2. 海军军医大学(第二军医大学)基础医学院人体解剖学教研室,上海 200433;
3. 海军军医大学(第二军医大学)转化医学研究中心干细胞与再生医学研究室,上海 200433
2. Department of Anatomy, College of Basic Medical Sciences, Naval Medical University (Second Military Medical University), Shanghai 200433, China;
3. Laboratory of Stem Cell and Regenerative Medicine, Center of Translational Medicine, Naval Medical University (Second Military Medical University), Shanghai 200433, China
胰腺导管腺癌(pancreatic ductal adenocarcinoma,PDAC)是最常见的胰腺肿瘤,其恶性程度高,致死率居高不下,预后普遍较差,5年生存率仅为11%~13%[1-3]。在中国,PDAC总人群发病率在所有肿瘤中位列第8位,死亡率位列第6位,并呈现出不断上升的趋势[4]。PDAC的发生和发展与其肿瘤微环境(tumor microenvironment,TME)密切相关[5],其中肿瘤相关巨噬细胞是胰腺TME中最丰富且最早浸润的免疫细胞群之一[6-7]。巨噬细胞在PDAC中倾向于极化为M2型,从而促进和支持肿瘤发生、免疫逃逸、转移和化疗耐药等恶性行为[7]。因此,深入研究PDAC中巨噬细胞的基因特征,尤其是其与预后相关的基因及其表达特征,对PDAC早期诊断和靶向治疗具有重要意义。
本研究收集了临床上PDAC患者肿瘤组织样本,利用单细胞测序技术分析组织转录组数据,结合癌症基因组图谱(The Cancer Genome Atlas,TCGA)数据库,获取早期与晚期PDAC差异表达的巨噬细胞相关预后基因,利用单细胞测序数据对预后基因在PDAC中的免疫浸润情况以及在不同时期不同亚群巨噬细胞中的表达情况进行解析,为深入发掘PDAC的早期诊断标志物和治疗靶点提供数据支撑。
1 材料和方法 1.1 数据收集与筛选从临床上收集7例PDAC患者肿瘤及其癌旁组织样本,包括早期(TNM分期Ⅰ~Ⅲ期)患者3例、晚期(TNM分期Ⅳ期)患者4例,其中1例晚期患者缺少相应癌旁组织样本(已通过海军军医大学第一附属医院伦理委员会审批),利用单细胞测序技术分析其转录组数据。从TCGA数据库中下载早期和晚期PDAC相关转录组数据。分别利用聚类图和火山图对临床数据和TCGA数据进行可视化处理,筛选早期与晚期PDAC中巨噬细胞相关差异表达基因。对差异表达基因进行单因素回归分析和对数秩检验,评价其与预后的关系(若P<0.05,则该基因与预后有关)。利用维恩图将临床标本和TCGA数据库中筛选得到的与预后有关的巨噬细胞基因取交集,获得早期与晚期差异表达的与预后有关的巨噬细胞相关基因。进一步利用基因表达谱交互分析2(Gene Expression Profiling Interactive Analysis 2,GEPIA2)在线生存分析工具(http://gepia2.cancer-pku.cn/#analysis)筛选出差异有统计学意义的预后风险基因。
1.2 预后风险评分模型的构建基于TCGA数据库,使用最小绝对收缩和选择算子(least absolute shrinkage and selection operator,LASSO)-Cox回归分析构建PDAC预后风险评分模型,把得到的系数和基因表达值相乘后求和则可获得风险评分。依据其中位数将PDAC患者分为高风险组和低风险组,使用Kaplan-Meier生存曲线评估PDAC预后模型的预测能力。
1.3 肿瘤组织中的免疫浸润情况分析基于TCGA中的肿瘤样本数据,采用通过估计RNA转录本相对子集进行细胞类型鉴定(cell-type identification by estimating relative subsets of RNA transcripts,CIBERSORT)方法,在挑选出的M0、M1、M2型巨噬细胞及调节性T细胞(regulatory T cell,Treg细胞)这几个免疫浸润细胞中,分析筛选出的预后相关基因的表达情况。
1.4 单细胞测序分析基于单细胞测序数据,采用Seurat包进行分析,通过t-分布随机近邻嵌入(t-distributed stochastic nearest-neighbour embedding,tSNE)聚类降维分析不同巨噬细胞聚类中预后相关基因的表达,不同的细胞亚群用SingleR包进行注释。利用Monocle包对巨噬细胞发育阶段及预后相关基因的表达进行拟时序分析,明确预后相关基因在早期和晚期PDAC巨噬细胞中的表达差异。
1.5 基因富集分析利用注释、可视化和集成发现数据库(Database for Annotation, Visualization and Integrated Discovery;DAVID)(https://david.ncifcrf.gov/),对早期和晚期PDAC巨噬细胞的差异表达基因进行基因本体(Gene Ontology,GO)功能富集分析,明确早期与晚期PDAC巨噬细胞生物学功能的改变。
1.6 蛋白质-蛋白质相互作用(protein-protein interaction,PPI)网络分析利用STRING数据库(https://cn.string-db.org/)获取预后相关基因的PPI网络相关数据,通过Cytoscape 3.7.1软件对PPI网络进行深入分析,获得预后相关基因之间的表达关系和其他共表达蛋白,从而推测其在PDAC中的作用。
1.7 免疫组织化学分析从人类蛋白质图谱(The Human Protein Atlas,HPA)数据库(https://www.proteinatlas.org/)中下载胰腺癌组织及正常胰腺组织预后相关蛋白表达的免疫组织化学染色图片,分析其在胰腺癌组织和正常胰腺组织中的表达情况。
1.8 统计学处理应用R 4.0软件对数据进行统计学分析。两组间比较采用Wilcoxon秩和检验,多组间比较采用单因素方差分析。检验水准(α)为0.05。
2 结果 2.1 早期与晚期PDAC患者巨噬细胞预后相关差异表达基因的获取基于临床13组PDAC患者组织的数据,利用Seurat包,在tSNE降维分析后,共聚类出15个细胞亚群,利用SingleR包对细胞亚群进行注释,其中亚群5注释为巨噬细胞(图 1A)。通过堆叠柱状图,将15个细胞亚群在13组PDAC患者组织中的占比进行可视化处理(图 1B)。基于3例早期、4例晚期PDAC患者的相关数据,分析比较早期与晚期PDAC患者巨噬细胞相关基因的表达差异,共发现了583个与预后有关的差异表达基因。基于TCGA数据库中早期与晚期PDAC的差异表达基因,绘制火山图(图 1C),并进一步分析发现了2 019个与预后有关的差异表达基因。将其与583个从临床患者组织样本中获取的差异表达基因取交集,获得48个与预后有关的早期与晚期PDAC患者巨噬细胞差异表达基因(图 1D)。
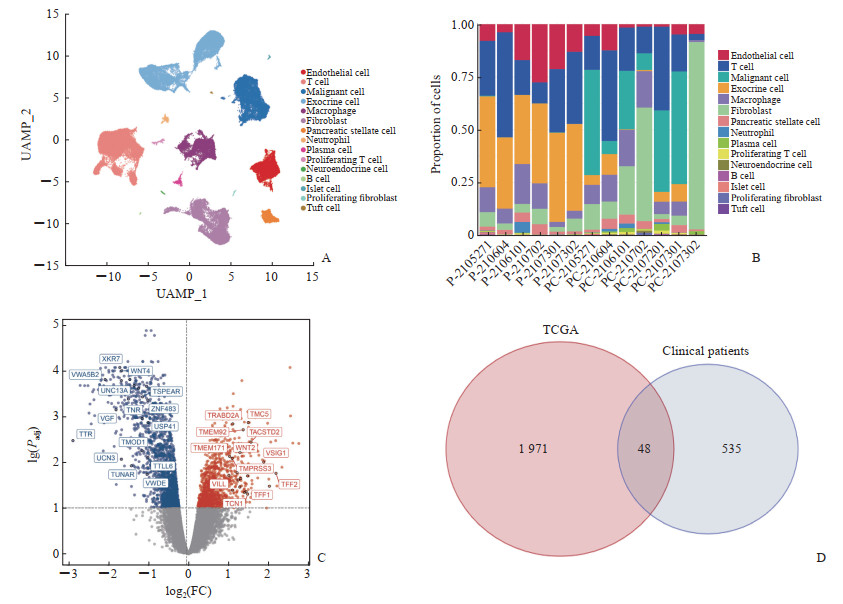
|
图 1 PDAC患者样本数据的可视化分析与预后相关基因的获取 Fig 1 Visual analysis of PDAC sample data and acquisition of prognosis-related genes A: The tSNE diagram showed the different cell types in PDAC patients; B: The stacked bar chart showed the percentage of different cells in PDAC patients (P indicates paracancerous tissue, and PC indicates cancerous tissue); C: Volcano map based on early versus late PDAC differentially expressed genes in the TCGA database; D: Venn diagram showed early versus late PDAC prognostic genes shared by TCGA and patients. PDAC: Pancreatic ductal adenocarcinoma; tSNE: t-distributed stochastic neighbor embedding; UAMP: Uniform manifold approximation and projection; TCGA: The Cancer Genome Atlas; FC: Fold change; Padj: Adjusted P value. |
2.2 基于巨噬细胞相关差异表达基因的PDAC预后风险评分模型的构建与评价
采用LASSO-Cox回归分析方法,筛选出11个关键巨噬细胞相关差异表达基因:红细胞膜条带4.1蛋白样3(erythrocyte membrane protein band 4.1 like 3, EPB41L3),谷胱甘肽S-转移酶κ1(glutathione S-transferase kappa 1,GSTK1),犬尿氨酸酶(kynureninase,KYNU),乳酸脱氢酶A(lactate dehydrogenase A,LDHA),丝裂原活化蛋白激酶激酶激酶8(mitogen-activated protein kinase kinase kinase 8,MAP3K8),2’-5’-寡腺苷酸合成酶2(2’-5’-oligoadenylate synthetase 2,OAS2),过氧化物酶2(peroxiredoxin 2,PRDX2),Serpin家族B成员1(Serpin family B member 1,SERPINB1),转酮酶(transketolase,TKT),Tweety家族成员3(Tweety family member 3,TTYH3),ZFP36无名指蛋白样1(ZFP36 ring finger protein like 1,ZFP36L1)。以这11个关键基因构建PDAC预后风险评分模型:风险评分=-0.026 8×EPB41L3+0.015 0×GSTK1+0.080 2×KYNU+0.332 0×LDHA-0.030 4×MAP3K8+0.097 2×OAS2-0.148 3×PRDX2+0.176 1×SERPINB1+0.019 6×TKT-0.144 6×TTYH3+0.216 3×ZFP36L1。依据风险评分的中位数将PDAC患者分为高风险组和低风险组,使用Kaplan-Meier生存曲线对预后模型进行评估,结果显示高风险组患者总生存期低于低风险组患者(P<0.000 1,图 2A)。利用GEPIA2数据库分析所获得的全部48个与预后有关的基因,尤其是8个正向相关基因中,发现OAS2和人类异常纺锤体样小头畸形相关蛋白(assembly factor for spindle microtubules,ASPM)基因在PDAC组织中的表达量显著高于正常组织,并且高表达组的总生存期低于低表达组,是PDAC的不良预后基因(图 2B~2E)。
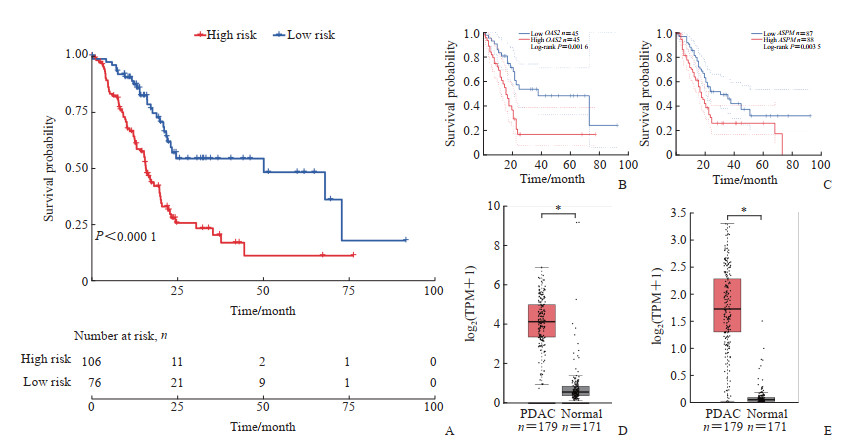
|
图 2 PDAC巨噬细胞相关差异表达基因预后风险评分模型的评价与预后相关风险基因的筛选 Fig 2 Evaluation of a prognostic risk scoring model for PDAC macrophage-associated differentially expressed genes and screening of prognostically relevant risk genes A: Kaplan-Meier survival curves of PDAC patients with different risks in TCGA database; B: GEPIA2 analysis showed the correlation between OAS2 and overall survival of patients; C: GEPIA2 analysis showed the correlation between ASPM and overall survival of patients; D: GEPIA2 analysis showed that OAS2 expression was higher in PDAC tissues than in normal tissues; E: GEPIA2 analysis showed that ASPM expression was higher in PDAC tissues than in normal tissues. *P<0.05. PDAC: Pancreatic ductal adenocarcinoma; TCGA: The Cancer Genome Atlas; GEPIA2: Gene Expression Profiling Interactive Analysis 2; OAS2: 2’-5’-oligoadenylate synthetase 2; ASPM: Assembly factor for spindle microtubules; TPM: Transcripts per million. |
2.3 PDAC预后基因OAS2与ASPM在TCGA肿瘤组织中的免疫浸润情况
基于TCGA数据库中PDAC样本数据,通过CIBERSORT方法研究OAS2和ASPM基因在Treg细胞及M0、M1、M2型巨噬细胞中的表达情况,结果显示OAS2和ASPM在4种细胞中的表达趋势基本一致,在Treg细胞和M2型巨噬细胞中表达水平均较高(图 3)。
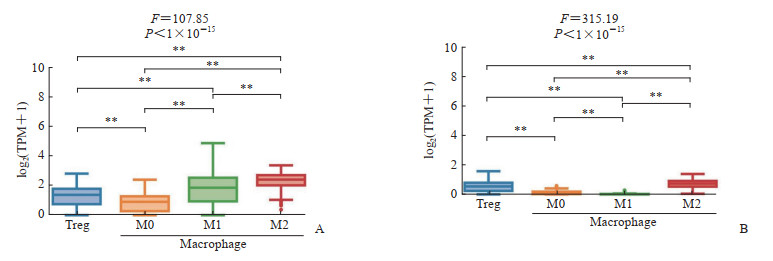
|
图 3 PDAC预后基因在TCGA肿瘤组织中的免疫浸润情况 Fig 3 Immune infiltration of PDAC prognostic genes in TCGA tumor tissue A: Immune infiltration of OAS2 in TCGA tumor tissue; B: Immune infiltration of ASPM in TCGA tumor tissue. **P<0.01. PDAC: Pancreatic ductal adenocarcinoma; TCGA: The Cancer Genome Atlas; OAS2: 2’-5’-oligoadenylate synthetase 2; ASPM: Assembly factor for spindle microtubules; TPM: Transcripts per million; Treg: Regulatory T cell. |
2.4 PDAC巨噬细胞分群与预后基因定位
利用Seurat包对PDAC巨噬细胞进行聚类,共聚类出6个亚群,并通过SingleR包对其进行注释(图 4A)。其中,表达溶酶体相关膜蛋白3(lysosomal associated membrane protein 3,LAMP3)的树突状细胞、表达纤维胶蛋白1(ficolin 1,FCN1)的巨噬细胞、表达丝氨酸蛋白酶(serine protease,PRSS)的巨噬细胞和朗格汉斯细胞更倾向于M1型巨噬细胞,发挥抗肿瘤免疫作用;而表达分泌型磷蛋白1(secreted phosphoprotein 1,SPP1)的巨噬细胞和表达增殖标志物Ki-67(marker of proliferation Ki-67,MKI67)的巨噬细胞更倾向于M2型巨噬细胞,起到促进肿瘤增殖和转移的作用[8-9]。通过聚类点图(dot plot)对OAS2和ASPM这2个预后基因在巨噬细胞不同亚群中的表达情况进行可视化分析,结果发现OAS2主要在SPP1+巨噬细胞中表达,ASPM主要在MKI67+巨噬细胞中表达(图 4B)。结合免疫浸润分析结果,提示OAS2和ASPM可能对PDAC的免疫微环境起到负向的调节作用,从而影响患者预后。
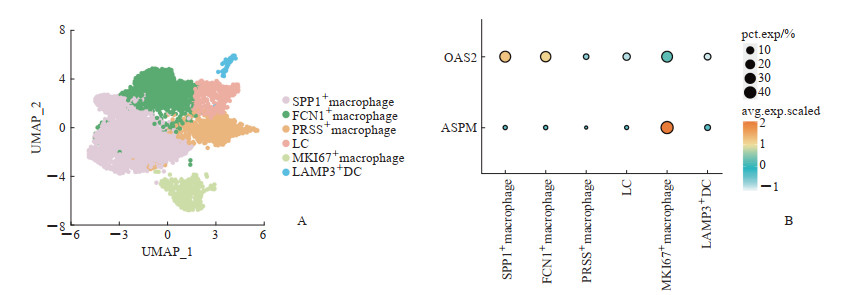
|
图 4 PDAC预后基因在巨噬细胞中的分布 Fig 4 Distribution of PDAC prognostic genes in macrophages A: The tSNE diagram showed the different macrophage types; B: Dot plot showed the expression levels of OAS2 and ASPM in different macrophage clusters. PDAC: Pancreatic ductal adenocarcinoma; tSNE: t-distributed stochastic neighbor embedding; SPP1: Secreted phosphoprotein 1; FCN1: Ficolin 1; PRSS: Serine protease; LC: Langerhans cell; MKI67: Marker of proliferation Ki-67; LAMP3: Lysosomal associated membrane protein 3; DC: Dendritic cell; UMAP: Uniform manifold approximation and projection; OAS2: 2’-5’-oligoadenylate synthetase 2; ASPM: Assembly factor for spindle microtubules; pct.exp: Percentage expression; avg.exp.scaled: Average expression scaled. |
2.5 巨噬细胞相关预后基因在不同PDAC时期的表达
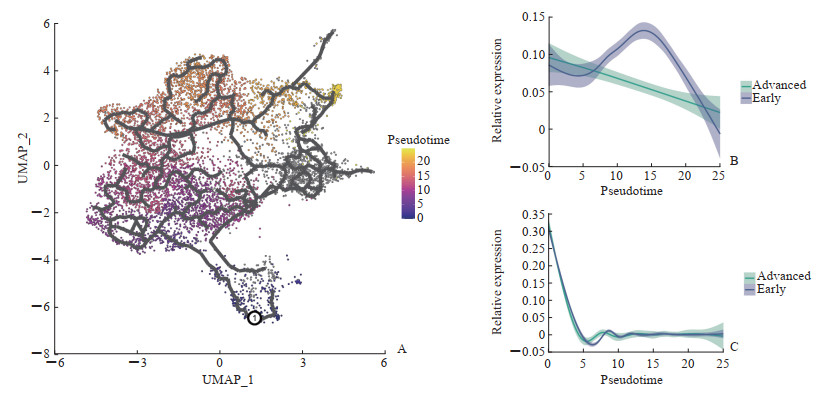
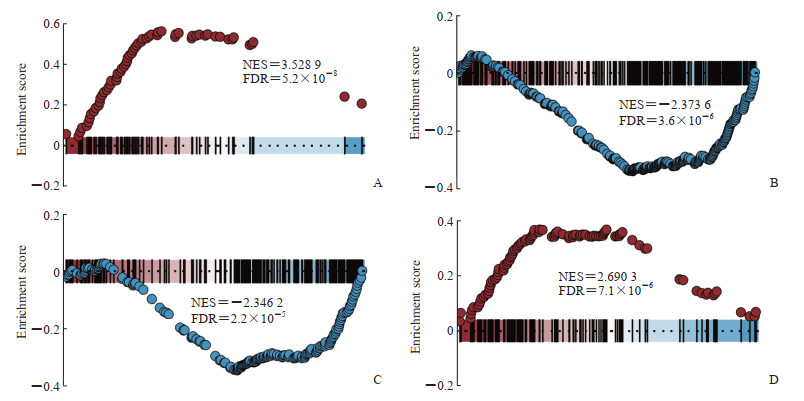
基于单细胞测序数据,运用拟时序分析解析不同巨噬细胞亚群的演化过程(图 5A)。结果表明随着PDAC的进展,巨噬细胞从M1倾向型向M2倾向型演化。对OAS2和ASPM这2个预后基因在早期和晚期PDAC巨噬细胞中的表达情况进行拟时序分析,结果显示OAS2在PDAC早期与晚期的表达趋势具有明显差异,即早期的表达量先升高后降低,晚期表达量逐渐降低(图 5B)。这提示OAS2可以作为PDAC的早期诊断标志物和治疗靶点。而ASPM在PDAC早期与晚期的表达趋势几乎无差异(图 5C)。对早期和晚期PDAC巨噬细胞相关差异表达基因进行GO富集分析发现,与早期巨噬细胞相比,晚期巨噬细胞免疫系统过程和细胞通讯功能下调,细胞质翻译和多肽代谢功能上调(图 6)。
|
图 5 巨噬细胞相关预后基因在不同PDAC时期的表达情况 Fig 5 Expression levels of macrophage-associated prognostic genes in different PDAC periods A: Pseudotime analysis showed the developmental trajectory of macrophages; B: Expression levels of OAS2 in different PDAC periods; C: Expression levels of ASPM in different PDAC periods. PDAC: Pancreatic ductal adenocarcinoma; UMAP: Uniform manifold approximation and projection; OAS2: 2’-5’-oligoadenylate synthetase 2; ASPM: Assembly factor for spindle microtubules. |
|
图 6 早期和晚期PDAC巨噬细胞相关差异表达基因GO功能富集分析 Fig 6 GO enrichment analysis of differentially expressed genes associated with early and late PDAC macrophages A: Enrichment analysis showed upregulation of cytoplasmic translation pathways; B: Enrichment analysis showed downregulation of cell communication pathways; C: Enrichment analysis showed downregulation of immune system process pathways; D: Enrichment analysis showed upregulation of peptide metabolic process pathways. PDAC: Pancreatic ductal adenocarcinoma; GO: Gene Ontology; NES: Normalized enrichment score; FDR: False discovery rate. |
2.6 预后基因的PPI网络分析及其在肿瘤组织中的表达
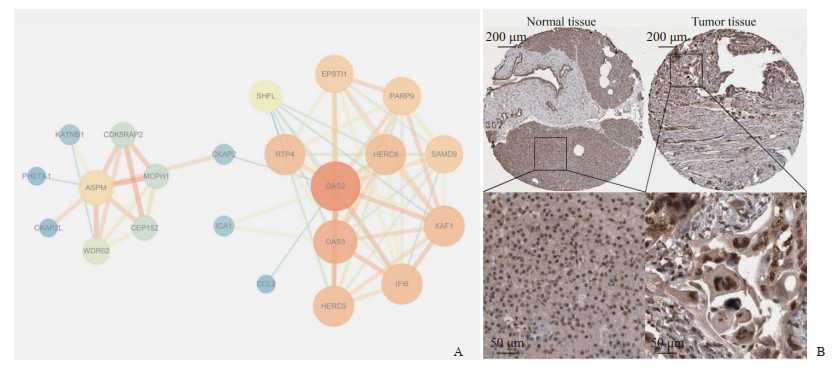
利用STRING数据库对OAS2和ASPM进行PPI网络分析,并借助Cytoscape软件进行网络可视化,获得OAS2和ASPM之间的表达关系及其他共表达蛋白(图 7A),大多数共表达蛋白与肿瘤关系密切。利用HPA数据库获取胰腺癌组织及正常胰腺组织OAS2蛋白表达的免疫组织化学染色图片(图 7B),对比发现胰腺癌组织中棕黄色染色更深,即OAS2蛋白在肿瘤组织中的表达高于正常组织。
|
图 7 PDAC预后基因的PPI网络分析及其在肿瘤组织中的表达 Fig 7 PPI network analysis of PDAC prognostic genes and their expression in tumor tissues A: The PPI network of OAS2 and ASPM; B: Immunohistochemical detection showed higher expression of OAS2 in tumor tissues than in normal tissues. PDAC: Pancreatic ductal adenocarcinoma; PPI: Protein-protein interaction; OAS2: 2’-5’-oligoadenylate synthetase 2; ASPM: Assembly factor for spindle microtubules. |
3 讨论
由于缺乏早期诊断的生物标志物及个体化治疗的有效靶点,PDAC仍然是一种难治性癌症[10]。近年来,高通量测序技术的不断发展与应用为深入挖掘PDAC免疫微环境、发生和发展机制、诊断标志物及治疗靶点提供了更多机会[11]。本研究基于单细胞测序技术,结合TCGA数据库和来自临床的PDAC患者数据,深入分析PDAC中肿瘤相关巨噬细胞的特征及关键预后基因的表达特征,为PDAC的早期诊断和靶向治疗提供新思路。
本研究共筛选出48个与预后有关的早期和晚期PDAC巨噬细胞相关差异表达基因。其中,有些基因对PDAC具有重要作用,如LDHA可促进胰腺癌肿瘤细胞的糖酵解效率、降低对氧的依赖性,从而促进肿瘤的生长、侵袭和转移[12];CD9介导了膜联蛋白A6(annexin A6,ANXA6)阳性细胞外囊泡的摄取,从而促进胰腺癌的转移[13]。有些基因具有重要的免疫调节作用,如白细胞介素1受体拮抗因子(interleukin-1 receptor antagonist,IL1RA)调节多种与IL-1相关的免疫和炎症反应,对胰腺癌具有抑制作用[14]。随后,从中确定11个关键基因构建PDAC预后风险评分模型,并通过生存曲线评估其预测能力。此模型对于预测PDAC的发生、发展及指导临床诊治具有一定意义。进一步利用GEPIA2在线生存分析工具筛选出OAS2和ASPM这2个与PDAC预后相关的风险基因。对其进行免疫浸润分析发现,OAS2和ASPM在Treg细胞和M2型巨噬细胞中高表达。基于单细胞测序数据,对PDAC巨噬细胞进行分型并分析预后基因的分布发现,OAS2主要在SPP1+巨噬细胞中表达,ASPM主要在MKI67+巨噬细胞中表达。SPP1+巨噬细胞和MKI67+巨噬细胞更倾向于M2型巨噬细胞,促进肿瘤增殖与转移[8-9]。Treg细胞和M2型巨噬细胞通过释放炎症细胞因子产生免疫抑制环境,起到促进PDAC进展的作用[15]。这再次印证了OAS2和ASPM是PDAC的潜在预后基因,并提示OAS2和ASPM可能对PDAC的免疫微环境起到负向调节作用,从而导致PDAC患者的不良预后。拟时序分析表明,随着PDAC的进展,巨噬细胞从M1倾向型向M2倾向型演化;OAS2在早期的表达量先升高后降低,在晚期的表达量逐渐降低。这提示OAS2可能是PDAC的潜在早期诊断标志物。GO富集分析结果显示,晚期PDAC巨噬细胞免疫系统过程功能下调,这可能是早期OAS2和ASPM高表达的结果。PPI网络分析获得了OAS2和ASPM之间的表达关系及其他共表达蛋白,它们大多数与肿瘤关系密切,这为未来深入研究提供了更多方向。免疫组织化学分析显示OAS2蛋白在肿瘤组织中的表达高于正常组织,这从实验角度提示OAS2可能是PDAC的潜在危险基因。
OAS2是2’-5’-寡腺苷酸合成酶中的一员,是一种由干扰素诱导表达的先天免疫应答的必需抗病毒蛋白,它可催化2’-5’-寡腺苷酸的合成以激活核糖核酸酶L(RNase L)并抑制病毒繁殖[16]。它在巨噬细胞中的功能包括抗病毒作用、免疫调节作用、炎症调节作用、细胞凋亡作用等。目前对OAS2的研究多集中于抗病毒免疫方面[17-18],在肿瘤方面的研究相对较少。OAS2在肿瘤中的作用表现出两面性,如在口腔鳞状细胞癌中,OAS2高表达的患者预后差[19];在复发性胶质母细胞瘤中,OAS2促进免疫抑制微环境而导致患者不良预后[20];而在乳腺癌中,OAS2高表达与患者良好预后相关[21]。本研究结果显示OAS2是PDAC的不良预后基因,在SPP1+巨噬细胞中特异性表达并在Treg细胞和M2型巨噬细胞中高表达,推测OAS2可能对PDAC的免疫微环境起到负向调控作用,促进抑制性免疫细胞的极化和产生,进而促进肿瘤的进展和转移。另外,本研究发现OAS2在PDAC早期表达量很高,这提示OAS2可能是潜在的PDAC早期诊断标志物。对于OAS2在PDAC中的可能机制,本研究基于PPI网络发现OAS2与C-C基序趋化因子配体2(C-C motif chemokine ligand 2,CCL2)关联程度较为紧密。最近一项研究指出,CCL2可通过CCL2/C-C基序趋化因子受体2(C-C motif chemokine receptor 2,CCR2)轴招募免疫抑制性巨噬细胞和髓系来源的其他抑制性免疫细胞,促进免疫抑制性微环境的形成,随后驱动胰腺上皮内瘤变(pancreatic intraepithelial neoplasia,PanIN)进展为PDAC[22]。由此推测,这可能是OAS2促进PDAC免疫抑制微环境形成和作为潜在早期标志物的机制。这突显出OAS2在PDAC诊治中的潜在研究价值,需要进行更深入的实验验证和临床研究。
综上所述,本研究为PDAC中肿瘤相关巨噬细胞的特征及相关预后基因OAS2和ASPM的表达特征提供了新的数据支持,为PDAC的早期诊断和靶向治疗提供了新思路,但具体机制和信号通路仍需要进一步的实验探索与研究。
| [1] |
SIEGEL R L, GIAQUINTO A N, JEMAL A. Cancer statistics, 2024[J]. CA Cancer J Clin, 2024, 74(1): 12-49. DOI:10.3322/caac.21820 |
| [2] |
SIEGEL R L, MILLER K D, WAGLE N S, et al. Cancer statistics, 2023[J]. CA Cancer J Clin, 2023, 73(1): 17-48. DOI:10.3322/caac.21763 |
| [3] |
KHALAF N, EL-SERAG H B, ABRAMS H R, et al. Burden of pancreatic cancer: from epidemiology to practice[J]. Clin Gastroenterol Hepatol, 2021, 19(5): 876-884. DOI:10.1016/j.cgh.2020.02.054 |
| [4] |
XIA C, DONG X, LI H, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants[J]. Chin Med J, 2022, 135(5): 584-590. DOI:10.1097/CM9.0000000000002108 |
| [5] |
SHERMAN M H, BEATTY G L. Tumor microenvironment in pancreatic cancer pathogenesis and therapeutic resistance[J]. Annu Rev Pathol, 2023, 18: 123-148. DOI:10.1146/annurev-pathmechdis-031621-024600 |
| [6] |
POH A R, ERNST M. Tumor-associated macrophages in pancreatic ductal adenocarcinoma: therapeutic opportunities and clinical challenges[J]. Cancers, 2021, 13(12): 2860. DOI:10.3390/cancers13122860 |
| [7] |
YANG S, LIU Q, LIAO Q. Tumor-associated macrophages in pancreatic ductal adenocarcinoma: origin, polarization, function, and reprogramming[J]. Front Cell Dev Biol, 2021, 8: 607209. DOI:10.3389/fcell.2020.607209 |
| [8] |
JIN H, CHOI H, KIM E S, et al. Natural killer cells inhibit breast cancer cell invasion through downregulation of urokinase-type plasminogen activator[J]. Oncol Rep, 2021, 45(1): 299-308. DOI:10.3892/or.2020.7840 |
| [9] |
GEORGOUDAKI A M, PROKOPEC K E, BOURA V F, et al. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis[J]. Cell Rep, 2016, 15(9): 2000-2011. DOI:10.1016/j.celrep.2016.04.084 |
| [10] |
DUCREUX M, SEUFFERLEIN T, VAN LAETHEM J L, et al. Systemic treatment of pancreatic cancer revisited[J]. Semin Oncol, 2019, 46(1): 28-38. DOI:10.1053/j.seminoncol.2018.12.003 |
| [11] |
TIBILETTI M G, CARNEVALI I, PENSOTTI V, et al. OncoPan®: an NGS-based screening methodology to identify molecular markers for therapy and risk assessment in pancreatic ductal adenocarcinoma[J]. Biomedicines, 2022, 10(5): 1208. DOI:10.3390/biomedicines10051208 |
| [12] |
COMANDATORE A, FRANCZAK M, SMOLENSKI R T, et al. Lactate dehydrogenase and its clinical significance in pancreatic and thoracic cancers[J]. Semin Cancer Biol, 2022, 86(Pt 2): 93-100. DOI:10.1016/j.semcancer.2022.09.001 |
| [13] |
NIGRI J, LECA J, TUBIANA S S, et al. CD9 mediates the uptake of extracellular vesicles from cancer-associated fibroblasts that promote pancreatic cancer cell aggressiveness[J]. Sci Signal, 2022, 15(745): eabg8191. DOI:10.1126/scisignal.abg8191 |
| [14] |
YUAN S, MIAO Y, RUAN X, et al. Therapeutic role of interleukin-1 receptor antagonist in pancreatic diseases: mendelian randomization study[J]. Front Immunol, 2023, 14: 1240754. DOI:10.3389/fimmu.2023.1240754 |
| [15] |
VELASCO R M, GARCíA A G, SáNCHEZ P J, et al. Tumour microenvironment and heterotypic interactions in pancreatic cancer[J]. J Physiol Biochem, 2023, 79(1): 179-192. DOI:10.1007/s13105-022-00875-8 |
| [16] |
FANG D C, JASS J R, WANG D X, et al. Infrequent loss of heterozygosity of APC/MCC and DCC genes in gastric cancer showing DNA microsatellite instability[J]. J Clin Pathol, 1999, 52(7): 504-508. DOI:10.1136/jcp.52.7.504 |
| [17] |
ELLWANGER J H, CHIES J A B. Host immunogenetics in tick-borne encephalitis virus infection--the CCR5 crossroad[J]. Ticks Tick Borne Dis, 2019, 10(4): 729-741. DOI:10.1016/j.ttbdis.2019.03.005 |
| [18] |
LEISCHING G, WIID I, BAKER B. OAS1, 2, and 3: significance during active tuberculosis?[J]. J Infect Dis, 2018, 217(10): 1517-1521. DOI:10.1093/infdis/jiy084 |
| [19] |
WANG J, WANG Y, KONG F, et al. Identification of a six-gene prognostic signature for oral squamous cell carcinoma[J]. J Cell Physiol, 2020, 235(3): 3056-3068. DOI:10.1002/jcp.29210 |
| [20] |
TATARI N, KHAN S, LIVINGSTONE J, et al. The proteomic landscape of glioblastoma recurrence reveals novel and targetable immunoregulatory drivers[J]. Acta Neuropathol, 2022, 144(6): 1127-1142. DOI:10.1007/s00401-022-02506-4 |
| [21] |
ZHANG Y, YU C. Prognostic characterization of OAS1/OAS2/OAS3/OASL in breast cancer[J]. BMC Cancer, 2020, 20(1): 575. DOI:10.1186/s12885-020-07034-6 |
| [22] |
LIU Y, DEGUCHI Y, WEI D, et al. Rapid acceleration of KRAS-mutant pancreatic carcinogenesis via remodeling of tumor immune microenvironment by PPARδ[J]. Nat Commun, 2022, 13(1): 2665. DOI:10.1038/s41467-022-30392-7 |
 2025, Vol. 46
2025, Vol. 46


