文章信息
- 李树强
- Li Shuqiang
- 影响肝细胞癌5年生存期的关键基因分析
- Analysis of hub genes affecting five-year survival in liver cancer
- 中国医科大学学报, 2020, 49(10): 949-953
- Journal of China Medical University, 2020, 49(10): 949-953
-
文章历史
- 收稿日期:2019-03-20
- 网络出版时间:2020-10-07 15:48
原发性肝癌是目前我国第四位常见的恶性肿瘤,在肿瘤致死疾病中排第三位,严重威胁人们的生命和健康。肝细胞癌(hepatocellular carcinoma,HCC)占原发性肝癌的80%以上,手术切除和肝移植是根治原发性HCC的主要手段,但是由于早期发现困难,放化疗效果不佳,5年生存率仅为15%[1]。因此,更好地了解HCC发生的分子机制,发现影响HCC 5年生存期的关键基因,为HCC治疗提供新的靶点已成当务之急。
1 材料与方法1.1癌症基因组图谱(the cancer genome atlas,TCGA)数据下载
登陆TCGA数据库(https://cancergenome.nih.gov)网站下载公开的HCC转录组数据和肿瘤样本临床数据。其中,生存期 > 5年(无论存活与否)的癌症样本43例,生存期 < 5年(已死亡)的癌症样本107例。本研究旨在探讨影响HCC 5年生存率的关键基因,故排除随访时间不到5年且存活的样本。
1.2 差异表达基因分析使用转录组数据中的reads count评估基因表达水平,应用R软件(3.5.1版本)中的limma及edgeR软件包[2]对数据进行标准化及差异表达分析,gplots软件包对数据进行图形可视化。筛选log FC绝对值> 2,FDR < 0.01的基因为差异表达基因。
1.3 功能和通路富集分析DAVID(The Database for Annotation,Visualization and Integrated Discovery)是一个在线数据库[3](https://david.ncifcrf.gov),为研究人员提供了一套全面的功能注释工具,以便理解大量基因背后的生物学意义。应用DAVID分别对上调和下调差异表达基因进行GO(gene ontology)和KEGG(kyoto encyclopedia of genes and genomes)富集分析,P < 0.05为差异有统计学意义。
1.4 候选关键基因筛选将差异基因上传到STRING数据库[4](https://string-db.org),选择置信度 > 0.4构建蛋白-蛋白互作(PPI)网络。基因在PPI网络的连接度(degree)≥8且排名前10位的基因定义为候选关键基因。
1.5 结果验证应用在线网站http://kmplot.com/analysis[5],绘制Kaplan-Meier生存曲线,计算95%置信区间的危险比和logrank P值,比较不同基因表达水平患者的生存期,筛选并验证候选关键基因是否为真正关键基因。
2 结果 2.1 识别差异表达基因下载数据后进行归一化、对数化,去掉没有对应基因注释信息的探针,如果同一个基因有多个探针表达值,取平均数,去掉重复探针,最终得到包含19 754个基因,150个样本的表达谱。通过R中edgeR包,log FC绝对值> 2,FDR < 0.01为表达差异基因筛选条件,共得到383个差异基因。其中表达上调基因253个,表达下调基因130个,见图 1、2。
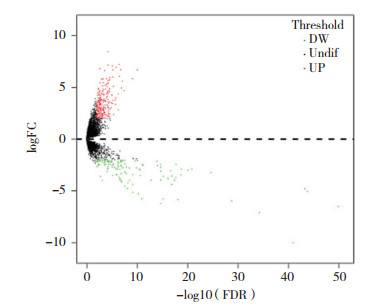
|
| 图 1 差异表达基因火山图 Fig.1 Volcano plots of differentially expressed genes |
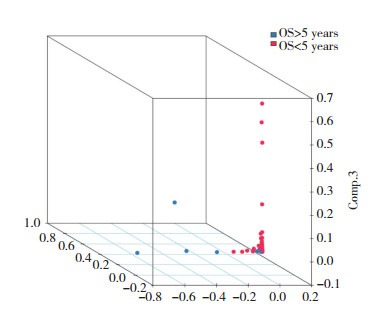
|
| 图 2 差异表达基因主成分分析 Fig.2 Principal component analysis of differentially expressed genes |
2.2 功能和通路富集分析
用DAVID分别对上调和下调差异表达基因进行GO和KEGG富集分析。得到了Biological process(BP)和KEGG pathway结果(表 1、2)。上调基因BP功能聚类主要集中在“positive regulation of cAMP biosynthetic process”,“synaptic transmission,cholinergic”,“cation transmembrane transport”,“regulation of membrane potential”等。有3种典型的KEGG通路在上调基因中过表达,包括“Neuroactive ligand-receptor interaction”,“Nicotine addiction”和“GABAergic synapse”。下调基因BP功能主要富集在“potassium ion transport”,“energy reserve metabolic process”和“oxygen transport”等,下调基因在KEGG通路中主要富集在“Salivary secretion”,“cAMP signaling pathway”和“Cardiac muscle contraction”等。由此可见,差异表达基因功能主要富集在与某些神经内分泌调节有关的生物学过程和通路中,通过膜电位改变,跨膜离子的转运和突触传递等影响患者5年生存率。
| Category | Term | ID | Count [n(%)] | P |
| BP | ||||
| Positive regulation of cAMP biosynthetic process | GO:0030819 | 7(2.85) | < 0.001 | |
| Synaptic transmission,cholinergic | GO:0007271 | 6(2.44) | < 0.001 | |
| Cation transmembrane transport | GO:0098655 | 6(2.44) | < 0.001 | |
| Regulation of membrane potential | GO:0042391 | 7(2.85) | < 0.001 | |
| Sequestering of zinc ion | GO:0032119 | 3(1.22) | < 0.001 | |
| Response to pain | GO:0048265 | 4(1.63) | < 0.001 | |
| Regulation of blood pressure | GO:0008217 | 6(2.44) | < 0.001 | |
| Chloride transport | GO:0006821 | 5(2.03) | < 0.001 | |
| Embryo implantation | GO:0007566 | 5(2.03) | < 0.001 | |
| Response to ethanol | GO:0045471 | 7(2.85) | < 0.001 | |
| Kegg | ||||
| Neuroactive ligand-receptor interaction | hsa04080 | 23(9.35) | < 0.001 | |
| Nicotine addiction | hsa05033 | 4(1.63) | 0.013 8 | |
| GABAergic synapse | hsa04727 | 5(2.03) | 0.022 2 |
| Category | Term | ID | Count [n(%)] | P |
| BP | ||||
| Potassium ion transport | GO:0006813 | 6(4.72) | < 0.001 | |
| energy reserve metabolic process | GO:0006112 | 3(2.36) | < 0.001 | |
| Oxygen transport | GO:0015671 | 3(2.36) | < 0.001 | |
| Regulation of cardiac conduction | GO:1903779 | 4(3.15) | < 0.001 | |
| Potassium ion import | GO:0010107 | 3(2.36) | 0.014 6 | |
| Negative regulation of smooth muscle cell proliferation | GO:0048662 | 3(2.36) | 0.015 7 | |
| Negative regulation of signal transduction | GO:0009968 | 3(2.36) | 0.021 2 | |
| Photoreceptor cell maintenance | GO:0045494 | 3(2.36) | 0.021 2 | |
| Cellular calcium ion homeostasis | GO:0006874 | 4(3.15) | 0.023 7 | |
| Activation of adenylate cyclase activity | GO:0007190 | 3(2.36) | 0.028 7 | |
| Kegg | ||||
| Salivary secretion | hsa04970 | 5(3.94) | < 0.001 | |
| cAMP signaling pathway | hsa04024 | 6(4.72) | < 0.001 | |
| Cardiac muscle contraction | hsa04260 | 4(3.15) | < 0.001 | |
| cGMP-PKG signaling pathway | hsa04022 | 5(3.94) | < 0.001 | |
| Regulation of lipolysis in adipocytes | hsa04923 | 3(2.36) | 0.036 1 | |
| Adrenergic signaling in cardiomyocytes | hsa04261 | 4(3.15) | 0.037 4 | |
| Oxytocin signaling pathway | hsa04921 | 4(3.15) | 0.046 0 | |
| Bile secretion | hsa04976 | 3(2.36) | 0.052 7 | |
| Thyroid hormone synthesis | hsa04918 | 3(2.36) | 0.054 1 | |
| Gastric acid secretion | hsa04971 | 3(2.36) | 0.058 3 |
2.3 候选关键基因筛选
差异表达基因在PPI网络的连接度≥8且排名前10位的基因定义为候选关键基因,包括GCG、LEP、PTH、HTR5A、CRH、CRHR1、CALCA、ADCY2、GAST和CHGA(图 3)。除LEP为表达下调基因外,其余均为表达上调基因。
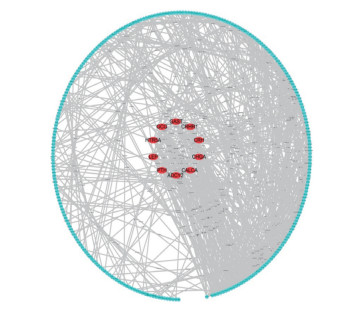
|
| The genes highlighted in red were among the top 10 candidate key genes for degree ranking. 图 3 蛋白-蛋白互作网络(PPI)图 Fig.3 Protein-protein interaction network |
2.4 生存曲线分析
绘制Kaplan-Meier生存曲线,结果(图 4)显示,除了LEP和CHGA,HCC患者中GCG、PTH、HTR5A、CRH、CRHR1、CALCA、ADCY2和GAST基因的高表达,其总生存率明显升高,故认为这些差异表达基因可能为HCC患者长期生存的关键基因。
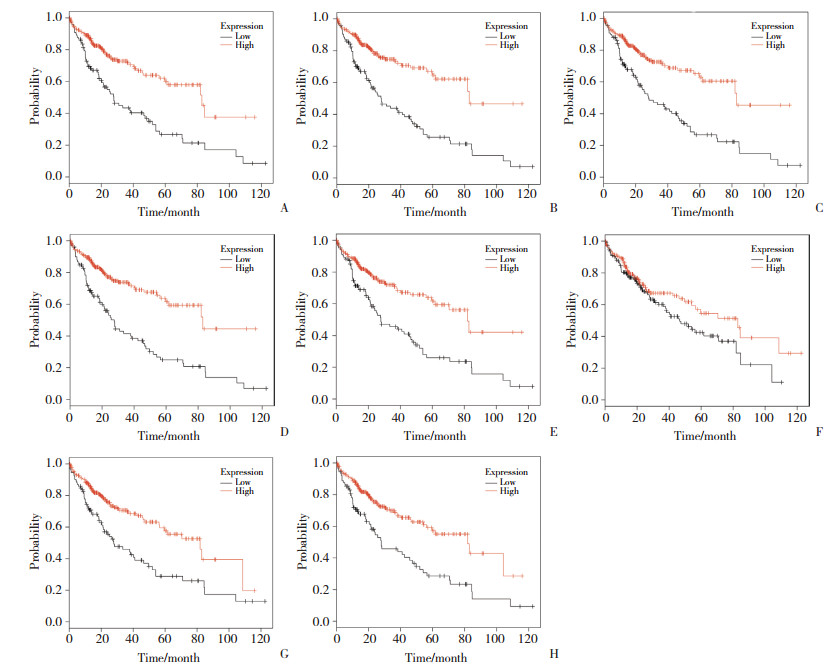
|
| A, GCG; B, PTH; C, HTR5A; D, CRH; E, CRHR1; F, CALCA; G, ADCY2; H, GAST. 图 4 肝细胞癌关键基因预后分析 Fig.4 Prognostic analysis of key genes in hepatocellular carcinoma |
3 讨论
HCC是全球第五大常见癌症,每年约有85万新发病例[6],占肝癌的80%以上[7]。HCC通常发生在肝硬化患者中,越来越多的HCC病例伴有非酒精性脂肪肝,这是肥胖和胰岛素抵抗的结果[8]。常用的治疗方法有器官移植、手术切除、经动脉化疗栓塞、局部射频消融和局部微波消融等[9]。由于早期没有临床症状,大多数HCC病例发现即为晚期,治疗疗效差。索拉非尼作为不可切除HCC病例的临床替代药物,疗效一般[10]。因此,从基因水平更好地了解HCC发生机制,发现新的治疗靶点已成当务之急。
本研究利用TCGA数据库中HCC mRNA基因及临床表型信息,通过一系列分析,确定了8个与HCC 5年生存期密切相关的关键基因,GCG在糖代谢和体内平衡中起着重要作用[11],PTH通过溶解骨骼中的盐分和防止肾脏排泄来提高血钙水平,与前列腺癌侵袭程度相关[12]。HTR5A是一种生物激素,起神经递质、激素和有丝分裂原的作用。这种受体的活性是由G蛋白介导的,可能通过调节细胞内Ca2+水平而发挥作用。CRH所编码的前蛋白经蛋白水解处理后产生成熟的神经肽激素。应激反应时,这种激素由下丘脑室旁核分泌,与促肾上腺皮质激素释放受体结合,刺激垂体释放促肾上腺皮质激素。这种蛋白的显著减少与阿尔茨海默病有关。CRHR1编码一种G蛋白耦联受体,对激活信号转导通路至关重要,这些通路调节包括压力、繁殖、免疫反应和肥胖在内的多种生理过程。最近研究[13]报道,CRH/CRHR1可通过IL-6/JAK2/STAT3信号通路和VEGF诱导的肿瘤血管生成,促进结肠癌细胞增殖。CALCA表达水平与多种肿瘤有关,如胃癌、肺癌和睾丸肿瘤等[14-16]。ADCY2是一种膜相关酶,催化二级信使环腺苷单磷酸的形成。GAST能刺激胃黏膜产生和分泌盐酸,胰腺分泌其消化酶。还能刺激平滑肌收缩,促进胃和肠的血液循环和水分分泌。
综上所述,本研究应用TCGA数据库,发现了8个与HCC 5年生存期有关的关键基因,为今后HCC的研究提供了新的方向。本研究仅是通过数据分析初步得到可能有价值的关键基因,作用机制尚不清楚,需要后续实验加以验证。
| [1] |
MOMIN BR, PINHEIRO PS, CARREIRA H, et al. Liver cancer survival in the United States by race and stage (2001-2009):findings from the CONCORD-2 study[J]. Cancer, 2017, 123: 5059-5078. DOI:10.1002/cncr.30820 |
| [2] |
DAI ZY, SHERIDAN JM, GEARING LJ, et al. edgeR:a versatile tool for the analysis of shRNA-seq and CRISPR-Cas9 genetic screens[J]. F1000Research, 2014, 3: 95. DOI:10.12688/f1000research.3928.2 |
| [3] |
HUANG DW, SHERMAN BT, LEMPICKI RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources[J]. Nat Protoc, 2009, 4(1): 44. DOI:10.1038/nprot.2008.211 |
| [4] |
SZKLARCZYK D, GABLE AL, LYON D, et al. STRING v11:protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets[J]. Nucleic Acids Res, 2019, 47(D1): D607-D613. DOI:10.1093/nar/gky1131 |
| [5] |
LÁNCZKY A, NAGY Á, BOTTAI G, et al. miRpower:a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients[J]. Breast Cancer Res Treat, 2016, 160(3): 439-446. DOI:10.1007/s10549-016-4013-7 |
| [6] |
LLOVET JM, ZUCMAN-ROSSI J, PIKARSKY E, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2016, 2: 16018. DOI:10.1038/nrdp.2016.18 |
| [7] |
ZHU RX, SETO WK, LAI CL, et al. Epidemiology of hepatocellular carcinoma in the Asia-Pacific region[J]. Gut Liver, 2016, 10(3): 332-339. DOI:10.5009/gnl15257 |
| [8] |
SEYDEL GS, KUCUKOGLU O, ALTINBAS A, et al. Economic growth leads to increase of obesity and associated hepatocellular carcinoma in developing countries[J]. Ann Hepatol, 2016, 15(5): 662-672. DOI:10.5604/16652681.1212316 |
| [9] |
GOMAA AI, WAKED I. Recent advances in multidisciplinary management of hepatocellular carcinoma[J]. World J Hepatol, 2015, 7(4): 673-687. DOI:10.4254/wjh.v7.i4.673 |
| [10] |
NIU LL, LIU LP, YANG SL, et al. New insights into sorafenib resistance in hepatocellular carcinoma:responsible mechanisms and promising strategies[J]. Biochim et Biophys Acta BBA-Rev Cancer, 2017, 1868(2): 564-570. DOI:10.1016/j.bbcan.2017.10.002 |
| [11] |
PINTO LC, FALCETTA MR, RADOS DV, et al. Glucagon-like peptide-1 receptor agonists and pancreatic cancer:a meta-analysis with trial sequential analysis[J]. Sci Rep, 2019, 9: 2375. DOI:10.1038/s41598-019-38956-2 |
| [12] |
BRÄNDSTEDT J, ALMQUIST M, ULMERT D, et al. Vitamin D, PTH, and calcium and tumor aggressiveness in prostate cancer:a prospective nested case-control study[J]. Cancer Causes Control, 2016, 27(1): 69-80. DOI:10.1007/s10552-015-0684-3 |
| [13] |
FANG XJ, HONG YL, DAI L, et al. CRH promotes human colon cancer cell proliferation via IL-6/JAK2/STAT3 signaling pathway and VEGF-induced tumor angiogenesis[J]. Mol Carcinog, 2017, 56(11): 2434-2445. DOI:10.1002/mc.22691 |
| [14] |
CHU AN, LIU JW, YUAN Y, et al. Comprehensive analysis of aberrantly expressed CeRNA network in gastric cancer with and without H.pylori infection[J]. J Cancer, 2019, 10(4): 853-863. DOI:10.7150/jca.27803 |
| [15] |
YANG ZP, QI WB, SUN L, et al. DNA methylation analysis of selected genes for the detection of early-stage lung cancer using circulating cell-free DNA[J]. Adv Clin Exp Med, 2018, 28(3): 355-360. DOI:10.17219/acem/84935 |
| [16] |
WANG Y, GRAY DR, ROBBINS AK, et al. Subphenotype meta-analysis of testicular cancer genome-wide association study data suggests a role for RBFOX family genes in cryptorchidism susceptibility[J]. Hum Reprod, 2018, 33(5): 967-977. DOI:10.1093/humrep/dey066 |
 2020, Vol. 49
2020, Vol. 49




