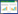文章信息
- 刘铭霄, 吴丽红, 江千舟, 谢微, 王羽仪, 于淼
- LIU Mingxiao, WU Lihong, JIANG Qianzhou, XIE Wei, WANG Yuyi, YU Miao
- Er:YAG激光辅助治疗广泛型Ⅲ期C级牙周炎前后龈下菌斑菌群变化
- Shift of subgingival plaque community after Er: YAG laser adjunctive treatment of generalized stage Ⅲ, grade C periodontitis
- 中国医科大学学报, 2022, 51(4): 336-343
- Journal of China Medical University, 2022, 51(4): 336-343
-
文章历史
- 收稿日期:2021-05-24
- 网络出版时间:2022-04-29 14:53
牙周炎是以菌斑生物膜为始动因素的口腔慢性感染性疾病,可破坏患者的牙周支持组织,造成附着丧失及牙槽骨吸收,最终导致牙脱落,为中国成年人失牙的首位原因[1]。由于牙周炎进展速度较慢,早期症状不明显,就诊时多已发展为Ⅲ期[2]。牙周炎患者的主要治疗目标是清除菌斑生物膜。传统牙周基础治疗主要采用龈下刮治和根面平整术(scaling and root planning,SRP)。但单纯运用传统牙周基础治疗难以清理到较为狭窄的区域,形成的玷污层不利于牙周组织愈合[3-4],且无法有针对性地清除存留于牙周袋内的牙周致病菌[5-6],常辅助应用全身抗生素治疗,但易产生耐药性[7]。
铒:钇铝-石榴石(erbium:yttrium aluminum garnet,Er:YAG)激光可在不造成热损伤的前提下清除软硬组织[3],能较好地清洁根分叉区等狭窄区域[8],具有杀菌潜能和延缓细菌再定植进程的作用[9]。目前一般采用探诊深度(probing depth,PD)、附着丧失水平(attachment loss,AL)、菌斑指数(plaque index,PLI)等评估Er:YAG激光的临床疗效,缺乏微生物学指标,而降低牙周致病菌在龈下菌斑中的比例是牙周炎治疗成功的标准之一[10]。本研究从微生物学角度出发,拟通过观察对比广泛型Ⅲ期C级牙周炎(generalized stage Ⅲ grade C periodontitis,GPⅢ-C)患者治疗前后的龈下菌斑菌群多样性及相对丰度等指标,探讨Er:YAG激光辅助传统牙周基础治疗的微生物学疗效,为Er:YAG激光的临床应用提供科学指导依据。
1 材料与方法 1.1 研究对象本研究招募了2019年9月至2020年8月于广州医科大学附属口腔医院牙周科就诊的GPⅢ-C患者10例(男5例,女5例),年龄(45.8±11.36)岁,采用欧洲牙周病联合会与美国牙周病学会2018年制定的牙周病新分类中GPⅢ-C的诊断标准[11]。排除标准:近3个月内进行牙周治疗,服用抗生素、非激素类抗炎药物及免疫抑制剂,有影响牙周治疗的全身疾病等。本研究已获得广州医科大学附属口腔医院伦理委员会审批(LCYJ2021010),所有研究对象均签署知情同意书。
1.2 研究方法 1.2.1 临床处理传统牙周基础治疗均进行龈上洁治术,再行超声龈下洁治术,随后用龈下刮治器行SRP,清除根面病变牙骨质及龈下牙石;Er:YAG治疗应用Er:YAG激光(德国Fotona公司,模式为SP,30 mJ,30 Hz,0.90 W,水8,气4;LP,120 mJ,15 Hz,1.80 W,水4,气4)照射经传统牙周基础治疗后的牙根表面及牙周袋内壁。
1.2.2 取样及分组本研究采用单盲(对临床指标评价者设盲)随机对照研究。每例受试者行自身对照。G1为治疗前对照组,G2为牙周基础治疗组,G3为Er:YAG激光治疗组。牙周基础治疗与Er:YAG激光辅助治疗均于初诊时完成。将患者口腔分为4个区段,选取牙周PD≥6 mm、临床AL≥5 mm、探诊出血阳性患牙,采集每个区段内PD最深位点的龈下菌斑,并将4个位点的集合菌斑迅速放入1.8 mL冻存管,-80 ℃冰箱保存,G1、G2、G3 3次采集位点相同。
1.2.3 16S rDNA测序采用PowerSoil® DNA Isolation Kit(美国MoBio公司)提取样本细菌的全部DNA,随后对16S rDNA基因V3~V4区进行PCR扩增,使用高通量测序平台Illumina Miseq(美国Illumina公司)对扩增序列进行测序。
1.2.4 菌群数据的生物信息分析测序数据通过自编的生物信息工具进行低质量过滤,并采用FLASH软件v1.2.11(http://ccb.jhu.edu/software/FLASH/index.shtml)进行序列拼接,再通过USEARCH聚合成OTUs(可分类元件)比对细菌库(Greengene v13.5)进行细菌分类注释,从而获得所有样本的菌群组成数据。随后,对各组样本的细菌丰度在门、纲、目、科和属水平进行差异检验分析。
1.3 统计学分析运用R软件(v3.6.0)进行统计学分析。对3组样本的菌群结构进行差异检验,2组比较采用Wilcoxon秩和检验,3组间比较采用Kruskal-Wallis秩和检验。P < 0.05为差异有统计学意义。
2 结果 2.1 3组样本菌群整体结构变化3组样本alpha多样性分析显示,G1组Shannon指数(2.84)与G2组(4.17)比较差异显著(P = 0.009),G2组菌群多样性(4.17)明显高于G1组(2.84),G3组(4.01)菌群多样性较G2组略有下降(图 1A)。菌斑组成的主成分分析(principal components analysis,PCA)结果提示,G1组与G3组的样本呈现各自聚集趋势,而G2组则为G1和G3组的中间过渡形式(图 1B)。
|
| A, alpha diversity analysis of G1, G2 and G3;B, PCA analysis of G1, G2 and G3. 图 1 龈下菌斑菌群分布PCA分析及alpha多样性分析 Fig.1 PCA and alpha diversity analysis of subgingival plaque |
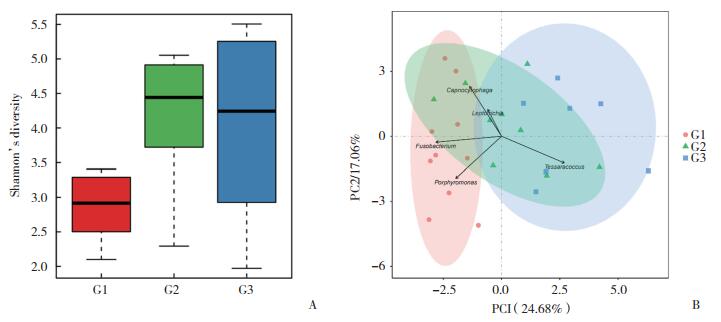
2.2 属水平优势菌及组成差异分析
对各组进行菌属结构分析(表 1,图 2),G1组优势菌属分别为梭形杆菌属(25.3%)、卟啉单胞菌属(17.7%)、二氧化碳嗜纤维菌属(9.6%)、普雷沃氏菌属(8.4%)、纤毛菌属(7.2%);G2组分别为未分类属(29.3%)、梭形杆菌属(19.3%)、二氧化碳嗜纤维菌属(7.3%)、放线菌属(6.8%)、卟啉单胞菌属(4.8%);G3组分别为未分类属(29.9%)、四合球菌属(12.1%)、放线菌属(10.2%)、罗斯氏菌属(7.9%)、梭形杆菌属(7.3%)。对3组菌属相对丰度差异性比较(表 2),梭形杆菌属(G1组25.38%,G2组19.29%,G3组7.31%)、卟啉单胞菌属(G1组17.72%,G2组4.79%,G3组3.09%)、普雷沃氏菌属(G1组8.44%,G2组2.54%,G3组2.13%)、坦纳氏菌属(G1组1.83%,G2组1.67%,G3组0.52%)的相对丰度有统计学差异(P < 0.05),在G1、G2、G3组的比例呈逐渐下降趋势。
| No. | G1 group | G2 group | G3 grouup | |||||
| Bacteria | Proportion(%) | Bacteria | Proportion(%) | Bacteria | Proportion(%) | |||
| 1 | Fusobacterium | 25.378±10.597 | Unclassified | 29.265±12.316 | Unclassified | 29.865±17.372 | ||
| 2 | Porphyromonas | 17.721±13.115 | Fusobacterium | 19.294±12.844 | Tessaracoccus | 12.083±24.917 | ||
| 3 | Capnocytophaga | 9.638±19.370 | Capnocytophaga | 7.258±9.066 | Actinomyces | 10.220±14.496 | ||
| 4 | Prevotella | 8.443±6.701 | Actinomyces | 6.842±6.070 | Rothia | 7.948±13.706 | ||
| 5 | Leptotrichia | 7.206±11.766 | Porphyromonas | 4.793±2.672 | Fusobacterium | 7.312±7.186 | ||
| 6 | Unclassified | 5.627±2.493 | Leptotrichia | 4.640±5.625 | Porphyromonas | 3.092±3.455 | ||
| 7 | Rothia | 2.954±4.873 | Tessaracoccus | 4.077±8.156 | Prevotella | 2.128±2.146 | ||
| 8 | Campylobacter | 2.895±3.313 | Rothia | 3.544±4.113 | Leptotrichia | 2.081±1.942 | ||
| 9 | Actinomyces | 2.748±3.122 | Prevotella | 2.543±1.950 | Bacteroides | 1.799±1.904 | ||
| 10 | Filifactor | 2.670±1.859 | Tannerella | 1.672±1.802 | Arcobacter | 1.490±3.205 | ||
| 11 | Saccharibacteria genera incertae sedis | 2.104±3.463 | Filifactor | 1.318±0.949 | Olsenella | 1.015±1.590 | ||
| 12 | Olsenella | 1.851±4.784 | Campylobacter | 1.177±0.891 | Capnocytophaga | 0.983±1.064 | ||
| 13 | Tannerella | 1.827±1.868 | Olsenella | 0.860±1.070 | Phocaeicola | 0.980±1.532 | ||
| 14 | Phocaeicola | 1.243±2.626 | Parvimonas | 0.858±0.703 | Barnesiella | 0.903±1.976 | ||
| 15 | Fretibacterium | 1.137±1.583 | Fretibacterium | 0.807±0.564 | Filifactor | 0.871±0.674 | ||
| G1,before treatment; G2,after conventional mechanical treatment; G3,after both conventional mechanical treatment and Er: YAG laser adjunctive treatment. | ||||||||
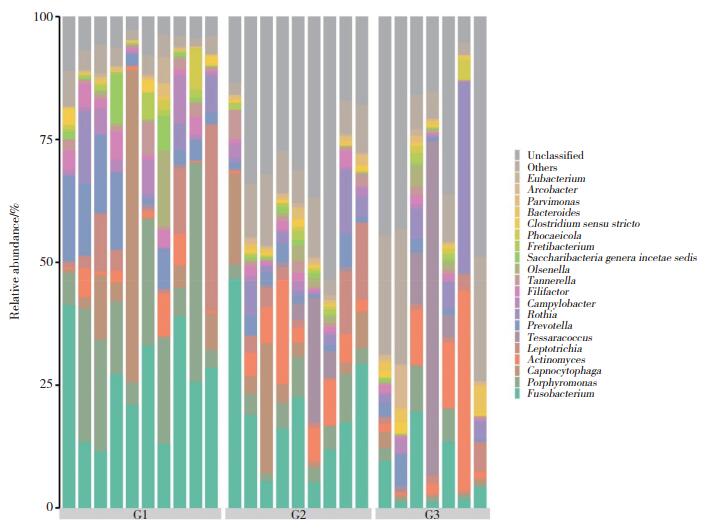
|
| 图 2 3组龈下菌斑菌群在属水平的组成柱状图 Fig.2 Histogram of composition of subgingival plaque flora at genus level |
| Comparison by groups | Genus level | G1 group | G2 group | G3 group | P | Enriched | |||||
| Mean | Standard deviation | Mean | Standard deviation | Mean | Standard deviation | ||||||
| G1 vs G2 vs G3 | |||||||||||
| Fusobacterium | 25.378 | 10.597 | 19.294 | 12.844 | 7.312 | 7.186 | 0.009 9 | ||||
| Porphyromonas | 17.721 | 13.115 | 4.793 | 2.672 | 3.092 | 3.455 | 0.003 5 | ||||
| Prevotella | 8.443 | 6.701 | 2.543 | 1.950 | 2.128 | 2.146 | 0.039 1 | ||||
| Tannerella | 1.827 | 1.868 | 1.672 | 1.802 | 0.519 | 0.572 | 0.026 4 | ||||
| G1 vs G2 | |||||||||||
| Porphyromonas | 17.721 | 13.115 | 4.793 | 2.672 | 0.007 6 | G1 | |||||
| Prevotella | 8.443 | 6.701 | 2.543 | 1.950 | 0.043 5 | G1 | |||||
| G1 vs G3 | |||||||||||
| Fusobacterium | 25.378 | 10.597 | 7.312 | 7.186 | 0.003 1 | G1 | |||||
| Porphyromonas | 17.721 | 13.115 | 3.092 | 3.455 | 0.004 6 | G1 | |||||
| Prevotella | 8.443 | 6.701 | 2.128 | 2.146 | 0.033 0 | G1 | |||||
| Filifactor | 2.670 | 1.859 | 0.871 | 0.674 | 0.043 1 | G1 | |||||
| Tannerella | 1.827 | 1.868 | 0.519 | 0.572 | 0.006 8 | G1 | |||||
| Tessaracoccus | 0.256 | 0.314 | 12.083 | 24.917 | 0.025 0 | G3 | |||||
| G2 vs G3 | |||||||||||
| Fusobacterium | 19.294 | 12.844 | 7.312 | 7.186 | 0.031 1 | G2 | |||||
| Capnocytophaga | 7.358 | 9.066 | 0.983 | 1.064 | 0.022 9 | G2 | |||||
| Tannerella | 1.672 | 1.802 | 0.519 | 0.572 | 0.041 8 | G2 | |||||
经Er:YAG激光治疗后,G3组与G2组相比(表 2),二氧化碳嗜纤维菌属(7.36% vs 0.98%)、梭形杆菌属(19.29% vs 7.31%)、坦纳氏菌属(1.67% vs 0.52%)等革兰氏阴性菌属的比例均显著下降(P < 0.05),而四合球菌属、放线菌属、罗斯氏菌属、产线菌属等革兰氏阳性菌属的相对丰度无显著差异,甚至存在罗斯氏菌属(G2组3.54%,G3组7.95%)和四合球菌属(G2组4.08%,G3组12.08%)的相对丰度升高的现象。G1组与G3组相比,菌属相对丰度差异大于仅经传统牙周基础治疗(G2组),主要体现在卟啉单胞菌属(G2组下降73.0%,G3组下降82.6%)和普雷沃氏菌属(G2组下降69.9%,G3组下降74.8%)。且G3组的梭形杆菌属(下降71.2%)、坦纳氏菌属(下降71.6%)、产线菌属(下降67.4%)较G1组显著下降,四合球菌属(上升46.20倍)较G1组显著上升(P < 0.05),但上述菌属在G1组与G2组间均无统计学差异。
2.3 种水平优势菌及组成差异分析各组前10位优势菌种的相对丰度见表 3和图 3。3组之间存在显著统计学差异的菌种见表 4,具核梭杆菌(G1组24.89%,G2组18.99%,G3组6.93%)、牙髓卟啉单胞菌(G1组3.97%,G2组1.96%,G3组0.48%)、福赛坦氏菌(G1组1.83%,G2组1.67%,G3组0.52%)、中间普氏菌(G1组3.21%,G2组0.55%,G3组0.28%)、直肠弯曲杆菌(G1组2.20%,G2组0.95%,G3组0.24%)等牙周致病菌比例呈逐渐下降趋势。牙龈卟啉单胞菌的相对丰度(G1组13.12%,G2组2.59%,G3组2.47%)虽呈降低趋势,但Er:YAG激光治疗前后其相对丰度无明显变化,差异无统计学意义(P > 0.05)。
| No. | G1 group | G2 group | G3 group | |||||
| Bacteria | Proportion(%) | Bacteria | Proportion(%) | Bacteria | Proportion(%) | |||
| 1 | Fusobacterium_nucleatum | 24.888±10.973 | Unclassified | 44.941±16.333 | Unclassified | 49.674±17.910 | ||
| 2 | Unclassified | 24.767±8.335 | Fusobacterium nucleatum | 18.994±12.937 | Tessaracoccus lapidicaptus | 12.083±24.917 | ||
| 3 | Porphyromonas gingivalis | 13.120±12.603 | Tessaracoccus lapidicaptus | 4.077±8.156 | Rothia aeria | 7.924±13.718 | ||
| 4 | Porphyromonas endodontalis | 3.967±4.053 | Rothia aeria | 3.502±4.447 | Fusobacterium nucleatum | 6.927±6.807 | ||
| 5 | Capnocytophaga sputigena | 3.419±8.100 | Capnocytophaga sputigena | 2.955±4.279 | Porphyromonas gingivalis | 2.475±3.191 | ||
| 6 | Leptotrichia_hongkongensis | 3.243±8.617 | Porphyromonas gingivalis | 2.591±2.030 | Arcobacter ellisii | 1.174±2.439 | ||
| 7 | Prevotella intermedia | 3.207±3.719 | Leptotrichia hongkongensis | 2.373±4.152 | Leptotrichia hongkongensis | 1.026±1.730 | ||
| 8 | Rothia aeria | 2.873±4.705 | Porphyromonas endodontalis | 1.955±1.903 | Phocaeicola abscessus | 0.980±1.532 | ||
| 9 | Campylobacter rectus | 2.200±2.736 | Tannerella forsythia | 1.672±1.802 | Olsenella uli | 0.978±1.610 | ||
| 10 | Tannerella forsythia | 1.827±1.868 | Actinomyces israelii | 1.268±2.930 | Cetobacterium somerae | 0.566±1.284 | ||
| 11 | Olsenella uli | 1.786±4.795 | Actinomyces dentalis | 1.042±1.462 | Roseburia intestinalis | 0.553±0.787 | ||
| 12 | Leptotrichia buccalis | 1.259±2.126 | Campylobacter rectus | 0.950±0.870 | Prevotella copri | 0.552±0.787 | ||
| 13 | Phocaeicola abscessus | 1.243±2.626 | Capnocytophaga leadbetteri | 0.922±1.018 | Tannerella forsythia | 0.519±0.572 | ||
| 14 | Fretibacterium fastidiosum | 1.137±1.583 | Parvimonas micra | 0.858±0.703 | Fretibacterium fastidiosum | 0.519±0.748 | ||
| 15 | Capnocytophaga leadbetteri | 1.114±2.682 | Fretibacterium fastidiosum | 0.807±0.564 | Propionibacterium acnes | 0.518±0.586 | ||
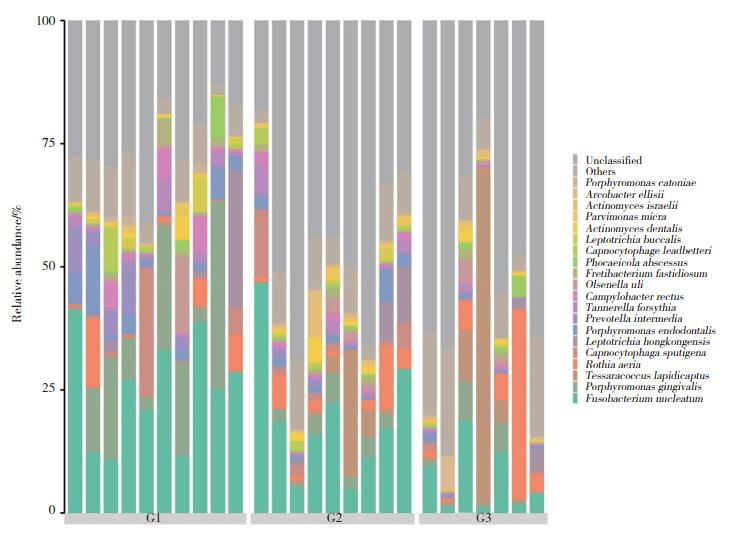
|
| 图 3 3组龈下菌斑菌群在种水平的组成柱状图 Fig.3 Histogram of composition of subgingival plaque at species level |
| Comparison by groups | Species level | G1 group | G2 group | G3 group | P | Enriched | |||||
| Mean | Standard deviation | Mean | Standard deviation | Mean | Standard deviation | ||||||
| G1 vs G2 vs G3 | |||||||||||
| Fusobacterium nucleatum | 24.888 | 10.973 | 18.994 | 12.937 | 6.927 | 6.807 | 0.009 9 | ||||
| Porphyromonas endodontalis | 3.967 | 4.053 | 1.955 | 1.903 | 0.477 | 0.432 | 0.004 8 | ||||
| Prevotella intermedia | 3.207 | 3.719 | 0.547 | 0.483 | 0.277 | 0.344 | 0.024 1 | ||||
| Tannerella forsythia | 1.827 | 1.868 | 1.672 | 1.802 | 0.519 | 0.572 | 0.026 4 | ||||
| Campylobacter rectus | 2.200 | 2.736 | 0.950 | 0.870 | 0.244 | 0.197 | 0.033 0 | ||||
| Campylobacter gracilis | 0.525 | 0.598 | 0.119 | 0.112 | 0.098 | 0.142 | 0.048 7 | ||||
| G1 vs G2 | |||||||||||
| Campylobacter gracilis | 0.525 | 0.598 | 0.119 | 0.112 | 0.045 1 | G1 | |||||
| G1 vs G3 | |||||||||||
| Fusobacterium nucleatum | 24.888 | 10.973 | 6.927 | 6.807 | 0.003 1 | G1 | |||||
| Porphyromonas endodontalis | 3.967 | 4.053 | 0.477 | 0.432 | 0.002 0 | G1 | |||||
| Prevotella intermedia | 3.207 | 3.719 | 0.277 | 0.344 | 0.021 7 | G1 | |||||
| Campylobacter rectus | 2.200 | 2.736 | 0.244 | 0.197 | 0.018 5 | G1 | |||||
| Tannerella forsythia | 1.827 | 1.868 | 0.519 | 0.572 | 0.006 8 | G1 | |||||
| Campylobacter gracilis | 0.525 | 0.598 | 0.098 | 0.142 | 0.045 3 | G1 | |||||
| G2 vs G3 | |||||||||||
| Fusobacterium nucleatum | 18.994 | 12.937 | 6.927 | 6.807 | 0.031 1 | G2 | |||||
| Porphyromonas endodontalis | 1.955 | 1.903 | 0.477 | 0.432 | 0.007 9 | G2 | |||||
| Tannerella forsythia | 1.672 | 1.802 | 0.519 | 0.572 | 0.041 8 | G2 | |||||
| Campylobacter rectus | 0.950 | 0.870 | 0.244 | 0.197 | 0.031 1 | G2 | |||||
| Capnocytophaga leadbetteri | 0.922 | 1.018 | 0.120 | 0.199 | 0.024 1 | G2 | |||||
G2组与G3组相比,经Er:YAG激光治疗后,G3组具核梭杆菌(18.99% vs 6.93%)、牙髓卟啉单胞菌(1.96% vs 0.48%)、福赛坦氏菌(1.67% vs 0.52%)、直肠弯曲杆菌(0.95% vs 0.24%)、利氏二氧化碳嗜纤维菌(0.92% vs 0.12%)等革兰氏阴性菌的比例均显著下降(P < 0.05),但Tessaracoccus lapidicaptus(4.08% vs 12.08%)、空间罗氏菌(3.5% vs 7.92%)等革兰氏阳性菌的比例上升。且G3组中间普氏菌(下降91.4%)较G1组显著下降(P < 0.05),但中间普氏菌的相对丰度在G1与G2组间无统计学差异。
3 讨论 3.1 Er:YAG激光可降低牙周致病菌的相对丰度进而削弱致病能力牙周炎在多种牙周致病菌的作用下发生发展,有效清除牙周致病菌是牙周治疗的关键[10, 12]。多项研究[13-15]结果表明,增设Er:YAG激光辅助治疗的临床疗效参数优于单纯传统牙周基础治疗。虽然牙龈卟啉单胞菌被认为是关键牙周致病菌,但本研究从微生物学角度发现,Er:YAG激光辅助治疗取得良好疗效的作用机制为它能显著降低具核梭杆菌、牙髓卟啉单胞菌、福赛坦氏菌等牙周致病菌的相对丰度(P < 0.05),而不在于清除牙龈卟啉单胞菌(P = 0.76)。ZENGIN等[16]的研究也发现,Er:YAG激光辅助治疗虽然能取得良好的临床疗效,但并不会显著降低龈下菌斑中牙龈卟啉单胞菌的丰度。这可能是由于牙龈卟啉单胞菌对波长为2 940 nm的Er:YAG激光吸收系数较低,Er:YAG激光无法使牙龈卟啉单胞菌内部压力升高产生微爆破作用和显著降低脂多糖(lipopolysaccharide,LPS),从而清除牙龈卟啉单胞菌。本研究中,丰度最高、致病性较强的具核梭杆菌(下降63.5%)为主要牙周致病菌,它存在的部位常伴随着牙龈卟啉单胞菌[17]。因其具有较强的黏附和入侵细胞能力,能使牙龈卟啉单胞菌黏附于其表面从而助长牙龈卟啉单胞菌入侵牙龈上皮细胞和主动脉内皮细胞的能力[18],增强了牙龈卟啉单胞菌的致病性。不仅如此,DIAZ等[19]研究发现,在缺乏具核梭杆菌的环境下牙龈卟啉单胞菌无法存活,提示具核梭杆菌可以为牙龈卟啉单胞菌提供必要的生存条件。因此,Er:YAG激光虽然不能选择性作用于牙龈卟啉单胞菌,但其能显著降低具核梭杆菌的相对丰度,进而间接降低牙龈卟啉单胞菌的致病能力,对牙周炎的治疗具有积极意义。
3.2 Er:YAG激光对龈下菌斑微生物膜的改善作用牙周炎发病过程中龈下微生物群发生了从革兰氏阳性菌占优势向革兰氏阴性菌占优势的转变[20],慢性牙周炎患者龈下微生物群中革兰氏阴性菌占75%左右[21]。因此,牙周炎的治疗目标不仅在于清除菌斑和牙石,还在于实现将龈下菌斑生物膜转变为革兰氏阳性菌占优势的微生物膜。本研究发现,Er:YAG激光缺乏对放线菌属、四合球菌属、罗斯氏菌属、产线菌属等革兰氏阳性菌属以及Tessaracoccus lapidicaptus、空间罗氏菌等革兰氏阳性菌种的清除作用,因而表现为优势菌属中Er:YAG激光治疗前后革兰氏阳性菌的相对丰度升高和革兰氏阴性菌的相对丰度下降。这与Er:YAG激光的杀菌机制密切相关,Er:YAG激光(波长2.94 μm)通过作用于革兰氏阴性菌细胞壁上的LPS(波长2.92 μm)而达到灭菌的目的[22-23],而上述革兰氏阳性菌细胞壁缺乏LPS。不仅如此,具核梭杆菌通过为密螺旋体、福赛坦氏菌、牙龈卟啉单胞菌附着于龈下菌斑提供结合位点[24],促进龈下菌斑由革兰氏阳性菌占优势的微生物膜向革兰氏阴性菌占优势的微生物膜转变。而Er:YAG激光对具核梭杆菌的杀灭作用能一定程度上阻断转变过程。故Er:YAG激光辅助治疗不仅有利于杀灭牙周致病菌(多为革兰氏阴性菌),还能促进龈下菌斑生物膜向革兰氏阳性菌占优势的微生物膜转变。提示传统牙周基础治疗存在局限性,单独运用无法选择性降低革兰氏阴性菌的相对丰度,而Er:YAG激光辅助治疗具有优越性。
本研究纳入了10例GPⅢ-C患者,共采集30个龈下菌斑样本,G2组、G3组分别有1个和3个样本扩增失败,可能是由于经传统牙周基础治疗及Er:YAG激光辅助治疗后,龈下菌斑余留量较少,采集量不足,也从侧面反映了Er:YAG激光辅助治疗能进一步降低龈下菌斑菌群丰度,尚需扩大样本量进一步深入研究。本研究探讨了Er:YAG激光作用后的即刻效应,未来可增设定期随访研究,同时,今后将采用平行对照设计,扩大研究的样本量,以验证本研究结果。
综上所述,Er:YAG激光辅助治疗虽对牙龈卟啉单胞菌的作用有限,但能在单纯应用传统牙周基础治疗的基础上,进一步选择性清除具核梭杆菌、福赛坦氏菌、牙髓卟啉单胞菌等牙周致病菌,且有利于龈下菌斑向革兰氏阳性菌占优势的微生物膜转变,值得临床上作为辅助治疗应用和推广。
| [1] |
孟焕新. 牙周病学[M]. 5版. 北京: 人民卫生出版社, 2020: 333.
|
| [2] |
郭淑娟, 刘倩, 丁一. 牙周病和植体周病国际新分类简介[J]. 国际口腔医学杂志, 2019, 46(2): 125-134. DOI:10.7518/gjkq.2019001 |
| [3] |
AGOOB ALFERGANY M, NASHER R, GUTKNECHT N. Calculus removal and root surface roughness when using the Er: yag or Er, Cr: ysgg laser compared with conventional instrumentation method: a literature review[J]. Photobiomodul Photomed Laser Surg, 2019, 37(4): 197-226. DOI:10.1089/photob.2018.4465 |
| [4] |
GRZECH-LEŚNIAK K, SCULEAN A, GAŠPIRC B. Laser reduction of specific microorganisms in the periodontal pocket using Er: yag and Nd: yag lasers: a randomized controlled clinical study[J]. Lasers Med Sci, 2018, 33(7): 1461-1470. DOI:10.1007/s10103-018-2491-z |
| [5] |
TAKAMATSU N, YANO K, HE T, et al. Effect of initial periodontal therapy on the frequency of detecting Bacteroides forsythus, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans[J]. J Periodontol, 1999, 70(6): 574-580. DOI:10.1902/jop.1999.70.6.574 |
| [6] |
PATEL PV, PATEL A, KUMAR S, et al. Effect of subgingival application of topical ozonated olive oil in the treatment of chronic periodontitis: a randomized, controlled, double blind, clinical and microbiological study[J]. Minerva Stomatol, 2012, 61(9): 381-398. |
| [7] |
COSTA JV, PORTUGAL J, NEVES CB, et al. Should local drug delivery systems be used in dentistry?[J]. Drug Deliv Transl Res, 2021. DOI:10.1007/s13346-021-01053-x |
| [8] |
SAĞLAM M, KÖSEOĞLU S, TAŞDEMIR I, et al. Combined application of Er: yag and Nd: yag lasers in treatment of chronic periodontitis. A split-mouth, single-blind, randomized controlled trial[J]. J Periodontal Res, 2017, 52(5): 853-862. DOI:10.1111/jre.12454 |
| [9] |
DOMÍNGUEZ A, GÓMEZ C, GARCÍA-KASS AI, et al. IL-1beta, TNF-alpha, total antioxidative status and microbiological findings in chronic periodontitis treated with fluorescence-controlled Er: yag laser radiation[J]. Lasers Surg Med, 2010, 42(1): 24-31. DOI:10.1002/lsm.20873 |
| [10] |
TELES RP, HAFFAJEE AD, SOCRANSKY SS. Microbiological goals of periodontal therapy[J]. Periodontol 2000, 2006, 42: 180-218. DOI:10.1111/j.1600-0757.2006.00192.x |
| [11] |
PAPAPANOU PN, SANZ M, BUDUNELI N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-Implant diseases andconditions[J]. J Periodontol, 2018, 89(Suppl 1): S173-S182. DOI:10.1002/jper.17-0721 |
| [12] |
FERES M, RETAMAL-VALDES B, FERMIANO D, et al. Microbiome changes in young periodontitis patients treated with adjunctive metronidazole and amoxicillin[J]. J Periodontol, 2021, 92(4): 467-478. DOI:10.1002/JPER.20-0128 |
| [13] |
MA L, ZHANG XL, MA Z, et al. Clinical effectiveness of Er: yag lasers adjunct to scaling and root planing in non-surgical treatment of chronic periodontitis: a meta-analysis of randomized controlled trials[J]. Med Sci Monit, 2018, 24: 7090-7099. DOI:10.12659/MSM.911863 |
| [14] |
曹盈, 吴文蕾, 薛雅琴, 等. Nd: YAG激光和Er: YAG激光治疗重度牙周炎的效果观察[J]. 口腔医学, 2018, 38(6): 514-517. DOI:10.13591/j.cnki.kqyx.2018.06.008 |
| [15] |
SUMRA N, KULSHRESTHA R, UMALE V, et al. Lasers in non-surgical periodontal treatment-a review[J]. J Cosmet Laser Ther, 2019, 21(5): 255-261. DOI:10.1080/14764172.2018.1525744 |
| [16] |
ZENGIN CELIK T, SAGLAM E, ERCAN C, et al. Clinical and microbiological effects of the use of erbium: yttrium-aluminum-garnet laser on chronic periodontitis in addition to nonsurgical periodontal treatment: a randomized clinical trial-6 months follow-up[J]. Photobiomodul Photomed Laser Surg, 2019, 37(3): 182-190. DOI:10.1089/photob.2018.4510 |
| [17] |
SOCRANSKY SS, HAFFAJEE AD, CUGINI MA, et al. Microbial complexes in subgingival plaque[J]. J Clin Periodontol, 1998, 25(2): 134-144. DOI:10.1111/j.1600-051x.1998.tb02419.x |
| [18] |
KOLENBRANDER PE. Oral microbial communities: biofilms, interactions, and genetic systems[J]. Annu Rev Microbiol, 2000, 54: 413-437. DOI:10.1146/annurev.micro.54.1.413 |
| [19] |
DIAZ PI, ZILM PS, ROGERS AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments[J]. Microbiology (Reading), 2002, 148(Pt 2): 467-472. DOI:10.1099/00221287-148-2-467 |
| [20] |
DARVEAU RP. Periodontitis: a polymicrobial disruption of host homeostasis[J]. Nat Rev Microbiol, 2010, 8(7): 481-490. DOI:10.1038/nrmicro2337 |
| [21] |
边专. 口腔生物学[M]. 4版. 北京: 人民卫生出版社, 2012.
|
| [22] |
WANG CY, LEE BS, JHANG YT, et al. Er: YAG laser irradiation enhances bacterial and lipopolysaccharide clearance and human gingival fibroblast adhesion on titanium discs[J]. Sci Rep, 2021, 11: 23954. DOI:10.1038/s41598-021-03434-1 |
| [23] |
ISHIKAWA I, AOKI A, TAKASAKI AA. Potential applications of Erbium: yag laser in periodontics[J]. J Periodontal Res, 2004, 39(4): 275-285. DOI:10.1111/j.1600-0765.2004.00738.x |
| [24] |
KOLENBRANDER PE, ANDERSEN RN, BLEHERT DS, et al. Communication among oral bacteria[J]. Microbiol Mol Biol Rev, 2002, 66(3): 486-505. DOI:10.1128/MMBR.66.3.486-505.2002 |
 2022, Vol. 51
2022, Vol. 51