因分子标记的鉴定
利用本课题组创建的与抗病基因Gsb-1和Gsb-6紧密连锁的SSR标记CMCT505和SCAR标记SGSB1800筛选471-145聚合单株,含有基因Gsb-1的材料可以扩增出190 bp的特异性片段(图 1-A),而含有基因Gsb-6的材料可以扩增出1 800 bp的特异性片段(图 1-B)。因此,聚合基因471-145单株可以同时扩增出190和1 800 bp 2条特异性条带。
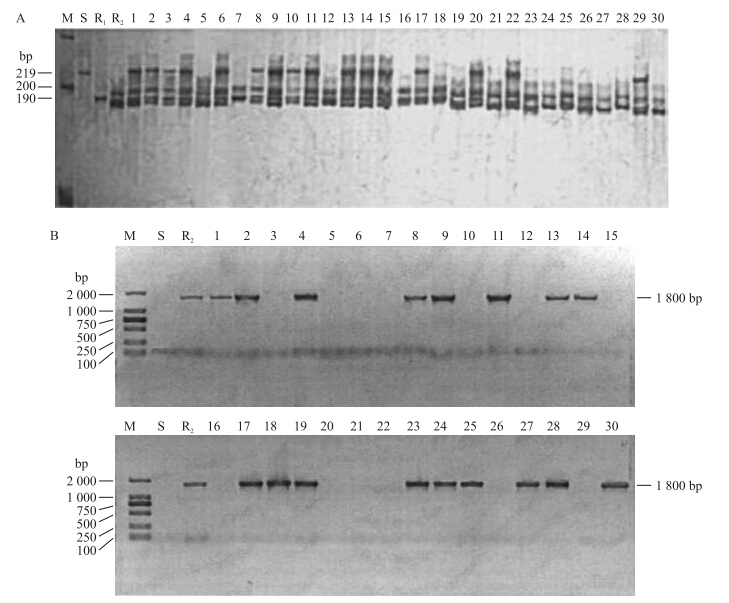 |
图 1 PCR检测BC3F3部分单株的抗病基因 Fig. 1 PCR detection of resistance genes in parts of BC3F3 individuals
1)A.CMCT505标记检测BC3F3部分单株含抗病基因Gsb-1的结果The result of detection about resistance gene Gsb-1 in parts of BC3F3 individuals by marker CMCT505;B.SGSB1800标记检测BC3F3部分单株含抗病基因Gsb-6的结果The result of detection about resistance gene Gsb-6 in parts of BC3F3 individuals by marker SGSB1800. 2)M:标准Marker;S:白皮脆Baipicui;R1:PI140471;R2:PI420145;1~30:BC3F3 3)编号2、4、8、17、18、24、25和28可扩增出190和1 800 bp两条特异性条带,为聚合单株。Number 2,4,8,17,18,24,25 and 28 were pyramided individuals which could amplify two bands of 190 and 1 800 bp. |
由表 2可见:A型菌株接种浓度为5×105 mL-1时,回交亲本‘白皮脆’的平均病级RI为3.60,表现为感病,单基因抗源材料和聚合基因材料BC3F3均表现为高抗;当接种浓度为5×107 mL-1时,‘白皮脆’平均病级RI为4.50,表现为高感,单基因抗源PI140471和聚合基因材料BC3F3的平均病级RI分别为1.90和1.66,表现为抗,PI420145的RI为2.30,表现中抗;当接种浓度为5×109 mL-1时,‘白皮脆’平均病级RI为4.90,表现高感,单基因抗源PI140471和PI420145均表现为感病,而聚合基因材料BC3F3的平均病级RI为2.70,表现中抗。
| 菌株Strain | 材料Material | 5×105 mL-1 | 5×107 mL-1 | 5×109 mL-1 | |||
| RI | 抗性Resistance | RI | 抗性Resistance | RI | 抗性Resistance | ||
| A | 白皮脆Baipicui | 3.60a | S | 4.50a | HS | 4.90a | HS |
| PI140471 | 0.00d | HR | 1.90d | R | 3.10c | S | |
| PI420145 | 0.12d | HR | 2.30c | MR | 3.30b | S | |
| BC3F3 | 0.75c | HR | 1.66e | R | 2.70d | MR | |
| A1 | 白皮脆Baipicui | 3.40b | S | 4.20b | HS | 4.80a | HS |
| PI140471 | 0.00d | HR | 1.84d | R | 2.81d | MR | |
| PI420145 | 0.00d | HR | 1.92d | R | 3.10c | S | |
| BC3F3 | 0.69c | HR | 1.58e | R | 2.60d | MR | |
| 注:1)同一列内不同小写字母表示差异性显著(P<0.05)。2)HR:高抗;R:抗;MR:中抗;S:感病;HS:高感。 Note: 1)Values followed by different lowercase letters are significant differentce at 0.05 level.2)HR:High resistance;R:Resistance;MR:Moderate resistance;S:Susceptible;HS:High sensitive.The same as follows. | |||||||
A1型菌株接种浓度为5×105 mL-1时,‘白皮脆’平均病级RI为3.40,表现为感病,单基因抗源和聚合基因材料BC3F3均表现为高抗;当接种浓度为5×107 mL-1时,‘白皮脆’平均病级RI为4.20,表现为高感,单基因抗源和聚合基因材料均表现为抗;当接种浓度为5×109 mL-1时,‘白皮脆’平均病级RI为4.80,表现为高感,单基因抗源PI140471的RI为2.81,表现中抗,PI420145的RI为3.10,表现感病,而聚合基因材料BC3F3的平均病级RI为2.60,表现为中抗。 2.3 田间抗性观察与果实品质测定
由表 3可以看出:改良‘白皮脆’聚合了双亲的优良性状,田间表现高抗甜瓜蔓枯病,除果实维生素C含量高于‘白皮脆’外,在果型、单果质量、果实脆度、果肉厚度及可溶性固形物含量方面与回交亲本‘白皮脆’并无显著差异。
| 材料Material | 抗性Resistance | 果型Fruitshape | 单果质量/kgFruit weight | 果实脆度/(kg·cm-2)Fruit crispness | 果肉颜色Fleshcolor | 果肉厚度/cmFleshthickness | 果肉质地Fleshtexture | 可溶性固形物含量/%Soluble solidcontent | 维生素C含量/(mg·100 g-1)Vitamin C content |
| 白皮脆Baipicui | S | 橄榄型Oval | 1.50±0.216 | 1.14±0.082 | 橙色Orange | 3.21±0.135 | 脆Fragile | 12.10±0.516 | 10.31±0.495 |
| 471-145 | HR | 圆柱型Cylindrical | 0.61±0.107 | 0.75±0.053 | 白色White | 1.50±0.101 | 软Soft | 5.32±0.185 | 13.82±0.570 |
| BC3F3 | HR | 橄榄型Oval | 1.33±0.190 | 1.06±0.071 | 浅橙色Light orange | 3.10±0.124 | 脆Fragile | 10.91±0.460 | 12.49±0.513 |
研究表明,利用与抗病基因紧密连锁的分子标记辅助聚合多个抗性基因,可以提高选择的准确性,是培育具有持久抗性和综合抗性品种(系)的有效策略[15, 16, 17, 18]。目前,分子标记辅助育种研究在水稻、玉米和番茄等作物上已有较多的报道,但在甜瓜抗蔓枯病育种方面还鲜有报道,主要是因为与抗性基因紧密连锁的分子标记还比较少。
利用分子标记辅助选择抗病基因的可靠性主要取决于连锁程度,标记与基因连锁的越紧密,辅助选择的准确性就越高[19, 20, 21]。本研究所利用的分子标记CMCT505和SGSB1800是由本课题组创建的,与抗病基因Gsb-1和Gsb-6的遗传连锁距离分别为5.2和2.0 cM,理论上可以用来准确检测甜瓜所含有的抗蔓枯病基因。另外,从抗性鉴定结果分析表明,改良‘白皮脆’材料均因携带抗病基因Gsb-1和Gsb-6而提高了对蔓枯病的抗性水平,说明分子标记CMCT505和SGSB1800可以准确检测Gsb-1和Gsb-6两个抗性基因。为提高分子标记辅助聚合育种的效率和准确度,理想的方法是选用目标基因内标记,但要求对基因已经精细定位甚至克隆[22, 23]。本课题组对甜瓜不同抗蔓枯病基因正在进一步的研究中,希望可以在抗性基因两侧找到更加紧密连锁的分子标记,完善检测体系,提高分子标记辅助甜瓜抗蔓枯病聚合育种的效率。
传统的甜瓜抗蔓枯病苗期接种鉴定孢子液浓度为5×105 mL-1,可以有效区分出抗、感材料的差异,但不能准确区分抗性材料之间的差异[9, 24]。因此,本试验采用不同梯度浓度接种法,以准确鉴定单基因抗源和聚合基因材料抗性能力的差异。结果显示:单基因抗源PI140471和PI420145对不同蔓枯病菌表现出一定程度的选择性抗性,而聚合基因材料却一直表现为抗病,说明抗病基因Gsb-1和Gsb-6的聚合可以提高甜瓜对蔓枯病的抗性和抗谱,这与Matsumoto等[12]在黄瓜上和朱明涛等[13]在番茄上的研究结果相一致。甜瓜蔓枯病抗性是由多基因控制的数量性状,并受其他微效基因修饰增加作用贡献[7, 8],果实形状及果肉颜色受主要基因控制,表现出较高的遗传力,受环境影响较小[25, 26, 27, 28]。因此,对甜瓜品种进行蔓枯病抗性改良时,在早代对蔓枯病抗性及果实性状可进行定向选择,对表现高抗且果实性状与轮回亲本一致或相近的单株继续回交或自交留种。本研究在BC3F3世代即获得蔓枯病抗性显著提高且果实性状良好的改良‘白皮脆’材料,对甜瓜抗蔓枯病品种的选育具有重要的意义。
| [1] | Keinath A P, Farnham M W, Zitter T A. Morphological, pathological, and genetic differentiation of Didymella bryoniae and Phoma spp.isolated from cucurbits[J]. Phytopathology, 1995, 85(3):364-369 |
| [2] | 吴明珠, 伊鸿平, 冯炯鑫, 等. 哈密瓜南移东进生态育种与有机生态型无土栽培技术研究[J]. 中国工程科学, 2000, 2(8):83-88[Wu M Z, Yi H P, Feng J X, et al. Ecobreeding of hamimelon and soilless cultivation of organic ecotype[J]. Engineering Science, 2000, 2(8):83-88(in Chinese with English abstract)] |
| [3] | 陈秀蓉, 魏永良, 张建文. 甜瓜蔓枯病菌及其生物学特性的研究[J]. 甘肃农业大学学报, 1993, 28(1):56-61[Chen X R, Wei Y L, Zhang J W. Studies on the muskmelon black rot fungus and its biological characters[J]. Journal of Gansu Agricultural University, 1993, 28(1):56-61(in Chinese with English abstract)] |
| [4] | Tsutsumi C Y, Silva N. Screening of melon populations for resistance to Didymella bryonia [J]. Braz Arch Biol Technol, 2004, 47(2):171-177 |
| [5] | Frantz J D, Jahn M M. Five independent loci each control monogenic resistance to gummy stem blight in melon( Cucumis melo L.)[J]. Theor Appl Genet, 2004, 108(6):1033-1038 |
| [6] | Joseph N W, Zhou X H, Chen J F. Identification of amplified fragment length polymorphism markers linked to gummy stem blight( Didymella bryoniae )resistance in melon( Cucumis melo L.)PI420145[J]. HortScience, 2009, 44(1):32-34 |
| [7] | 刘龙洲, 翟文强, 陈亚丽, 等. 甜瓜蔓枯病抗性QTL定位的研究[J]. 果树学报, 2013, 30(5):748-752[Liu L Z, Zhai W Q, Chen Y L, et al. QTL molecular marker location of gummy stem blight resistance in melon( Cucumis melo )[J]. Journal of Fruit Science, 2013, 30(5):748-752(in Chinese with English abstract)] |
| [8] | Keinath A P, Holmes G J, Everts K L, et al. Evaluation of combinations of chlorothalonil with azoxystrobin, harpin, and disease forecasting for control of downy mildew and gummy stem blight on melon[J]. Crop Protection, 2007, 26:83-88 |
| [9] | Zhang Y P, Molly K, Anagnostou K. Screening melon for resistance to gummy stem blight in the greenhouse and field[J]. HortScience, 1997, 32(1):117-121 |
| [10] | Wolukau J N, Zhou X H, Li Y, et al. Resistance to gummy stem blight in melon( Cucumis melo L.)germplasm and inheritance of resistance from plant introductions 157076, 420145, and 323498[J]. HortScience, 2007, 42(2):215-221 |
| [11] | 邹明学, 许勇, 张海英, 等. 葫芦科瓜类作物分子标记辅助育种研究进展[J]. 生物技术通报, 2007(4):72-78[Zou M X, Xu Y, Zhang H Y, et al. Progress in molecular marker-assisted breeding of Cucurbitaceae[J]. Biotechnology Bulletin, 2007(4):72-78(in Chinese with English abstract)] |
| [12] | Matsumoto Y, Watanabe N, Kuboyama T, et al. Cross-species transferability of 86 cucumber( Cucumis sativus L.)microsatellite markers to gherkin( C.anguria L.)[J]. Scientia Horticulturae, 2012, 136(4):110-114 |
| [13] | 朱明涛, 孙亚林, 郑莎, 等. 分子标记辅助聚合番茄抗病基因育种[J]. 园艺学报, 2010, 37(9):1416-1422[Zhu M T, Sun Y L, Zheng S, et al. Pyramiding disease resistance genes by molecular marker-assisted selection in tomato[J]. Acta Horticulturae Sinica, 2010, 37(9):1416-1422(in Chinese with English abstract)] |
| [14] | 马庆华, 王贵禧, 梁丽松. 质构仪穿刺试验检测冬枣质地品质方法的建立[J]. 中国农业科学, 2011, 44(6):1210-1217[Ma Q H, Wang G X, Liang L S. Establishment of the detecting method on the fruit texture of Dongzao by puncture test[J]. Scientia Agricultura Sinica, 2011, 44(6):1210-1217(in Chinese with English abstract)] |
| [15] | 岳欢, 吴星波, 郝俊杰, 等. 黄瓜抗白粉病分子育种研究现状与展望[J]. 植物遗传资源学报, 2014, 15(1):120-128[Yu H, Wu X B, Hao J J, et al. Status and prospects in molecular breeding of powdery mildew resistance in cucumber[J]. Journal of Plant Genetic Resources, 2014, 15(1):120-128(in Chinese with English abstract)] |
| [16] | Behera T K, Staub J E, Behera S, et al. Marker-assisted backcross selection in an interspecific cucumis population broadens the genetic base of cucumber( Cucumis sativus L.)[J]. Euphytica, 2011, 178(2):261-272 |
| [17] | Joobeur T, Nolin S J, Dean R A, et al. The fusarium wilt resistance locus Fom-2 of melon contains a single resistace gene with complex features[J]. Plant Journal, 2004, 39:283-297 |
| [18] | 许勇, 张海英, 康国斌, 等. 西瓜抗枯萎病育种分子标记辅助选择的研究[J]. 遗传学报, 2000, 27(2):151-157[Xu Y, Zhang H Y, Kang G B, et al. Studies of molecular marker-assisted-selection for resistance to Fusarium wilt in watermelon( Citrullus lanatus )breeding[J]. Acta Genetica Sinica, 2000, 27(2):151-157(in Chinese with English abstract)] |
| [19] | Francisco J, Palomares-Rius M A, Viruel F J, et al. Simple sequence repeat markers linked to QTL for resistance to watermelon mosaic virus in melon[J]. Theor Appl Genet, 2011, 123:1207-1214 |
| [20] | Zalapa J E, Staub J E, McCreight J D, et al. Detection of QTL for yield-related traits using re-combinant inbred lines derived from exotic and elite US western shipping melon germplasm[J]. Theor Appl Genet, 2007, 114(7):1185-1201 |
| [21] | 毕研飞, 徐兵划, 钱春桃, 等. 甜瓜抗蔓枯病育种研究进展[J]. 中国蔬菜, 2013(20):10-16[Bi Y F, Xu B H, Qian C T, et al. Research progress in melon resistance breeding to gummy stem blight[J]. China Vegetables, 2013(20):10-16(in Chinese with English abstract)] |
| [22] | Perin C, Hagen L, Katzir N, et al. A reference map of Cucumis melo based on two recombinant inbred line populations[J]. Theoretical and Applied Genetics, 2002, 104(617):1017-1034 |
| [23] | Yuan X J, Pan J S, Cai R, et al. Genetic mapping and QTL analysis of fruit and flower related traits in cucumber( Cucumis sativus L.)using recombinant inbred lines[J]. Euphytica, 2008, 164(2):473-491 |
| [24] | 徐兵划, 钱春桃, 王红英, 等. 甜瓜蔓枯病抗性聚合材料中防卫基因的表达分析[J]. 南京农业大学学报, 2014, 37(5):63-68. doi:10.7685/j.issn.1000-2030.2014.05.010[Xu B H, Qian C T, Wang H Y, et al. The expression analysis of defense genes in the genes pyramided melon( Cucumis melo L.)resistance to gummy stem blight[J]. Journal of Nanjing Agricultural University, 2014, 37(5):63-68(in Chinese with English abstract)] |
| [25] | 杨光华, 范荣, 杨小锋, 等. 甜瓜果实颜色3个质量性状基因的定位[J]. 园艺学报, 2014, 41(5):898-906[Yang G H, Fan R, Yang X F, et al. Construction of a highly dense genetic map using SNP and mapping of three qualitative traits in Cucumis melo [J]. Acta Horticulturae Sinica, 2014, 41(5):898-906(in Chinese with English abstract)] |
| [26] | 张宁, 张显, 张勇, 等. 甜瓜远缘群体果实性状遗传分析[J]. 西北农业学报, 2014, 23(5):121-128[Zhang N, Zhang X, Zhang Y, et al. Genetic analysis of fruit traits in interspecific population of melon[J]. Acta Agriculturae Boreali-occidentalis Sinica, 2014, 23(5):121-128(in Chinese with English abstract)] |
| [27] | Harel-Beja R, Tzuri G, Portnoy V, et al. A genetic map of melon highly enriched with fruit quality QTLs and EST markers, including sugar and carotenoid metabolism genes[J]. Theor Appl Genet, 2010, 121:511-533 |
| [28] | Esteras C, Formisano G, Roig C, et al. SNP genotyping in melons:genetic variation, population structure, and linkage disequilibrium[J]. Theor Appl Genet, 2013, 126(5):1285-1303 |




