文章信息
- 张薇, 崔为体, 段星亮, 汪瑾, 沈文飚, 谢彦杰. 2015.
- ZHANG Wei, CUI Weiti, DUAN Xingliang, WANG Jin, SHEN Wenbiao, XIE Yanjie. 2015.
- UV-B胁迫下紫花苜蓿qRT-PCR内参基因的筛选
- Reference gene selection for qRT-PCR normalization in alfalfa under UV-B irradiation
- 南京农业大学学报, 38(2): 248-254
- Journal of Nanjing Agricultural University, 38(2): 248-254.
- http://dx.doi.org/10.7685/j.issn.1000-2030.2015.02.011
-
文章历史
- 收稿日期:2014-05-20
2. 南京农业大学生物学实验教学中心, 江苏 南京 210095
2. Biological Experiment Teaching Center, Nanjing Agricultural University, Nanjing 210095, China
在生物合成、代谢和降解速率的研究中,判定相关代谢通路中关键调控基因的表达量非常重要。通常有2种手段研究相关基因表达量:RNA印迹(Northern blotting)和反转录多聚酶链式反应(RT-RCR)。RNA印迹是通过溴乙锭染色或者rRNA探针杂交进行定量,通常用于检测表达水平较高的基因。实时定量RT-PCR(qRT-PCR)因为具有灵敏度高、特异性强、重复性好等特点,因此应用范围更加广泛[1, 2]。因此,qRT-PCR仍然是研究基因表达量过程中接受度最高的方法之一。
qRT-PCR过程中,通常需要选取表达稳定的内参基因对待检测基因进行标准化,因此内参基因的选择变得尤为重要。近几年研究表明,几乎不存在表达量一直稳定的基因[2],常用的内参基因有Actin 2、GAPDH、18 S RNA、β-tublin和UBQ等[3]。其中:Actin 2 编码细胞骨架结构蛋白[4];GADPH为糖酵解过程中的关键酶[5]; 18 S RNA与胞质核糖体小亚基及翻译相关[6];β-tublin参与细胞生长[7];UBQ与蛋白质修饰、结合和降解有关[8]。一般来说,在不同的细胞、组织,不同的生理阶段,它们的表达量可能存在较大差异[9]。因此在试验过程中,通常由所研究的对象和需要来寻找适合的特异性稳定表达的内参基因。
迄今已对拟南芥、土豆、番茄、花生和香蕉等在不同的胁迫(如重金属、缺铁等)以及不同的生长阶段(如成熟期等)进行内参基因的筛选[2, 10, 11, 12, 13, 14, 15]。辐射条件下对微藻内参基因筛选的结果表明,β-actin和GAPDH可以作为双内参,而 18 S RNA稳定性则较差[16]。紫花苜蓿(Medicago Sativa L.)由于蛋白质含量高而作为优质牧草在全世界广泛种植。研究紫花苜蓿对环境适应的机制具有重要的农业价值。目前,臭氧层空洞使植物面临UV-B辐射的威胁增强。研究UV-B辐射胁迫下紫花苜蓿内参基因的筛选,对 探究紫花苜蓿逆境适应变化过程中的关键调控基因具有重要意义,也对如何提高牧草种植的产量提供一定理论基础。 1 材料与方法 1.1 材料培养
本试验所用紫花苜蓿为商业品种‘维多利亚’,种子使用前经5% NaClO表面消毒10 min,随后蒸馏水冲洗15 min,然后于25 ℃的培养箱中黑暗培养1 d。选择长势一致的幼苗转移到塑料盒中,每2 d更换1/4 Hoagland营养液,pH 6.0。光/暗培养时间为14 h/10 h,光照强度为200 μmol · m-2 · s-1,培养温度为(25±1)℃。培养14 d后,进行10.8 kJ · m-2强度UV-B处理,分别在处理后0、1、2和5 d时对根、茎和叶进行取样。 1.2 方法
以Trizol(Invitrogen,USA)提取RNA,通过分光光度计(NanoDrop 2000,Thermo Scientific,USA)测定RNA质量,选取A260/A280为1.9~2.1的RNA为模板,通过AMV反转录酶[宝生物工程(大连)有限公司]将RNA反转录为cDNA,对所得cDNA进行qRT-PCR分析。qRT-PCR体系:2×SYBR Premix Ex TaqTM 7.5 μL[宝生物工程(大连)有限公司],上、下游引物(10 μmol · L-1)各0.12 μL,cDNA 0.5 μL,ddH2O 6.76 μL。反应条件为:95 ℃ 10 min;94 ℃ 20 s,50 ℃ 20 s,68 ℃ 20 s(40个循环);60~94 ℃每隔0.5 s升高0.5 ℃,检测熔解曲线。所有引物见表 1。
| 基因 Gene | GenBank序列号 GenBank accession No. | 引物序列(5′→3′) Primer sequcence | PCR产物大小/bp PCR product size |
| Actin2 | JQ028730 | F:AAAAGGATGCCTATGTTGGTG;R:TAAGTGGAGCCTCAGTTAGAAGTA | 186 |
| GAPDH | GQ398120 | F:TCATTCCGTGTCCCAACCG;R:CCACATCATCTTCAGTGTAACCCA | 151 |
| MSC27 | X63872 | F:AGAATGGAATGTTGTGGGAGG;R:GTCATCAACACCCTCATCTTCTC | 113 |
| 18S RNA | KJ507198 | F:GCTCTGCCCGTTGCTCTGATGAT;R:CCTTGGATGTGGTAGCCGTTTCT | 195 |
| β-tublin | AJ319667 | F:CACATTGGTCAAGCCGGTAT;R:ACCGGTCTCACTGAAGAACG | 157 |
| bZIP | HQ911778 | F:TGCTTCACCAACTCCGCAT;R:CAGGTCCCTTCCCTTCAAACT | 179 |
| UBQ | XM_003629847 | F:GCAGCAACCAACGAAGCAAGA;R:CACCACGAAGACGTAGGACAAGG | 246 |
| PPPrep | XM_003620228 | F:GGAAAACTGGAGGATGCACGTA;R:ACAAGCCCTCGACACAAAACC | 112 |
| Ms03_50f03 | XM_003613901 | F:ATGCACTGGAGGAAGAGCAC;R:TCCTCCGACTCTGACTCTGC | 133 |
| Ms03_69f07 | XM_003601618 | F:GAAGGTTCGCGTGAAGTGC;R:CACCGAGAGTGATGTGATCC | 140 |
为了更好地挑选稳定的内参基因,对UV-B处理后不同时间每个内参候选基因的相关CT值求平均值后进行后续分析。所有结果均为3次以上独立试验结果,所有结果均为恢复生长5 d采集数据的整合数据分析。将所得CT值进行转换后,通过geNorm、NormFinder等分析软件进行分析。其中geNorm是由Vandesompele等[17]于2002年开发的软件,一般默认阈值M等于1.5,当某个基因M大于1.5时,剔除该基因;只有M值小于1.5的基因可以考虑作为内参基因。因此,geNorm输出M值折线图的横坐标从左至右代表了基因依次筛选和逐个删除的过程,最终保留下的即为最稳定的内参基因。 2 结果与分析 2.1 10个候选内参基因在不同组织中CT值的比较
即使是在不同组织或同一组织不同生长阶段中,同一个基因的表达丰度都可能存在差异。当某一个基因表达量高时,其CT值反而低。 本试验中,分别对紫花苜蓿10个候选内参基因在根、茎和叶组织中的表达进行分析(图 1)。各内参基因CT值为10~35。其中传统的内参基因 18 S RNA表达丰度最高,其CT值范围为11.58~14.58;UBQ、Ms 03_69f07、GAPDH、Actin2和Ms03_50f03表达丰度居中;而候选内参基因PPPrep表达丰度在各组织均为最低。在根和茎组织中,除18 S RNA和PPPrep之外,所有检测基因的表达量均相近(CT值差异为0.3~1.0);在叶片中,各内参基因表达量与根或茎 组织中的差异度较大(CT值差异为1.2~2.4)。因此,需要分别对根、茎、叶的候选内参基因进行筛选分析。
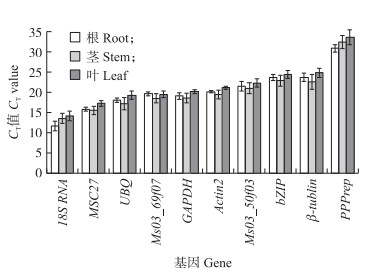 | 图 1 10个候选内参基因在不同苜蓿组织中的CT值 Fig. 1 Average cycle threshold(CT)values for the 10 candidate reference genes in different alfalfa tissues |
从图 2可知:非UV-B胁迫下,MSC 27和Ms03_69f07 的稳定性较高(图 2-A),UBQ次之;但UV-B胁迫下,MSC 27 和UBQ较稳定,Ms 03_69f07 次之(图 2-B)。β-tublin和 18 S RNA在2种情况下稳定性都较差。为了更准确地挑选UV-B处理下合适的内参基因,于是将对照组和UV-B处理组的M值合并整合,进行总M值分析。结果(图 2-C)显示:MSC 27和UBQ较稳定,Ms03_69f07次之,而β-tublin和18 S RNA的稳定性依然较差,因此可以将这2个基因排除在候选内参基因之外。
 |
图 2 UV-B胁迫下紫花苜蓿根部qRT-PCR内参基因的geNorm分析 Fig. 2 Ranking of candidate reference genes based on stability values calculated by geNorm for UV-B irradiation of alfalfa rootsA.非UV-B胁迫下苜蓿根部候选内参基因M值的geNorm分析;B.UV-B胁迫下苜蓿根部候选内参基因M值的geNorm分析;C.非UV-B和UV-B胁迫下苜蓿根部候选内参基因总M值的geNorm分析;D.苜蓿根部候选内参基因V值的geNorm分析 A.Average expression M values analysis of candidate reference genes of alfalfa roots without UV-B irradiation by geNorm;B.Average expression M values analysis of candidate reference genes of alfalfa roots with UV-B irradiation by geNorm;C.Average expression M values analysis of candidate reference genes of alfalfa roots with and without UV-B irradiation by geNorm;D.Average expression pairwise variation(V)analysis of candidate reference genes of alfalfa roots by geNorm |
为了进一步确定合适的内参基因,geNorm软件引入新的变量——配对差异值(V值),只有当Vn/n+1小于阈值0.15时,可以不用引入新的基因作为内参基因。从根部内参结果(图 2-D)分析可得知:在3种情况下V2/3均小于0.15,这表明只需使用稳定性较高的2个内参基因可以满足试验要求。 2.3 UV-B胁迫下苜蓿茎部qRT-PCR内参基因的筛选分析
从图 3-A、B、C可知:对照组和UV-B处理组紫花苜蓿茎部候选内参基因稳定性分析结果略有差异,但内参基因MSC 27、Actin2和Ms03_50f03均较为稳定,而β-tublin、18 S RNA和PPPrep的稳定性都较差。UV-B胁迫下UBQ稳定性较高,但它在正常条件下并不稳定,因此UBQ可能不适合作为内参基因使用。V值检测(图 3-D)表明:与根部内参基因分析结果相似,2个稳定性较高的内参基因已经符合筛选条件可以作为内参基因使用,但是从总和统计的V值结果可以发现,引入第3个内参基因,V值进一步下降,这表明在试验过程中选用3个内参基因,可以使结果更加符合实际情况。
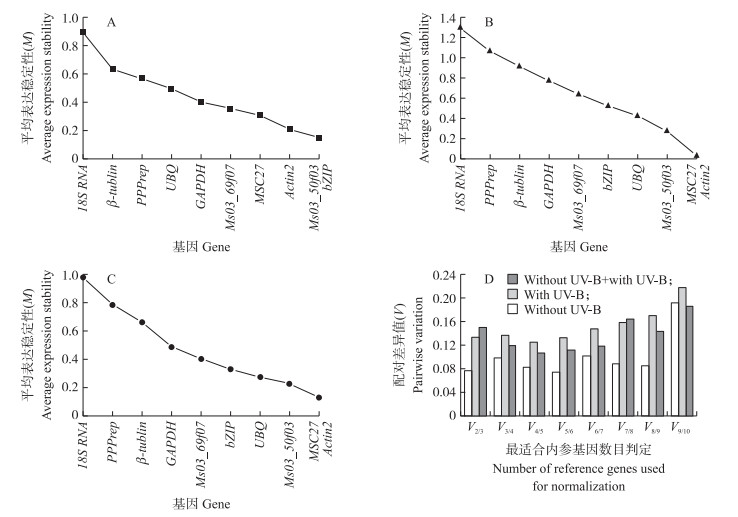 |
图 3 UV-B胁迫下苜蓿茎部qRT-PCR内参基因的geNorm分析 Fig. 3 Ranking of candidate reference genes based on stability values calculated by geNorm for UV-B irradiation of alfalfa stemsA.非UV-B胁迫下苜蓿茎部候选内参基因M值的geNorm分析;B.UV-B胁迫下苜蓿茎部候选内参基因M值的geNorm分析;C.非UV-B和UV-B胁迫下苜蓿茎部候选内参基因总M值的geNorm分析;D.茎部候选内参基因V值的geNorm分析 A.Average expression M values analysis of candidate reference genes of alfalfa stems without UV-B irradiation by geNorm;B.Average expression M values analysis of candidate reference genes of alfalfa stems with UV-B irradiation by geNorm;C.Average expression M values analysis of candidate reference genes of alfalfa stems with and without UV-B irradiation by geNorm;D.Average expression pairwise variation(V)analysis of candidate reference genes of alfalfa stems by geNorm |
从图 4-A、B、C可知:GAPDH和Actin 2的稳定性都较高,而β-tublin、18 S RNA和PPPrep的稳定性都较差,可以排除这3个基因作为内参基因的可能。相应的V值结果表明:选取稳定性较高的2个内参基因组合符合筛选条件,而引入第3个内参基因时,3种情况下V值均上升,因此仅使用2个内参基因在本试验条件下最佳(图 4-D)。
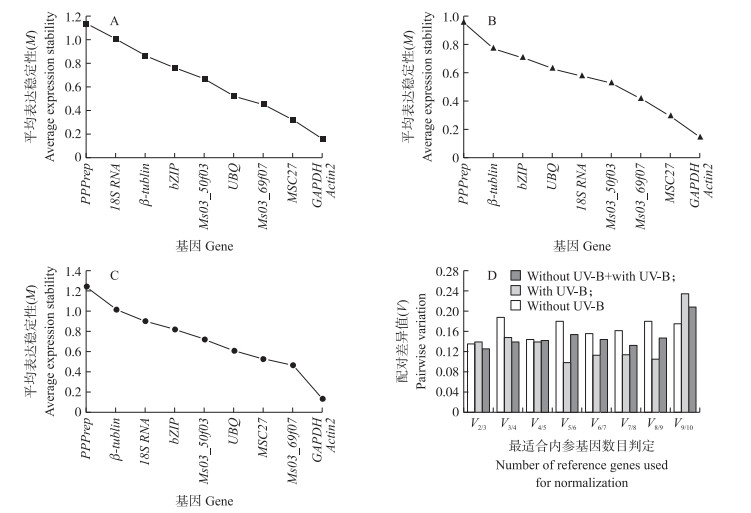 |
图 4 UV-B胁迫下苜蓿叶片qRT-PCR内参基因的geNorm分析 Fig. 4 Ranking of candidate reference genes based on stability values calculated by geNorm for UV-B irradiation of alfalfa leavesA.非UV-B胁迫下苜蓿叶片候选内参基因M值的geNorm分析;B.UV-B胁迫下苜蓿叶片候选内参基因M值的geNorm分析;C.非UV-B和UV-B胁迫下苜蓿叶片候选内参基因总M值的geNorm分析;D.苜蓿叶片候选内参基因V值的geNorm分析 A.Average expression M values analysis of candidate reference genes of alfalfa leaves without UV-B irradiation by geNorm;B.Average expression M values analysis of candidate reference genes of alfalfa leaves with UV-B irradiation by geNorm;C.Average expression M values analysis of candidate reference genes of alfalfa leaves with and without UV-B irradiation by geNorm;D.Average expression pairwise variation(V)analysis of candidate reference genes of alfalfa leaves by geNorm |
geNorm软件是基于内参基因互不影响的前提下,当内参基因在UV-B胁迫下存在相互影响的可能时,上述结果的准确性就有待进一步研究。因此,本试验还使用了Andersen等[18]设计的分析软件NormFinder(该软件得到的值越小代表了该基因越稳定)。NormFinder分析结果(表 2)与geNorm结果并不完全一致。在紫花苜蓿根和茎组织中,Actin 2和MSC27 稳定性都较高,其中茎组织的结果与geNorm软件结果一致,但根部的结果存在差异,这可能是2个软件算法不同造成的;叶片中稳定性较高的基因为Actin 2 和GAPDH,该结果与geNorm的分析结果一致。
|
基因 Gene |
|
|
|
| 18S RNA | 0.376 | 0.105 | 0.498 | 0.134 | 0.237 | 0.071 |
| MSC27 | 0.110 | 0.050 | 0.049 | 0.039 | 0.230 | 0.069 |
| UBQ | 0.170 | 0.059 | 0.156 | 0.050 | 0.205 | 0.064 |
| Ms03_69f07 | 0.163 | 0.057 | 0.143 | 0.047 | 0.276 | 0.080 |
| GAPDH | 0.290 | 0.085 | 0.200 | 0.060 | 0.123 | 0.048 |
| Actin2 | 0.138 | 0.053 | 0.035 | 0.046 | 0.131 | 0.049 |
| Ms03_50f03 | 0.254 | 0.076 | 0.149 | 0.049 | 0.192 | 0.061 |
| bZIP | 0.225 | 0.070 | 0.105 | 0.041 | 0.199 | 0.062 |
| β-tublin | 0.397 | 0.111 | 0.180 | 0.055 | 0.332 | 0.093 |
| PPPrep | 0.338 | 0.096 | 0.148 | 0.048 | 0.369 | 0.103 |
近几年,qRT-PCR成为研究基因相对表达量的主要方法,关于内参基因的研究也显得尤为重要。理想的内参基因需要满足以下条件:在不同组织、不同生长阶段、不同的处理下均有表达,且表达稳定;与目的基因表达水平相似[18, 19, 20]。传统的内参基因包括 18S RNA、MSC27、UBQ、GAPDH、Actin2 和β-tublin等。这些基因广泛存在于各种细胞,它们是维持生物结构和代谢所必需的。传统的观点认为在各种细胞和生理过程中上述内参基因的表达较稳定。然而实际情况往往并非如此。例如,β-tublin在桃[20]和黄瓜[21]的不同组织中表达存在较大差异,这与本试验结果一致;在桃[13]和拟南芥[1]中稳定性较高的UBQ,在本研究中却并不是合适的内参基因。
Gutierrez等[1]的研究结果表明,拟南芥内参基因在不同组织、不同生长时期表达量存在较大差异。李钱峰等[22]发现在不同的水稻品种中,内参基因表达量存在差异。本研究中,即使是同一种胁迫,紫花苜蓿根、茎、叶不同组织合适的内参基因也不同,需要将不同组织分别进行分析[23]。因此,对不同胁迫条件下的内参基因进行筛选是很有意义的工作。此外,已有报道表明可以通过对模式生物全基因组进行搜索并进行试验验证的方法来获取可靠的内参基因[24]。本试验中 18 S RNA CT值最小,即表达丰度最高,但在实际应用过程中,18 S RNA的表达量远大于所研究的目的基因,因此很难消除基线对试验结果的影响[17]。即使该基因的稳定性较高,也并不适合作为紫花苜蓿的内参基因。
目前有多款软件用于内参基因结果分析,包括geNorm、NormFinder和Bestkeeper等。在本试验中,通过geNorm和NormFinder两种软件对内参基因稳定性进行分析,结果表明,在不同的植物组织中内参基因稳定性并不相同,这与Kakar等[25]研究结果是一致的,尤其是叶片中内参基因稳定性与根部和茎部差异较大。在3个组织中,Actin 2稳定性均较高,而18S RNA以及β-tublin稳定性较差,因此18 S RNA和β-tublin并不适合作为内参基因。UV-B胁迫下,苜蓿幼苗根部使用MSC 27和UBQ较为适宜,而在茎部分析中,使用MSC27和Actin2较为适宜,而叶片分析则宜使用Actin2 和GAPDH为内参基因,这与蔡文凯等[16]结果一致。
| [1] | Gutierrez L, Mauriat M, Guénin S, et al. The lack of a systematic validation of reference genes:a serious pitfall undervalued in reverse transcription-polymerase chain reaction(RT-PCR)analysis in plants[J]. Plant Biotechnol J, 2008, 6(6):609-618 |
| [2] | Chi X, Hu R, Yang Q, et al. Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR[J]. Mol Genet Genom, 2012, 287(2):167-176 |
| [3] | 袁伟, 万红建, 杨悦俭. 植物实时荧光定量PCR内参基因的特点及选择[J]. 植物学报, 2013, 47(4):427-436 [Yuan W, Wan H J, Yang Y J. Characterization and selection of reference genes for real-time quantitative RT-PCR of plants[J]. Chin Bull Bot, 2013, 47(4):427-436(in Chinese with English abstract)] |
| [4] | Reid K E, Olsson N, Schlosser J, et al. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development[J]. BMC Plant Biol, 2006, 6:27 |
| [5] | Faccioli P, Ciceri GP, Provero P, et al. A combined strategy of "in silico"transcriptome analysis and web search engine optimization allows an agile identification of reference genes suitable for normalization in gene expression studies[J]. Plant Mol Biol, 2007, 63(5):679-688 |
| [6] | Kim B R, Nam H Y, Kim S U, et al. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice[J]. Biotechnol Lett, 2003, 25(21):1869-1872 |
| [7] | Brunner A M, Yakovlev I A, Strauss S H. Validating internal controls for quantitative plant gene expression studies[J]. BMC Plant Biol, 2004, 4:14 |
| [8] | 涂礼莉, 张献龙, 刘迪秋, 等. 棉花纤维发育和体细胞胚发生过程中实时定量PCR内对照基因的筛选[J]. 科学通报, 2007, 52(20):2379-2385 [Tu L L, Zhang X L, Liu D Q, et al. The selection of inner reference genes by real-time RT-PCR of Gossypium hirsutum during the development of fiber and somatic embryogenesis[J]. Chin Sci Bull, 2007, 52(20):2379-2385(in Chinese with English abstract)] |
| [9] | Suzuki T, Higgins P J, Crawford D R. Control selection for RNA quantitation[J]. Biotechniques, 2000, 29(2):332-337 |
| [10] | Nicot N, Hausman J F, Hoffmann L, et al. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress[J]. J Exp Bot, 2005, 56(421):2907-2914 |
| [11] | Dekkers B J, Willems L, Bassel GW, et al. Identification of reference genes for qRT-PCR expression analysis in Arabidopsis and tomato seeds[J]. Plant Cell Physiol, 2012, 53(1):28-37 |
| [12] | Stein R J, Waters B M. Use of natural variation reveals core genes in the transcriptome of iron-deficient Arabidopsis thaliana roots[J]. J Exp Bot, 2012, 63(2):1039-1055 |
| [13] | Remans T, Smeets K, Opdenakker K, et al. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations[J]. Planta, 2008, 227(6):1343-1349 |
| [14] | Han B, Yang Z, Samma M K, et al. Systematic validation of candidate reference genes for qRT-PCR normalization under iron deficiency in Arabidopsis[J]. Biometals, 2013, 26(3):403-413 |
| [15] | Chen L, Zhong H Y, Kuang J F, et al. Validation of reference genes for qRT-PCR studies of gene expression in banana fruit under different experimental conditions[J]. Planta, 2011, 234(2):377-390 |
| [16] | 蔡文凯, 胡金璐, 李双双, 等. 辐射条件下微藻基因表达内参基因的选择[J]. 空间科学学报, 2013, 33(6):651-658 [Cai W K, Hu J L, Li S S, et al. Selection of suitable in ternal control genes in microalgae under radiation condition[J]. Chin J Space Sci, 2013, 33(6):651-658(in Chinese with English abstract)] |
| [17] | Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biol, 2002, 3(7):research 0034.1-0034.11 |
| [18] | Andersen C L, Jensen J L, Ørntoft T F. Normalization of real-time quantitative reverse transcription-PCR data:a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Res, 2004, 64(15):5245-5250 |
| [19] | Schmittgen T D, Zakrajsek B A. Effect of experimental treatment on housekeeping gene expression:validation by real-time, quantitative RT-PCR[J]. Biochem Biophys Methods, 2000, 46(1):69-81 |
| [20] | Tong Z, Gao Z, Wang F, et al. Selection of reliable reference genes for gene expression studies in peach using real-time PCR[J]. BMC Mol Biol, 2009, 10(1):71 |
| [21] | Migocka M, Papierniak A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators[J]. Molecular Breeding, 2011, 28(3):343-357 |
| [22] | 李钱峰, 蒋美艳, 于恒秀, 等. 水稻胚乳RNA定量RT-PCR分析中参照基因选择[J]. 扬州大学学报:农业与生命科学版, 2008, 29(2):61-66 [Li Q F, Jiang M Y, Yu H X, et al. Selection of internal reference genes for quantitative RT-PCR analysis of total RNA from endosperm of rice(Oryza sativa L.)[J]. Journal of Yangzhou University:Agricultural and Life Science Edition, 2008, 29(2):61-66(in Chinese with English abstract)] |
| [23] | 陈艳, 乔璟, 沈益新. 春季紫花苜蓿根系性状与地上部生长性状的相关性分析[J]. 南京农业大学学报, 2012, 35(1):108-112. doi:10.7685/j.issn.1000-2030.2012.01.019 [Chen Y, Qiao J, Shen Y X. Analysis of the relationship between root traits and plant growth traits of alfalfa in spring[J]. Journal of Nanjing Agricultural University, 2012, 35(1):108-112(in Chinese with English abstract)] |
| [24] | Czechowski T, Stitt M, Altmann T, et al. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis[J]. Plant Physiol, 2005, 139(1):5-17 |
| [25] | Kakar K, Wandrey M, Czechowski T, et al. A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula[J]. Plant Methods, 2008, 4(1):18 |




