文章信息
- 刘军
- LIU Jun
- 运动强度对不同性别代谢综合征患者骨骼肌mRNA表达谱的影响
- Influence of Exercise Intensity on Skeletal Muscle mRNA Profiles in Cardiometabolic High-Risk Male/Female Population
- 中国医科大学学报, 2019, 48(12): 1106-1111
- Journal of China Medical University, 2019, 48(12): 1106-1111
-
文章历史
- 收稿日期:2019-01-24
- 网络出版时间:2019-12-06 10:32
肥胖、血脂异常及久坐生活方式是导致心血管疾病、糖尿病等多种疾病的高危因素,是亚健康人群的重要特征[1]。运动训练可通过增加去脂体质量、减少体脂量、改善体脂分布促进人体恢复健康,其效果与运动训练的类型(有氧/阻力运动)以及运动强度有关[2]。既往研究[2]表明有氧运动可减轻体质量和体脂含量,调节骨骼肌内的重要代谢通路(线粒体代谢等),运动量是影响血脂水平、胰岛素敏感性的重要因素。
基于高通量数据所进行的基因组学研究,已成为揭示疾病机制和基因生物功能的重要手段,基因富集分析(gene set enrichment analysis,GSEA)是解释基因表达谱数据的核心分析工具之一[3]。GSEA可对样本所表达的基因进行分类,把具有相同生物学功能和调节机制、处于同一信号通路的基因富集出来,通过富集的这些优势通路阐明其生物学特征及相关机制,进而提示疾病防控靶点或指导疾病进展监控与预后预测。已有研究[4]显示运动训练可增加胰岛素敏感性,通过对骨骼肌结构和功能的调节影响心血管代谢疾病的发生与发展,而心血管代谢的改善与有氧运动强度有关。
本研究利用基因数据集GSE48278,以具有慢性心血管代谢疾病危险因素的亚健康状态人群为研究对照,探讨不运动以及不同强度有氧运动6个月对男性和女性骨骼肌mRNA表达谱的改变,并利用生物信息学方法分析此种改变潜在的生物功能与分子机制。
1 材料与方法 1.1 原始数据集与分级训练方案从NCBI的GEO数据库(http://www.ncbi.nlm.nih.gov/geo)下载代谢综合征患者骨骼肌基因表达数据集GSE48278。共纳入112例,随机选取1998年至2003和2004年至2009年进行的第1次和第2次通过特定方式体育锻炼进行有针对性的降低风险干预措施的研究(studies of a targeted risk reduction intervention through sefined exercise,STRRIDE)试验。纳入标准:(1)年龄18~70岁;(2)不活动;(3)超重至轻度肥胖(体质量指数25~35 kg/m2)和血脂异常[低密度脂蛋白胆固醇:3.34~4.92 mmol/L或高密度脂蛋白胆固醇 < 1.04 mmol/L(男性)或1.17 mmol/L(女性)]。排除标准:(1)已确诊糖尿病或空腹血糖 > 6.99 mmol/L;(2)高血压(血压 > 160/90 mmHg);(3)已知患有心血管疾病,禁止运动训练的肌肉骨骼疾病;(4)吸烟;(5)服用影响糖/脂类代谢的药物;(6)应用减肥食谱。参与者随机分配到不运动或小量中、高有氧运动组,运动强度分别为小量中强度运动(5 021 kJ/周,40%~55%
基于研究目的,本研究选取不运动和有氧运动的参与者。剔除阻力训练者。另外,大量高强度运动量超过中国人常规耐受范围,参考价值有限,因此也被剔除。最终纳入者按性别划分,每个性别纳入者分为3组:不运动组(5例)、小量中强度组(5例)、小量高强度组(5例),共30例。
1.3 基因数据分析每例在入组时和系统运动6个月后,从骨骼肌活组织检查提取总RNA进行基因表达微阵列分析(Affymetrix U133 Plus 2.0)。本研究应用该数据集的series matrix数据。以探针为基本单位进行后续分析。
应用Gene Set Enrichment analysis 3.0版软件,以入组时与不运动、小量中强度运动、小量高强度运动6月后比较,以MSigDB在线基因集数据库的hallmark gene sets作为参考基因集,对基因表达谱进行GSEA。设置随机组合次数为1 000次。以P < 0.05,且FDR q < 0.25为差异具有统计学意义。对有统计学意义的基因集进行leading edge gene分析。使用GPL570的注释文件对leading edge gene分析所选定的探针进行注释,利用genecards网站(http://www.genecards.org)汇总各个相关基因的功能等信息。
2 结果 2.1 男性患者锻炼前后的GSEA比较结果显示,与入组时比较,6个月后不运动组的标本富集了“凋亡”,“炎症反应”,“上皮间质转化”,“KRAS信号”等通路,如IL6_JAK_STAT3_SIGNALING、ALLOGRAFT_REJECTION、EPITHELIAL_MESENCHYMAL_TRANSITION、COMPLEMENT、APOPTOSIS、INFLAMMATORY_RESPONSE、KRAS_SIGNALING_UP、APICAL_SURFACE、XENOBIOTIC_METABOLISM、COAGULATION和IL2_STAT5_SIGNALING等;小量中强度组未富集任何通路;而小量高强度组富集的通路为PANCREAS_BETA_CELLS。见表 1、图 1。
| Group | Hallmarks | Size | ES | P | FDR q | Leading edge |
| Male without exercise | IL6_JAK_STAT3_SIGNALING | 85 | -0.53 | < 0.01 | 0.04 | tags=49%, list=21%, signal=62% |
| ALLOGRAFT_REJECTION | 191 | -0.49 | < 0.01 | 0.07 | tags=36%, list=14%, signal=42% | |
| EPITHELIAL_MESENCHYMAL_TRANSITION | 192 | -0.56 | 0.08 | 0.08 | tags=55%, list=20%, signal=69% | |
| COMPLEMENT | 192 | -0.45 | 0.02 | 0.10 | tags=39%, list=21%, signal=49% | |
| APOPTOSIS | 159 | -0.47 | 0.04 | 0.13 | tags=45%, list=26%, signal=60% | |
| INFLAMMATORY_RESPONSE | 189 | -0.41 | 0.04 | 0.20 | tags=31%, list=17%, signal=37% | |
| KRAS_SIGNALING_UP | 192 | -0.41 | 0.07 | 0.20 | tags=43%, list=25%, signal=57% | |
| APICAL_SURFACE | 42 | -0.40 | 0.07 | 0.22 | tags=26%, list=13%, signal=30% | |
| XENOBIOTIC_METABOLISM | 194 | -0.33 | 0.05 | 0.23 | tags=38%, list=26%, signal=51% | |
| COAGULATION | 133 | -0.38 | 0.09 | 0.23 | tags=44%, list=26%, signal=59% | |
| IL2_STAT5_SIGNALING | 182 | -0.37 | 0.08 | 0.25 | tags=41%, list=25%, signal=55% | |
| Male with vigorous-intensity exercise | PANCREAS_BETA_CELLS | 36 | -0.51 | 0.03 | 0.16 | tags=31%, list=14%, signal=35% |
| Female with moderate-intensity exercise | OXIDATIVE_PHOSPHORYLATION | 188 | -0.45 | 0.09 | 0.24 | tags=49%, list=33%, signal=72% |
| PEROXISOME | 98 | -0.43 | < 0.01 | 0.21 | tags=47%, list=33%, signal=69% | |
| Female with vigorous-intensity exercise | OXIDATIVE_PHOSPHORYLATION | 188 | -0.57 | < 0.01 | 0.01 | tags=69%, list=28%, signal=96% |
| ADIPOGENESIS | 184 | -0.41 | < 0.01 | 0.13 | tags=45%, list=28%, signal=62% | |
| FATTY_ACID_METABOLISM | 150 | -0.40 | 0.03 | 0.17 | tags=45%, list=29%, signal=63% | |
| HEDGEHOG_SIGNALING | 35 | -0.47 | 0.03 | 0.21 | tags=40%, list=21%, signal=51% |
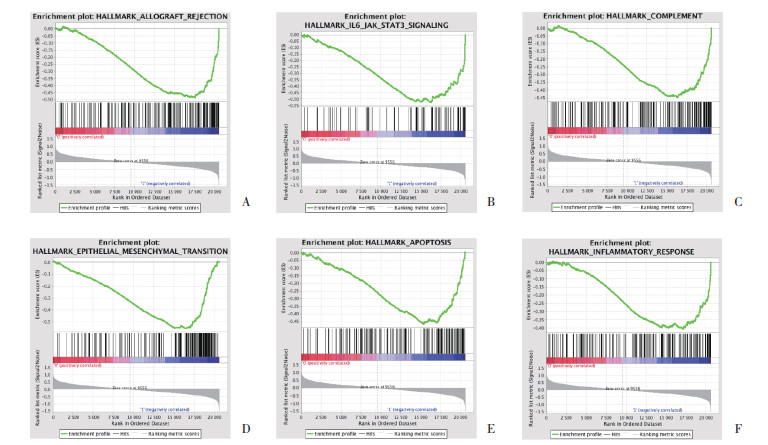
|
| A, ALLOGRAFT_REJECTION pathway; B, IL6-JAK-STAT3 pathway; C, COMPLEMENT pathway; D, EPITHELIAL_MESENCHYMAL_TRANSITION pathway; E, APOPTOSIS pathway; F, INFLAMMATORY_RESPONSE pathway. 图 1 与入组时比较,男性不运动组6个月后骨骼肌基因谱的GSEA结果 Fig.1 GSEA results of skeletal muscle gene profiles in males without exercise after 6 months compared with the gene profiles at enrollment |
2.2 女性锻炼前后的GSEA比较
与入组时比较,6个月后不运动组的标本未富集任何通路;小量中强度组富集了氧化磷酸化(OXIDATIVE_PHOSPHORYLATION)、过氧化物酶体(PEROXISOME)等通路;小量高强度组则富集了氧化磷酸化(OXIDATIVE_PHOSPHORYLATION)、脂肪发生(ADIPOGENESIS)、脂肪酸代谢(FATTY_ACID_METABOLISM)、HEDGEHOG信号等通路,见表 1、图 2。
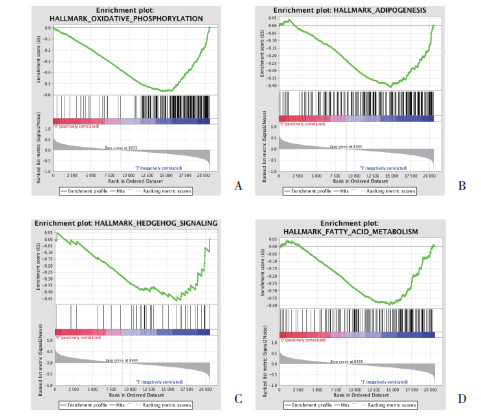
|
| A, OXIDATIVE_PHOSPHORYLATION pathway; B, ADIPOGENESIS pathway; C, FATTY_ACID_METABOLISM pathway; D, HEDGEHOG pathway. 图 2 与入组时比较,女性小量高强度运动组6个月后骨骼肌基因谱的GSEA结果 Fig.2 GSEA results of the skeletal muscle gene profiles in females with 6 months low-amount vigorous-intensity exercise compared with the gene profiles at enrollment |
2.3 男性、女性锻炼前后领头亚群基因分析
与入组时比较,男性不运动组6个月后的骨骼肌细胞中在所富集通路出现频次最多的领头亚群基因包括TIMP2、IL2RB、LYN、SERPINE1等,见图 3A。与入组时比较,女性小量高强度组6个月后骨骼肌细胞中高频度出现于富集通路中的领头亚群基因包括ECHS1、DLD、SDHC、ECH1、ACAA2、MDH2、ACADM、DECR1、RETSAT等,见图 3B。
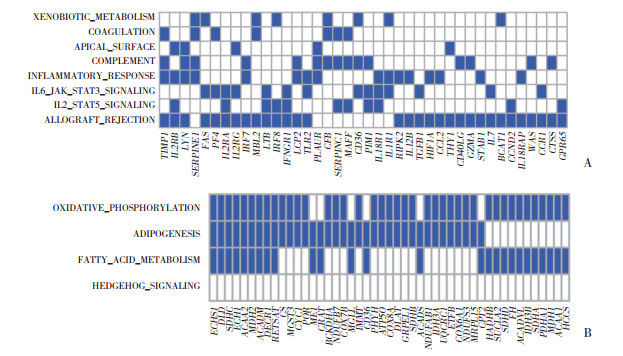
|
| A, most frequent leading edge genes of the skeletal muscles in metabolic syndrome males after 6 months without exercise; B, most frequent leading edge genes of the skeletal muscles in metabolic syndrome females after 6 months low-amount vigorous-intensity exercise. vertical axis, signal pathway; horizental axis, gene name. 图 3 与入组时比较,参与者6个月后骨骼肌中领头亚群基因分析结果 Fig.3 Leading edge genes of the skeletal muscles in participants after 6 months compared with those at enrollment |
3 讨论
本研究结果显示,男性与入组时比较,不运动组骨骼肌细胞富集了凋亡、炎症反应和异常增生与转化相关通路,在各强度运动组未发现此种改变;女性不运动组骨骼肌细胞无特殊变化,小量中高强度运动训练则增强骨骼肌有氧代谢、维持肌纤维结构以及促进肌细胞损伤修复等生物过程。
骨骼肌作为代谢、心血管相关疾病的重要靶器官,其结构与功能的稳态是维持人体健康的重要基石[4]。而人体由亚健康状态发展为代谢、心血管疾病的过程中,骨骼肌的代谢适应性下降,出现胰岛素敏感性下降/抵抗,从而导致多重代谢改变,其本质主要存在以下两方面变化:(1)线粒体氧化能力/三羟酸循环破坏;(2)免疫功能破坏、炎症/免疫反应增强、细胞死亡/凋亡增多。已有研究[5-6]显示代谢异常的小鼠模型或具有代谢综合征/2型糖尿病的患者中,可观察到骨骼肌发生胰岛素抵抗以及损伤相关性改变(线粒体功能失调、线粒体超氧产物以及凋亡、免疫相关信号通路激活)。
本研究结果显示,亚健康状态的男性,6个月不运动骨骼肌细胞出现了炎性细胞因子介导的免疫活性、排斥反应、补体系统、炎症反应增强,并存在凋亡、上皮间质转化、高凝和异常增生信号通路活化,即向更易于发生心血管、代谢疾病的方向发展。而运动情况下未出现这些不利的因素,在小量高强度运动组出现了胰岛β-细胞增强,即可能通过促进胰岛素分泌逆转亚健康状态。亚健康状态女性6个月不运动未见骨骼肌发生不利变化,而小量运动6个月无论中强度还是高强度均可促进骨骼肌细胞中的氧化磷酸化代谢、增强过氧化物及脂肪酸的代谢,从而促进亚健康状态逆转。另外,已有研究[7]显示小量高强度运动中活化的HEDGEHOG信号通路可能有利于促进高强度运动后骨骼肌发育、损伤修复,并负责调节快慢肌纤维特性。
根据领头亚群基因分析结果,男性不运动组中上调的TIMP2基因参与了上皮间质转化、免疫反应、炎症反应、氧化应激反应等多种机体变化,并与高血压、代谢综合征等疾病的发生发展密切相关[8];IL2RB基因主要参与机体内免疫反应,与2型糖尿病、高血压及心脑血管事件等相关[9];LYN基因为酪氨酸蛋白激酶,与多种恶性肿瘤的发生发展相关[10];SERPINE1基因上调使患者易于形成血栓,可见于肥胖、高脂血症、2型糖尿病等代谢相关疾病以及乳腺癌等恶性肿瘤和多种感染性疾病[11-12]。目前,这些基因改变认为是不运动导致亚健康状态男性骨骼肌发生不利变化、并促进其发生心血管、代谢疾病的核心因素。另一方面,在女性小量高强度运动组筛出的基因中,ECHS1基因属于水合酶超家族,负责线粒体脂肪酸代谢,其表达可抑制肿瘤细胞和病毒复制[13-14];DLD基因作为氧化还原酶或蛋白水解酶调节机体的能量代谢[15];SDHC基因作为参与线粒体能量代谢(三羧酸循环和有氧呼吸链)的关键酶,其表达缺失与多种恶性肿瘤发生相关[16];ECH1基因属于水解酶超家族,其功能与营养状况相关[17];ACAA2基因参与线粒体脂肪酸代谢,避免细胞发生凋亡[18];MDH2基因为线粒体内参与三羧酸循环的脱氢酶,其表达减低或缺失与恶性肿瘤密切相关[19];ACADM基因参与线粒体脂肪酸代谢,其异常与多种代谢性疾病相关[20];DECR1基因主要在线粒体内参与氧化反应和脂肪酸代谢[21];而RETSA基因则参与脂肪形成,在肥胖患者体内其表达下降[22]。本研究结果显示这些基因可能是运动使女性受益、逆转其亚健康状态的关键基因。
综上所述,本研究针对处于亚健康状态的心血管代谢疾病高危人群,基于在线数据GSE48278,通过生物信息学分析其骨骼肌细胞基因组学改变,明确了不运动对于男性骨骼肌结构与功能的不利影响;并发现小量运动训练可增强女性骨骼肌有氧代谢并维持骨骼肌的组成与功能。本研究从基因组学角度,解释了适量运动对亚健康人群的重要性,对明确如何逆转亚健康状态,降低心血管代谢疾病风险提供了重要依据,具有一定的科学价值。
| [1] |
BATEMAN LA, SLENTZ CA, WILLIS LH, et al. Comparison of aerobic versus resistance exercise training effects on metabolic syndrome (from the studies of a targeted risk reduction intervention through defined exercise-STRRIDE-AT/RT)[J]. Am J Cardiol, 2011, 108(6): 838-844. DOI:10.1016/j.amjcard.2011.04.037 |
| [2] |
WILLIS LH, SLENTZ CA, BATEMAN LA, et al. Effects of aerobic and/or resistance training on body mass and fat mass in overweight or obese adults[J]. J Appl Physiol, 2012, 113(12): 1831-1837. DOI:10.1152/japplphysiol.01370.2011 |
| [3] |
SUBRAMANIAN A, TAMAYO P, MOOTHA VK, et al. Gene set enrichment analysis:a knowledge-based approach for interpreting genome-wide expression profiles[J]. Proc Natl Acad Sci USA, 2005, 102(43): 15545-15550. DOI:10.1073/pnas.0506580102 |
| [4] |
HUFFMAN KM, KOVES TR, HUBAL MJ, et al. Metabolite signatures of exercise training in human skeletal muscle relate to mitochondrial remodelling and cardiometabolic fitness[J]. Diabetologia, 2014, 57(11): 2282-2295. DOI:10.1007/s00125-014-3343-4 |
| [5] |
WU C, XU G, TSAI SA, et al. Transcriptional profiles of type 2 diabetes in human skeletal muscle reveal insulin resistance, metabolic defects, apoptosis, and molecular signatures of immune activation in response to infections[J]. Biochem Biophys Res Commun, 2017, 482(2): 282-288. DOI:10.1016/j.bbrc.2016.11.055 |
| [6] |
DOTT W, WRIGHT J, CAIN K, et al. Integrated metabolic models for xenobiotic induced mitochondrial toxicity in skeletal muscle[J]. Redox Biol, 2018, 14: 198-210. DOI:10.1016/j.redox.2017.09.006 |
| [7] |
ANDERSON C, WILLIAMS VC, MOYON B, et al. Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity[J]. Genes Dev, 2012, 26(18): 2103-2117. DOI:10.1101/gad.187807.112 |
| [8] |
SABBATINI AR, BARBARO NR, DE FARIA AP, et al. Increased circulating tissue inhibitor of metalloproteinase-2 is associated with resistant hypertension[J]. J Clin Hypertens (Greenwich), 2016, 18(10): 969-975. DOI:10.1111/jch.12865 |
| [9] |
DURDA P, SABOURIN J, LANGE EM, et al. Plasma levels of soluble interleukin-2 receptor α: associations with clinical cardiovascular events and genome-wide association scan[J]. Arterioscler Thromb Vasc Biol, 2015, 35(10): 2246-2253. DOI:10.1161/ATVBAHA.115.305289 |
| [10] |
ROSEWEIR AK, QAYYUM T, LIM Z, et al. Nuclear expression of Lyn, a Src family kinase member, is associated with poor prognosis in renal cancer patients[J]. BMC Cancer, 2016, 16: 229. DOI:10.1186/s12885-016-2254-9 |
| [11] |
DE LA CRUZ-MOSSO U, ELENA RAMOS-ARELLANO L, FRANCISCO MUNOZ-VALLE J, et al. PAI-1 haplogenotype confers genetic susceptibility for obesity and hypertriglyceridemia in Mexican children[J]. Invest Clin, 2016, 57(3): 246-258. |
| [12] |
FAN QY, LI H, QIN YF, et al. Association of SERPINE1 rs6092 with type 2 diabetes and related metabolic traits in a Chinese population[J]. Gene, 2018, 661: 176-181. DOI:10.1016/j.gene.2018.04.011 |
| [13] |
ZHU XS, GAO P, DAI YC, et al. Attenuation of enoyl coenzyme A hydratase short chain 1 expression in gastric cancer cells inhibits cell proliferation and migration in vitro[J]. Cell Mol Biol Lett, 2014, 19(4): 576-589. DOI:10.2478/s11658-014-0213-5 |
| [14] |
ZHU XS, DAI YC, CHEN ZX, et al. Knockdown of ECHS1 protein expression inhibits hepatocellular carcinoma cell proliferation via suppression of Akt activity[J]. Crit Rev Eukaryot Gene Expr, 2013, 23(3): 275-282. DOI:10.1615/CritRevEukaryotGeneExpr.2013007531 |
| [15] |
AMBRUS A, ADAM-VIZI V. Human dihydrolipoamide dehydrogenase (E3) deficiency:novel insights into the structural basis and molecular pathomechanism[J]. Neurochem Int, 2018, 117: 5-14. DOI:10.1016/j.neuint.2017.05.018 |
| [16] |
GORSKA-PONIKOWSKA M, KUBAN-JANKOWSKA A, EISLER SA, et al. 2-methoxyestradiol affects mitochondrial biogenesis pathway and succinate dehydrogenase complex flavoprotein subunit A in osteosarcoma cancer cells[J]. Cancer Genomics Proteomics, 2018, 15(1): 73-89. DOI:10.21873/cgp.20067 |
| [17] |
ZHANG YK, QU YY, LIN Y, et al. Enoyl-CoA hydratase-1 regulates mTOR signaling and apoptosis by sensing nutrients[J]. Nat Commun, 2017, 8(1): 464. DOI:10.1038/s41467-017-00489-5 |
| [18] |
MILTIADOU D, HAGER-THEODORIDES AL, SYMEOU S, et al. Variants in the 3'untranslated region of the ovine acetyl-coenzyme A acyltransferase 2 gene are associated with dairy traits and exhibit differential allelic expression[J]. J Dairy Sci, 2017, 100(8): 6285-6297. DOI:10.3168/jds.2016-12326 |
| [19] |
CASCON A, COMINO-MENDEZ I, CURRAS-FREIXES M, et al. Whole-exome sequencing identifies MDH2 as a new familial paraganglioma gene[J]. J Natl Cancer Inst, 2015, 107(5): djv053. DOI:10.1093/jnci/djv053 |
| [20] |
DONG YX, LU HL, LI Q, et al. (5R)-5-hydroxytriptolide ameliorates liver lipid accumulation by suppressing lipid synthesis and promoting lipid oxidation in mice[J]. Life Sci, 2019, 232: 116644. DOI:10.1016/j.lfs.2019.116644 |
| [21] |
DE TONNAC A, LABUSSIERE E, VINCENT A, et al. Effect of α-linolenic acid and DHA intake on lipogenesis and gene expression involved in fatty acid metabolism in growing-finishing pigs[J]. Br J Nutr, 2016, 116(1): 7-18. DOI:10.1017/S0007114516001392 |
| [22] |
PANG XY, WANG SY, JURCZAK MJ, et al. Retinol saturase modulates lipid metabolism and the production of reactive oxygen species[J]. Arch Biochem Biophys, 2017, 633: 93-102. DOI:10.1016/j.abb.2017.09.009 |
 2019, Vol. 48
2019, Vol. 48




