2. 中国农业科学院农业环境与可持续发展研究所, 北京 100081;
3. 江西省科学院能源研究所, 南昌 330096
2. Institute of Environment and Sustainable Development in Agriculture, Chinese Academy of Agricultural Sciences, Beijing 100081;
3. Institute of Energy, Jiangxi Academy of Sciences, Nanchang 330096
畜禽养殖业是我国农业的支柱产业,在维持畜产品稳定供给、提高人民生活水平方面发挥着重要作用.随着畜禽养殖业的集约化、规模化发展,为提高动物生产性能、防治疾病,养殖过程添加了一定量的重金属与抗生素.据统计2006年我国兽用抗生素消耗9.7万吨,占全国抗生素总用量的54%(Luo et al., 2010).而不被机体吸收、降解的抗生素排放到环境中,据Zhou等(Zhou et al., 2013)估算我国每年生猪和奶牛养殖场抗生素排放量分别为3080和164 t.而养殖业每年重金属排放铜、锌分别为2397.23 t、4756.94 t(环保部等,2010).畜禽养殖粪污表现出重金属与抗生素复合污染特征(王瑞和魏源送,2013).Jindal等(2006)和Seiler和Berendonk(2012)研究发现畜禽养殖过程抗生素和重金属使用与养殖场及其周边环境抗性基因丰度的提高呈正相关关系.畜禽养殖粪便、污水成为抗性基因的重要蓄积库(张俊亚等,2015).抗性基因作为一种新型污染物,可能对公共健康、食品和饮用水安全构成威胁(Pruden et al., 2006).胡永飞等对162个健康人肠道微生物宏基因组(Metagenome)中的耐药基因进行了深入分析,发现四环素抗性基因的丰度最高,而人类肠道四环素抗性基因极有可能来自于兽用抗生素的使用以及抗性基因沿食物链的传播(Hu et al., 2013).
2014年世界卫生组织发布的《全球抗生素耐药报告》明确指出抗生素抗性是21世纪公共卫生的严峻挑战,针对动物生产应监督和促进畜禽业的合理用药,并强调了食用动物携带的抗生素抗性及其在食物链上的传播方面数据的缺乏,应加强此方面的研究(WHO,2014).我国和主要发达国家推行畜禽养殖废水的生物处理、农田利用等工艺模式,然而畜禽养殖废水携带的抗性基因在此过程的转归,以及抗性基因是否存在沿食物链的传播风险(Cheng et al., 2015),亟需开展相关研究.
因此,本研究通过查阅国内外文献,总结归纳了畜禽养殖废水含有的抗生素抗性基因在生物处理、农田利用过程的变化规律,并对今后的研究重点和方向提出建议和展望,以期为揭示抗性基因消减规律,降低畜禽养殖废水抗性基因传播风险提供借鉴.
2 畜禽养殖废水中抗生素抗性基因分布 (Occurrence of antibiotic resistance genes in animal wastewater)抗性基因根据其抗性机制不同分为3类,分别为降低细胞内抗生素浓度(包括降低细胞通透性或外排)、靶向改变(包括靶向保护或靶向突变)以及抗生素失活(Blair et al., 2015).畜禽养殖业抗生素的大量使用引起养殖环境抗性基因丰度的提高,抗性基因与抗生素之间存在相关关系(Wu et al., 2010;Zhu et al., 2013).Zhu等(2013)检测了我国3个省36份猪场环境样品(包括粪便、堆肥、土壤)中的149种抗性基因,结果表明检出的抗性基因对应的抗生素分别为大环内脂林可霉素链阳杀菌素B(macrolidelincosamidestreptogramin B,MLSB)、β内酰胺类、四环素类、喹诺酮氯霉素胺酰醇类、万古霉素等,按抗性机制分类抗生素失活检出率最高,其后依次为外排和细胞保护机制;而抗性基因丰度与转座酶基因丰度、铜、土霉素含量具有正相关关系.较高的抗性基因丰度可能由于在抗生素的选择压力下抗性基因宿主细菌的增殖,以及某些抗性基因通过移动基因元件(Mobile genetic elements)发生基因水平转移(Horizontal gene transfer)(Pruden et al., 2006; Rahube and Yost,2012).
在畜禽养殖废水方面,四环素类、磺胺类、大环内脂类抗生素的抗性基因研究较多,按抗性机制分类,畜禽养殖废水中抗性基因分布特征详见表 1.Cheng等(2013)测试了猪场废水中不同机制的四环素抗性基因,发现核糖体保护(靶向保护)抗性基因(tetQ、tetM、tetW、tetO)比外排泵机制抗性基因(tetA、tetB、tetC、tetL)、酶修饰(抗生素失活机制)抗性基因(tetX)丰度高,其在猪场废水中丰度分别为9.25×10-2、5.53×10-2、1.69×10-2和1.32×10-2 copies/16S rRNA.而McKinney等(2010)和Smith等(2004)研究也表明tetQ、tetM、tetW、tetO在猪场废水中具有较高的丰度.成卫孝(2014)研究了猪粪水厌氧发酵土壤生态系统中3种核糖体保护机制的四环素类抗性基因丰度tetQ>tetO>tetW,其中tetQ平均丰度最高1.84×10-1 copies/16S rRNA.冀秀玲等(2011)调查了上海地区猪场和牛场废水中磺胺类和四环素类抗性基因,含量最高的分别为sulA(108~1010 copies · mL-1)和tetW(106~107 copies · mL-1),而sulIII含量与磺胺类抗生素浓度的相关性较好,这可能与磺胺类抗生素易生物降解性有关;tetM含量与四环素类抗生素浓度相关性较弱.Ji等(2012)也指出TC与tet无显著相关性.除四环素类与磺胺类抗生素之外,泰乐菌素是应用最广泛的兽用抗生素之一(Chen et al., 2008),可能引起大环内脂类抗性基因以及MLSB的多重抗性基因丰度的提高.Chen等(2010)对3家猪场大环内脂抗性基因erm进行了定量检测,废水中ermB、ermF含量较高(在108~1010 copies · mL-1之间),而ermX在104~106 copies · mL-1范围.Jindal等(2006)通过寡聚糖杂交探针测试方法,发现猪粪水和氧化塘废水中50%的rRNA携带MLSB多重抗性基因.
| 表1 基于抗性机制分类畜禽养殖废水中抗性基因赋存特征 Table 1 Mechanism and abundance of antibiotic resistance genes of animal wastewater |
针对抗性基因与基因转移元件的相关性,sulI与intI1具有极显著的相关性(p<0.001;r=0.803)(成卫孝,2014),这可能由于sulI经常与一类整合子结合在一起(Sköld,2000).Roberts等(2005)指出tetM可能由转座子Tn916Tn1545和结合质粒介导.
3 畜禽养殖废水中重金属对抗生素抗性基因的影响 (Influence of heavy metal on antibiotic resistance genes in animal wastewater)畜禽养殖过程在饲料中添加铜、锌等重金属引起猪粪水中抗铜、抗锌细菌的增加,畜禽养殖废水存在抗生素与重金属复合污染特征.在重金属的选择压力下,畜禽养殖粪水中重金属抗性基因丰度较高.贡娇娜(2010)对猪饲料、肠道和粪便中抗铜细菌进行了分析鉴定,发现猪粪中抗铜大肠杆菌与饲料中硫酸铜添加量正相关,分离得到的239株抗铜细菌中携带抗铜基因pcoA、pcoC、pcoD,携带抗铜基因的细菌也同时携带链霉素和四环素的抗性基因(strA、strB、tetB).而窦秋燕(2010)研究了猪粪中抗锌细菌的分布规律,结果表明猪粪中普遍存在抗锌细菌,抗锌大肠杆菌的检出率与饲料中氧化锌的添加成正相关关系;抗锌菌株主要携带抗锌基因zntA.
畜禽养殖环境重金属的污染不仅引起重金属耐受菌及抗铜、抗锌基因丰度的提高,可能存在重金属与抗生素的协同选择作用(coselection),重金属的选择压力可能使抗生素抗性基因丰度维持在较高水平.欧盟国家已禁止抗生素饲料添加剂的使用,但减少抗生素使用并不会阻止抗性基因的传播(Salyers and AmábileCuevas,1997),养殖场重金属使用可能会通过协同选择增加抗生素抗性基因的传播.Ji等(2012)研究发现磺胺类sulA与重金属Hg、Cu、Zn具有显著相关关系.Hölzel等(2012)研究发现猪场废水中高浓度的Cu和Zn显著提高了耐β内酰胺大肠杆菌的丰度.
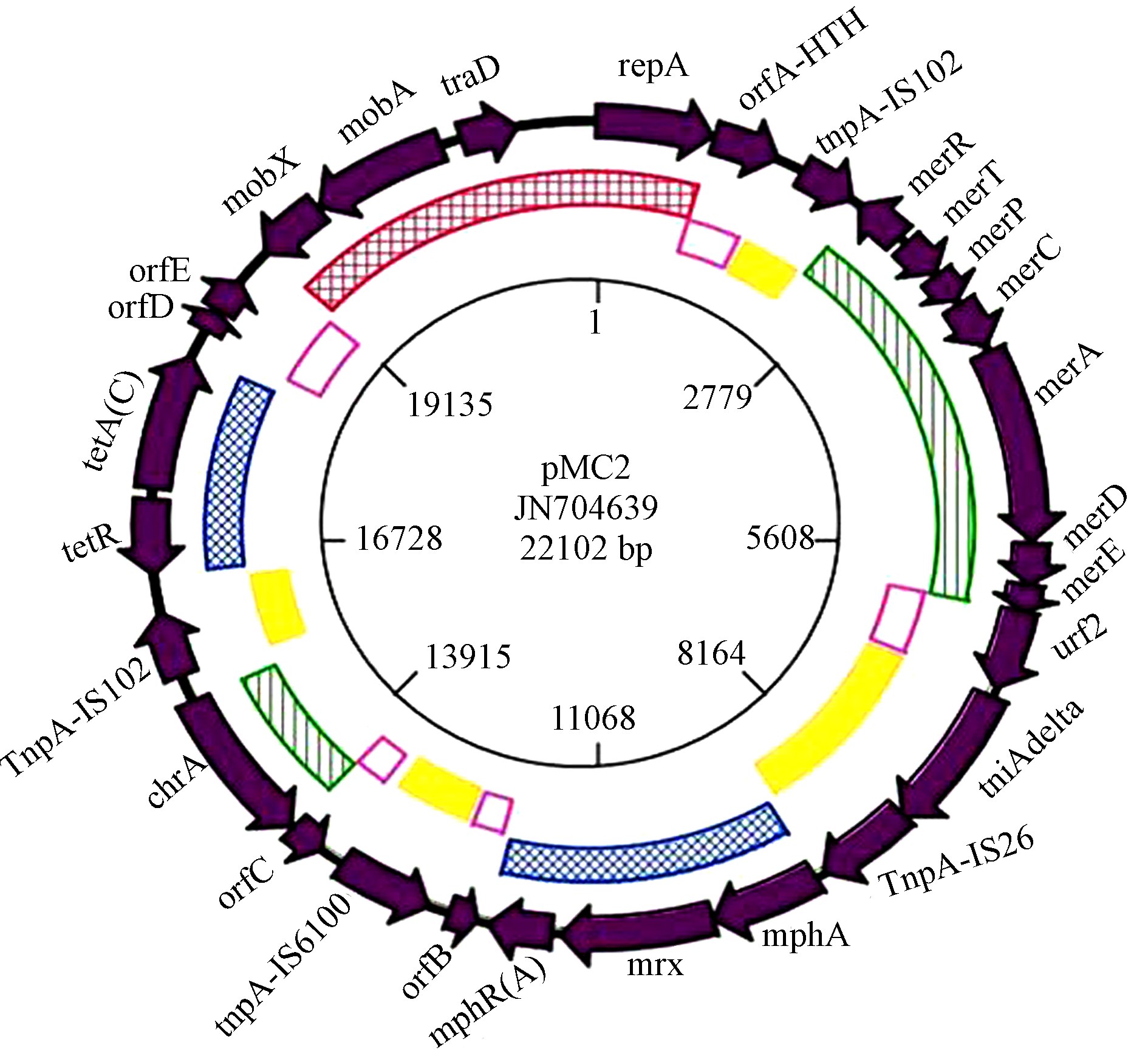 |
| 图1 猪粪分离的具有重金属与抗生素协同抗性的质粒pMC2(Rahube and Yost, 2012) Fig.1 The plasmid(pMC2)with antibiotic and heavy metal resistance genes isolated from swine manure(Rahube and Yost, 2012) |
Seiler和Berendonk(2012)指出重金属和抗生素抗性的协同选择机制主要是因为重金属和抗生素的抗性机制的耦合作用,包括交叉抗性(crossresistance)和协同抗性(coresistance).交叉抗性是某种抗性基因编码的酶或蛋白具有提高细胞耐受多种抑菌物质(如抗生素或重金属)的能力,如多重药剂外排泵(multi drug efflux pumps),其可以将毒性物质迅速排出细胞外(Martinez et al., 2009; BakerAustin et al., 2006).而协同抗性指的是具有两种或多种抗性功能的基因相互邻近并在一个移动基因元件上(Chapman,2003;Cantón and RuizGarbajosa, 2011).如猪粪中分离的质粒pMC2,携带大环内脂、四环素等抗生素抗性基因和汞、铬等重金属抗性基因,具有很强的移动和结合能力(Rahube and Yost, 2012).Seiler和Berendonk(2012)总结了畜禽粪便中重金属引起抗生素协同抗性的最小浓度(Minimum coselective concentration,MCC),Cu和Zn的MCC值分别为11.79和22.75 mg · kg-1 DM,但作者也指出非常缺乏畜禽养殖废水重金属对抗生素抗性基因协同选择的数据.另外,养殖废水复合污染的特性也增加了抗性基因研究的难度.
4 畜禽养殖废水处理工艺对抗性基因的消减 (Removal of antibiotic resistance genes during process of animal wastewater treatment) 4.1 常规生物处理工艺厌氧消化是畜禽养殖场采用最为广泛的废水处理工艺.Rysz等(2013)指出厌氧过程抑制细菌代谢,对抗性基因传播具有抑制作用.Ma等(2011)指出ARGs去除与厌氧菌群结构具有相关性,主要表现在抗性基因的宿主菌群在厌氧环境中的变化.
针对厌氧消化处理养殖废水抗性基因的变化,现场调研较多,参数优化的研究较少,针对猪场废水的研究较多,其他种类的养殖废水研究较少,不同生物处理工艺抗性基因赋存特征详见表 2.Tao等(2014)研究了不同规模猪场的废水生物处理系统抗性基因去除效果,结果表明厌氧消化和好氧生物处理对tetA、tetW、sul1、sul2、blaTEM抗性基因平均去除率在33.3%~97.56%.Chen等(2010)考察了环境温度下厌氧消化在不同季节的处理效果,夏季ermB、ermF、ermX的去除效果优于冬季,夏季厌氧消化出水较猪场原废水ermB、ermF、ermX和tetG平均降低1.2、0.8、0.7和1.1 log copies · mL-1,表明温度是厌氧消化去除抗性基因的重要控制指标.针对温度对厌氧消化抗性基因消减的影响,Diehl和Lapara(2010)指出高温厌氧消化对四环素类抗性基因tetA、tetO、tetW、tetX有显著去除,它们的去除符合一级反应动力学模型,而tetL只存在于革兰氏阴性菌,厌氧处理对其去除效果不明显,而在好氧高温处理(55 ℃)过程中tetL丰度表现出线性降低趋势.Beneragama等(2013)比较了高温和中温厌氧消化对牛粪中耐药菌的影响,结果表明高温可全部消灭多重耐药菌(抗头孢唑啉、新霉素、万古霉素、土霉素、氨苄西林等),而中温发酵只可以去除多重耐药菌1~2 log cfu · mL-1.
| 表2 猪场废水生物处理过程中抗性基因的赋存特性 Table 2 Occurrence of antibiotic resistance genes in the biological treatment of swine wastewater |
除厌氧消化工艺以外,氧化塘、人工湿地也是畜禽养殖场广泛使用的废水处理工艺.Joy等(Joy et al., 2013)调查了氧化塘储存猪场废水40 d抗性基因的变化,ermB和ermF的丰度分别降低了50%~60%和80%~90%,而tetX和tetQ丰度的消减符合一级反应动力学模型.Barkovskii等(2012)将氧化塘处理猪场废水后抗性基因的去除趋势归为两类,一类是相对丰度大幅降低甚至低于检测限,包括tetB、tetL;另一类为经处理后丰度不变甚至有所提高,包括tetG、tetM、tetO和tetX,可能因为这类基因常位于转移原件上,在废水中发生了基因的水平转移.郑加玉等(2013)采用水平流人工湿地处理猪场废水,结果表明tetW、tetM和tetO的浓度平均去除率分别为95.73%、92.21%和95.05%;可能由于土壤对抗性基因的吸附作用,湿地土壤中抗性基因的丰度有明显升高现象.Liu等(2013)模拟垂直流人工湿地中添加沸石研究抗性基因的消减规律,发现在HRT为30 h时猪场废水抗性基因去除效果较好.
4.2 膜生物反应器(Membrane bioreactor,MBR)工艺膜分离技术近年已在畜禽养殖废水处理领域得到了一定的研究与应用,并日益得到重视.例如,Padmasiri等(2007)采用厌氧MBR处理猪场废水,有机负荷为1.0 kg · m-3 · d-1高于其他厌氧消化工艺(Kim et al., 2013).Sui等(2014)采用好氧MBR处理猪场厌氧消化液TN负荷0.11 kg · m-3 · d-1较高.然而针对MBR处理畜禽养殖废水抗性基因去除规律的研究较少.Du等(2014)调研了污水处理厂采用A2OMBR工艺处理生活和工业混合废水对四环素类和磺胺类抗性基因的去除效果,结果表明MBR工艺对tetG、tetW、tetX、sul1和intI1分别去除了2.20、2.90、1.71、2.15和2.07 log copies · mL-1,膜出水抗性基因丰度仍然较高(2.85~4.97 log copies · mL-1),然而作者并未给出膜孔径等膜分离工艺参数.
同常规生物处理工艺相比,MBR的生物量高,可能存在较大的抗性基因水平转移风险.Yang等(2013)以RP4质粒作为水平转移研究对象,研究了MBR中抗性基因的水平转移效率,结果表明RP4在MBR中维持较高丰度104 copies/mg · biosolid,具有较高的水平转移效率(2.76×10-5/recipient),而RP4在常规活性污泥法的水平转移效率约4×10-6 /recipient(Geisenberger et al., 1999);尽管存在较高的水平转移效率,但由于微滤膜(PVDF,0.22 μm)的截留作用,出水检测不到携带抗性基因的RP4.由于膜的截留,一方面可消减膜出水的抗性基因浓度,另一方面导致反应器内污泥浓度高,可能使抗性基因在反应器内积累,提高了污泥中抗性基因的水平传播.污泥是重要的抗性基因蓄积库(佟娟和魏源送,2012),经过堆肥或厌氧消化处理后作为肥料土地利用,污泥的土地利用存在抗性基因的污染隐患(Rahube et al., 2014).
4.3 消毒工艺已有研究考察了消毒工艺(包括紫外、臭氧、加氯)处理畜禽养殖废水时对耐药菌的杀灭效果.Macauley等(2006)研究发现,加氯量和臭氧用量分别为30 mg · L-1和100 mg · L-1时,猪场氧化塘废水中细菌总数分别去除了2.2~3.4 log cfu · mL-1和3.3~3.9 log cfu · mL-1,然而林可酰胺、金霉素、磺胺甲恶唑耐药菌对加氯消毒不敏感,而四环素耐药菌对加氯消毒敏感,臭氧对耐药菌的影响并未给出相应结果.加氯对抗万古霉素肠球菌具有较好的灭杀作用(Rosenberg et al., 2014).而GomezAlvarez等(2012)研究加氯消毒对饮用水中抗性基因的影响,宏基因组数据表明加氯消毒后饮用水中仍含有编码β内酰胺酶(bla)、外排泵等抗生素抗性基因,表明耐受液氯氧化性的细菌同时携带抗生素抗性基因.关于紫外和臭氧对畜禽养殖废水抗性基因的去除研究较为缺乏,Zhang等(2015)研究了紫外灭菌对市政排水抗性基因消减的影响,结果表明紫外强度为249.5 mJ · cm-2时对抗性基因消减效果最佳,tetX和16S rRNA分别去除了0.58和0.60 log.Oh等(2014)采用模拟实验研究了臭氧对耐药性埃希氏大肠杆菌(Eschericia coli, E. coli)的去除,结果表明臭氧剂量为3 mg · L-1时耐药性E. coli去除了1 log.
4.4 组合工艺畜禽养殖废水通常采用厌氧好氧组合工艺进行处理.Chen等(2010)在监测某猪场夏季废水处理工艺对抗性基因去除效果时,发现经过厌氧消化好氧滤池处理,ermB丰度分别降低了1.2 log、0.9 log copies · mL-1,而ermB在出水储存池中已低于检测限;tetG在厌氧、好氧过程分别降低了1.1 log、3.4 log copies · mL-1.Cheng等(2013)对我国东部某猪场废水采用厌氧消化与氧化塘组合工艺去除抗性基因的效果进行了调查,发现tetO、tetQ、tetW有明显去除,丰度从10-1降至10-3 copies/16S rRNA,这可能由于tetQ和tetW宿主细菌多为厌氧菌,而tetO多为好氧菌携带,这些抗性基因无法在厌氧好氧交替环境中维持.而关于生物处理与消毒组合工艺对畜禽废水中抗性基因的去除作用,研究结果非常缺乏.
5 畜禽养殖废水农田利用对土壤和植物中抗性基因的影响 (Occurrence of antibiotic resistance genes in soil and plants during process of l and application of animal wastewater)由于畜禽养殖废水中富含有机质、氮、磷等营养物质,通常经过厌氧发酵、氧化塘等工艺处理后,作为肥水还田利用,这既节约了处理成本,也促进了养分循环利用,目前我国、美国、欧洲等国家都推行畜禽养殖废水的农田利用(李文哲等,2013;Gilley et al., 2007; Schröder and Neeteson, 2008).然而,畜禽养殖废水农田利用可能产生抗性基因从养殖场向农田土壤的传播风险.
土壤是重要的抗性基因储存库(Schmitt et al., 2006),其中主要的抗性基因来源包括土壤中固有的抗性微生物所携带的抗性基因,以及外源进入土壤中抗性微生物所携带的抗性基因,但有关土壤中抗性基因的研究较为缺乏.Heuer等(2008)指出猪粪施用于农田存在抗性基因的水平转移风险,由于粪源微生物与土壤微生物不同,粪源微生物进入土壤后在几个月中大量消失,但抗性基因可通过水平转移进入土壤本土微生物中,进而引起土壤微生物抗性基因丰度的增加.而UdikovicKolic等(2014)研究发现牛粪农田利用引起土壤中抗性基因blaCEP丰度的提高是由于携带抗性基因的假单胞菌(Pseudomonas sp.)和紫色杆菌(Janthinobacterium sp.)的增殖,而这两种细菌来自于土壤,而非粪便引入.粪便农田利用可引起抗性基因丰度提高,但其微生物学机制仍不明确.
畜禽养殖废水还田利用一定时间内会显著提高土壤中抗性基因丰度.吴楠等(2009)对北京某猪场周边土壤四环素抗性基因进行了定量检测,发现丰度较高的四环素类抗性基因为tetB/P、tetT、tetM、tetO和tetW,其基因拷贝数范围在106~108 copies · g-1 DM,并认为tet抗性基因存在由畜禽养殖向土壤的转移.Hong等(2013)的研究发现,猪场废水农田利用后土壤中抗性基因tetQ、tetZ和整合子intI1、intI2分别提高了500、9和6、123倍.成卫孝(2014)的研究发现,施用猪场厌氧消化液的土壤中四环素类抗性基因丰度为105~108 copies · g-1,显著高于未施用猪场废水的土壤,而作物类型对抗性基因的丰度影响较小.Huang等(2013)研究了抗性基因沿土壤深度的变化,结果表明tetO、tetW、tetM、tetA丰度沿土壤深度在0~80 cm逐渐降低.Heuer等(2008)发现,饲料中添加磺胺嘧啶显著影响猪粪还田后土壤中sul抗性基因的变化,添加磺胺处理组在第60 dsul1抗性基因丰度降低至10-3 copies/16S rRNA、而sul2升高至10-1 copies/16S rRNA,饲料未添加磺胺嘧啶处理组sul1和sul2均呈现降低趋势,丰度分别为10-6和10-5 copies/16S rRNA.Kopmann等(2013)研究了施用猪粪的玉米根际土壤与非根际土壤微生物群落变化,结果表明根际土壤sul1和sul2抗性基因略低于非根际土壤,可能与根际环境磺胺嘧啶降解速度快有关,而sul基因常与质粒结合,根际土壤是质粒发生结合转移的热点区域.Heuer和Smalla(2007)考察了土壤类型对抗性基因的影响,发现壤土中sul2基因丰度高于砂土.Su等(2014)采用宏基因组文库研究了土壤中不可培养细菌携带的抗性基因,结果表明猪粪还田的土壤携带四环素类、利福平、氨基糖胺类、氯霉素类抗性基因.同未施用畜禽粪便的土壤相比,Marti等(2013)发现施用猪粪的土壤中大环内脂类抗性基因(ermA、ermB、ermF等)和质粒(IncQ、IncW)丰度有提高.Heuer等(2007)发现携带多重抗性的质粒IncP-1ε在粪便施用后的土壤中扩散.
在畜禽养殖粪污还田利用时,不同种类抗性基因随时间的消减规律各不相同.Marti等(2013)指出施用猪粪后,土壤中抗性基因表现出先增加后降低趋势,但抗性基因相对丰度在1年的施肥间隔后无法回到本底值,尤其是sul1、ermB、strB、intI1、IncW repA在土壤中丰度较高.Fahrenfeld等(2014)的研究发现,猪粪还田后sul1、sul2、ermF快速升高,随后ermF消减速度最快,在施肥43~55 d后降至本底水平,而tetG、tetO、tetW在施肥土壤和控制土壤中无差异;并且作者指出粪便还田后1~2个月内土壤抗性基因丰度较高,应采取措施防止抗性基因进入水体或邻近土壤中.不同类型抗生素的抗性基因在土壤中恢复本底值的时间不同,例如,MLS抗性基因恢复到土壤本底值需要20 d(Zhou et al., 2010),sul1需要2个月(Heuer and Smalla, 2007),而四环素类抗性菌株需要6个月(Sengeløv et al., 2003).关于畜禽养殖废水对养殖场受纳水体的影响,CheeSanford等(2001)发现猪场氧化塘下游河流中250 m仍可得四环素抗性基因tetM.Chen等(2011)研究了福建闽江流域E. coli的耐药性,畜禽养殖废水可能是该流域抗生素耐药率高的重要因素,河水分离的E. coli中41%携带一类整合子,整合子介导的抗性基因包括aadA1、drfA1、drfA27、arr3等.
有关土壤环境中重金属与抗生素抗性基因的研究较少.Knapp等(2011)指出土壤中Cu含量(0~140 mg · kg-1DM)与tetM、tetW、ermB、ermF具有相关性,且blaOXA与Cu具有极显著相关性;Zn含量(0~38 mg · kg-1DM)与所测抗性基因的相关性不显著;因此畜禽养殖粪污在土壤环境中可能存在重金属与抗生素抗性基因的协同选择问题,需要进一步开展研究.
畜禽养殖废水浇灌蔬菜引起蔬菜携带抗性基因和耐药菌的研究非常少,然而该途径可能是畜禽养殖源抗性基因进入食物链的途径之一.Yang等(2014)对施用鸡粪种植的蔬菜内生菌进行了耐药性测试,发现芹菜、小白菜、黄瓜中头孢氨苄耐药菌的比例分别为16.9%~86.33%、21.76%~91.31%和0.21%~0.44%,蔬菜内生菌具有抗生素抗性的原因可能是耐药菌通过土壤进入植物,或者由于土壤中抗性基因被植物吸收,这需要进一步深入研究.Marti等(2013)的研究发现,施用猪粪的蔬菜表皮抗性基因的检出率较高,包括IncP oriV、sul2、tet(BT)、ermAF、qnrB、blaPSE和blaOXA20等抗性基因,并指出人类直接食用蔬菜是一种接触土壤耐药菌和抗性基因的途径.
6 结语与展望(Conclusion and prospect)尽管近年来畜禽养殖废水处理与利用过程抗性基因已开展了一定的研究,但现有研究较多采用现场调研方式,对抗性基因的转归机制和去除研究不足,缺乏畜禽养殖废水生物处理与农田利用全过程中抗性基因的系统性研究,难以提出抗性基因减控的有效策略.因此,本文提出如下研究展望:
1)已有研究大多针对畜禽养殖废水生物处理和农田利用过程中四环素类与磺胺类抗性基因的分布规律,但有关β内酰胺类、喹诺酮类抗性基因及其耐药菌的研究较为缺乏,而后者抗生素多用于人类疾病治疗,建议今后加强这方面的研究.
2)畜禽养殖废水抗性基因的消减机制尚不明确.现有畜禽养殖废水中抗性基因消减规律的研究不多,对抗性基因消减规律的解析不足.已有研究主要考察生物处理对抗性基因丰度消减的影响,较少关注功能菌群、工艺操作参数、环境参数与耐药菌群结构(抗性基因宿主细菌)的相互关系.
3)不同畜禽养殖废水和土壤类型、抗性基因类型对养殖废水农田利用抗性基因的传播规律不可一概而论,缺乏系统性的机制研究.需要从畜禽养殖废水生物处理和农田利用全过程对耐药菌、抗性基因转归和控制措施进行系统研究和综合评价.
| [1] | Baker-Austin C, Wright M, Stepanauskas R, et al. 2006. Co-selection of antibiotic and metal resistance[J]. Trends in Microbiology, 14(4):176-182 |
| [2] | Barkovskii A L, Manoylov K M, Bridges C. 2012. Positive and negative selection towards tetracycline resistance genes in manure treatment lagoons[J]. Journal of applied microbiology, 112(5):907-919 |
| [3] | Beneragama N, Iwasaki M, Lateef S, et al. 2013. The survival of multidrug-resistant bacteria in thermophilic and mesophilic anaerobic co-digestion of dairy manure and waste milk[J]. Animal Science Journal, 84(5):426-433 |
| [4] | Blair J M A, Webber M A, Baylay A J, et al. 2015. Molecular mechanisms of antibiotic resistance[J]. Nature Reviews Microbiology, 13(1):42-51 |
| [5] | Cantón R, Ruiz-Garbajosa P. 2011. Co-resistance:an opportunity for the bacteria and resistance genes[J]. Current Opinion in Pharmacology, 11(5):477-485 |
| [6] | Chapman J S. 2003. Disinfectant resistance mechanisms, cross-resistance, and co-resistance[J]. International Biodeterioration and Biodegradation, 51(4):271-276 |
| [7] | Chee-Sanford J C, Aminov R I, Krapac I J, et al. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities[J]. Applied and Environmental Microbiology, 67(4):1494-1502 |
| [8] | Chen B, Zheng W W, Yu Y, et al. 2011. Class 1 integrons, selected virulence genes, and antibiotic resistance in Escherichia coli Isolates from the Minjiang River, Fujian Province, China[J]. Applied and Environmental Microbiology, 77(1):148-155 |
| [9] | Chen J, Fluharty F, St-Pierre N, et al. 2008. Technical note:Occurrence in fecal microbiota of genes conferring resistance to both macrolide-lincosamide-streptrogramin B and tetracyclines concomitant with feeding of beef cattle with tylosin[J]. Journal of Animal Science, 86(9):2385-2391 |
| [10] | Chen J, Michel F C, Sreevatsan S, et al. 2010. Occurrence and persistence of erythromycin resistance genes (erm) and tetracycline resistance genes (tet) in waste treatment systems on swine farms[J]. Microbial Ecology, 60(3):479-486 |
| [11] | Cheng V C C, Wong S C Y, Ho P L, et al. 2015. Strategic measures for the control of surging antimicrobial resistance in Hong Kong and mainland of China[J]. Emerging Microbes and Infections, 4(2):e8 |
| [12] | Cheng W X, Chen H, Su C, et al. 2013. Abundance and persistence of antibiotic resistance genes in livestock farms:A comprehensive investigation in eastern China[J]. Environment International, 61:1-7 |
| [13] | 成卫孝. 2014.农业循环经济系统中抗生素和抗性基因的行为特征[D].杭州:浙江大学. 45 |
| [14] | Diehl D L, Lapara T M. 2010. Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids[J]. Environmental Science & Technology, 44(23):9128-9133 |
| [15] | 窦秋燕. 2010.猪粪中抗锌肠细菌及抗锌基因、抗生素抗性基因的和毒力基因研究[D].昆明:云南大学. 23 |
| [16] | Du J, Ren H Q, Geng J J, et al. 2014. Occurrence and abundance of tetracycline, sulfonamide resistance genes, and class 1 integron in five wastewater treatment plants[J]. Environmental Science and Pollution Research, 21(12):7276-7284 |
| [17] | Fahrenfeld N, Knowlton K, Krometis L A, et al. 2014. Effect of manure application on abundance of antibiotic resistance genes and their attenuation rates in soil:field-scale mass balance approach[J]. Environmental Science & Technology, 48(5):2643-2650 |
| [18] | Geisenberger O, Ammendola A, Christensen B B, et al. 1999. Monitoring the conjugal transfer of plasmid RP4 in activated sludge and in situ identification of the transconjugants[J]. FEMS Microbiology Letters, 174(9):9-17 |
| [19] | Gilley J E, Eghball B, Marx D B. 2007. Nutrient concentrations of runoff during the year following manure application[J]. Transactions of the ASABE, 50(6):1987-1999 |
| [20] | Gomez-Alvarez V, Pevetta R P, Domingo J W S. 2012. Metagenomic analyses of drinking water receiving different disinfection treatments[J]. Applied and Environmental Microbiology, 78(17):6095-6012 |
| [21] | 贡娇娜. 2010.猪场内外环境中抗铜肠细菌及其抗性基因的研究[D].昆明:云南大学. 58 |
| [22] | Heuer H, Smalla K. 2007. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months[J]. Environmental Microbiology, 9(3):657-666 |
| [23] | Heuer H, Focks A, Lamshöft M, et al. 2008. Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil[J]. Soil Biology and Biochemistry, 40(7):1892-1900 |
| [24] | Heuer H, Binh C T T, Jechalke S, et al. 2012. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems:diversification driven by class 1 integron gene cassettes[J]. Frontiers in Microbiology, 3:2 |
| [25] | Hölzel C S, Müller C, Harms K S, et al. 2012. Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance[J]. Environmental Research, 113:21-27 |
| [26] | Hong P Y, Yannarell A C, Dai Q H, et al. 2013. Monitoring the perturbation of soil and groundwater microbial communities due to pig production activities[J]. Applied and Environmental Microbiology, 79(8):2620-2629 |
| [27] | Hu Y F, Yang X, Qin J J, et al. 2013. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota[J]. Nature Communications, 4:2151 |
| [28] | 环保部,统计局,农业部. 2010.第一次全国污染源普查公报[R].北京:中华人民共和国环境保护部. 3 |
| [29] | Huang X, Liu C X, Li K, et al. 2013. Occurrence and distribution of veterinary antibiotics and tetracycline resistance genes in farmland soils around swine feedlots in Fujian Province, China[J]. Environmental Science and Pollution Research, 20(12):9066-9074 |
| [30] | Ji X L, Shen Q H, Liu F, et al. 2012. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China[J]. Journal of Hazardous Materials, 235-236:178-185 |
| [31] | 冀秀玲,刘芳,沈群辉,等. 2011.养殖场废水中磺胺类和四环素抗生素及其抗性基因的定量检测[J].生态环境学报, 20(5):927-933 |
| [32] | Jindal A, Kocherginskaya S, Mehboob A, et al. 2006. Antimicrobial use and resistance in swine waste treatment systems[J]. Applied and Environmental Microbiology, 72(12):7813-7820 |
| [33] | Joy S R, Bartelt-Hunt S L, Snow D D, et al. 2013. Fate and transport of antimicrobials and antimicrobial resistance genes in soil and runoff following land application of swine manure slurry[J]. Environmental Science & Technology, 47(21):12081-12088 |
| [34] | Joy S R, Li X, Snow D D, et al. 2014. Fate of antimicrobials and antimicrobial resistance genes in simulated swine manure storage[J]. Science of the Total Environment, 481:69-74 |
| [35] | Kim W, Cho K, Lee S, et al. 2013. Comparison of methanogenic community structure and anaerobic process performance treating swine wastewater between pilot and optimized lab scale bioreactors[J]. Bioresource Technology, 145:48-56 |
| [36] | Knapp C W, McCluskey S M, Singh B K, et al. 2011. Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils[J]. PLoS One, 6(11):e27300 |
| [37] | Koike S, Krapac I G, Oliver H D, et al. 2007. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period[J]. Applied and Environmental Microbiology, 73(15):4813-4823 |
| [38] | Kopmann C, Jechalke S, Rosendahl I, et al. 2013. Abundance and transferability of antibiotic resistance as related to the fate of sulfadiazine in maize rhizosphere and bulk soil[J]. FEMS Microbiology Ecology, 83(1):125-134 |
| [39] | 李文哲,徐名汉,李晶宇. 2013.畜禽养殖废弃物资源化利用技术发展分析[J].农业机械学报, 44(5):135-142 |
| [40] | Liu L, Liu C X, Zheng J Y, et al. 2013. Elimination of veterinary antibiotics and antibiotic resistance genes from swine wastewater in the vertical flow constructed wetlands[J]. Chemosphere, 91(8):1088-1093 |
| [41] | Luo Y, Mao D Q, Rysz M, et al. 2010. Trends in antibiotic resistance genes occurrence in the Haihe River, China[J]. Environmental Science & Technology, 44(19):7220-7225 |
| [42] | Ma Y J, Wilson C A, Novak J T, et al. 2011. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons[J]. Environmental Science & Technology, 45(18):7855-7861 |
| [43] | Macauley J, Qiang Z M, Adams C D, et al. 2006. Disinfection of swine wastewater using chlorine, ultraviolet light and ozone[J]. Water Research, 40(10):2017-2026 |
| [44] | Marti R, Scott A, Tien Y C, et al. 2013. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest[J]. Applied and Environmental Microbiology, 79(18):5701-5709 |
| [45] | Martinez J L, Sánchez M B, Martínez-Solano L, et al. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems[J]. FEMS Microbiology Reviews, 33(2):430-449 |
| [46] | McKinney C W, Loftin K A, Meyer M T, et al. 2010. Tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence[J]. Environmental Science & Technology, 44(16):6102-6109 |
| [47] | Oh J, Salcedo D E, Medriano C A, et al. 2014. Comparison of different disinfection processes in the effective removal of antibiotic-resistant bacteria and genes[J]. Journal of Environmental Sciences, 26(6):1238-1242 |
| [48] | Padmasiri S I, Zhang J Z, Fitch M, et al. 2007. Methanogenic population dynamics and performance of an anaerobic membrane bioreactor (AnMBR) treating swine manure under high shear conditions[J]. Water Research, 41(1):134-144 |
| [49] | Pruden A, Pei R T, Storteboom H, et al. 2006. Antibiotic resistance genes as emerging contaminants:Studies in northern Colorado[J]. Environmental Science & Technology, 40(12):7445-7450 |
| [50] | Rahube T O, Yost C K. 2012. Characterization of a mobile and multiple resistance plasmid isolated from swine manure and its detection in soil after manure application[J]. Journal of Applied Microbiology, 112(6):1123-1133 |
| [51] | Rahube T O, Marti R, Scott A, et al. 2014. Impact of fertilizing with raw or anaerobically digested sewage sludge on the abundance of antibiotic-resistant coliforms, antibiotic resistance genes, and pathogenic bacteria in soil and on vegetables at harvest[J]. Applied and Environmental Microbiology, 80(22):6898-6907 |
| [52] | Roberts M C. 2005. Update on acquired tetracycline resistance genes[J]. FEMS Microbiology Letters, 245(2):195-203 |
| [53] | Rosenberg R E, Micallef S A, Gibbs S G, et al. 2014. Detection of vancomycin-resistant enterococci (VRE) at four U. S. wastewater treatment plants that provide effluent for reuse[J]. Science of the Total Environment, 466-467:404-411 |
| [54] | Rysz M, Mansfield W R, Fortner J D, et al. 2013. Tetracycline resistance gene maintenance under varying bacterial growth rate, substrate and oxygen availability, and tetracycline concentration[J]. Environmental Science & Technology, 47(13):6995-7001 |
| [55] | Salyers A A, Amábile-Cuevas C F. 1997. Why are antibiotic resistance genes so resistant to elimination?[J]. Antimicrobial Agents and Chemotherapy, 41(11):2321-2325 |
| [56] | Schmitt H, Stoob K, Hamsher G, et al. 2006. Tetracyclines and tetracycline resistance in agricultural soils:Microcosm and field studies[J]. Microbial Ecology, 51(3):267-276 |
| [57] | Schröder J J, Neeteson J J. 2008. Nutrient management regulations in The Netherlands[J]. Geoderma, 144(3/4):418-425 |
| [58] | Seiler C, Berendonk T U. 2012. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture[J]. Frontiers in Microbiology, 3:399 |
| [59] | Sengeløv G, Agersø Y, Halling-Sørensen B, et al. 2003. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry[J]. Environment International, 28(7):587-595 |
| [60] | Sköld O. 2000. Sulfonamide resistance:mechanisms and trends[J]. Drug Resistance Updates, 3(3):155-160 |
| [61] | Smith M S, Yang R K, Knapp C W, et al. 2004. Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR[J]. Applied and Microbiology Biotechnology, 70(12):7372-7377 |
| [62] | Su J Q, Wei B, Xu C Y, et al. 2014. Functional metagenomic characterization of antibiotic resistance genes in agricultural soils from China[J]. Environment International, 65:9-15 |
| [63] | Sui Q W, Liu C, Dong H M, et al. 2014. Effect of ammonium nitrogen concentration on the ammonia-oxidizing bacteria community in a membrane bioreactor for the treatment of anaerobically digested swine wastewater[J]. Journal of Bioscience and Bioengineering, 118(3):277-283 |
| [64] | Tao C W, Hsu B M, Ji W T, et al. 2014. Evaluation of five antibiotic resistance genes in wastewater treatment systems of swine farms by real-time PCR[J]. Science of the Total Environment, 496:116-121 |
| [65] | 佟娟,魏源送. 2012.污水处理厂削减耐药菌与抗性基因的研究进展[J].环境科学学报, 32(11):2650-2656 |
| [66] | Udikovic-Kolic N, Wichmann F, Broderick N A, et al. 2014. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization[J]. Proceedings of the National Academy of Sciences of the United Stated of America, 111(42):15202-15207 |
| [67] | 王瑞,魏源送. 2013.畜禽粪便中残留的四环素类抗生素和重金属的污染特征及其控制[J].农业环境科学学报, 32(9):1705-1719 |
| [68] | WHO. 2014. Antimicrobial resistance:global report on surveillance[R]. Switzerland:World Health Organization. 1 |
| [69] | Wu N, Qiao M, Zhang B, et al. 2010. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China[J]. Environmental Science & Technology, 44(18):6933-6939 |
| [70] | 吴楠,乔敏,朱永官. 2009.猪场土壤中5种四环素抗性基因的检测和定量[J].生态毒理学报, 4(5):705-710 |
| [71] | Yang D, Wang J F, Qiu Z G, et al. 2013. Horizontal transfer of antibiotic resistance genes in a membrane bioreactor[J]. Journal of Biotechnology, 167(4):441-447 |
| [72] | Yang Q X, Ren S W, Niu T Q, et al. 2014. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables[J]. Environmental Science and Pollution Research, 21(2):1231-1241 |
| [73] | 张俊亚,魏源送,陈梅雪,等. 2015.畜禽粪便生物处理与土地利用全过程中抗生素和重金属抗性基因的赋存与转归特征研究进展[J].环境科学学报, 35(4):935-946, doi:10.13671/j.hjkxxb.2014.0843 |
| [74] | Zhang Y P, Zhang C Q, Parker D B, et al. 2013. Occurrence of antimicrobials and antimicrobial resistance genes in beef cattle storage ponds and swine treatment lagoons[J]. Science of the Total Environment, 463-464:631-638 |
| [75] | Zhang Y Y, Zhuang Y, Geng J N, et al. 2015. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection[J]. Journal of the Total Environment, 512-513:125-132 |
| [76] | 郑加玉,刘琳,高大文,等. 2013.四环素抗性基因在人工湿地中的去除及积累[J].环境科学, 34(8):3012-3017 |
| [77] | Zhou L J, Ying G G, Liu S, et al. 2013. Excretion masses and environmental occurrence of antibiotics in typical swine and dairy cattle farms in China[J]. Science of the Total Environment, 444:183-195 |
| [78] | Zhou Z, Raskin L, Zilles J L. 2010. Effects of swine manure on macrolide, lincosamide, and streptogramin B antimicrobial resistance in soils[J]. Applied and Environmental Microbiology, 76(7):2218-2224 |
| [79] | Zhu Y G, Johnson T A, Su J Q, et al. 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms[J]. Proceedings of the National Academy of Sciences of the United States of America, 110(9):3435-3440 |
 2016, Vol. 36
2016, Vol. 36


