文章信息
- 马珺, 姚一龙, 阮雪蕾, 沈书园, 邵连奇, 薛一雪
- MA Jun, YAO Yilong, RUAN Xuelei, SHEN Shuyuan, SHAO Lianqi, XUE Yixue
- circ-PTBP3对血肿瘤屏障通透性的影响
- Effect of circ-PTBP3 on the Permeability of Blood-Tumor Barrier
- 中国医科大学学报, 2019, 48(12): 1057-1062
- Journal of China Medical University, 2019, 48(12): 1057-1062
-
文章历史
- 收稿日期:2018-12-19
- 网络出版时间:2019-12-06 9:02
2. 中国医科大学附属盛京医院神经外科, 沈阳 110004
2. Deparment of Neurosurgery, Shengjing Hosipital, China Medical University, Shenyang 110004, China
脑胶质瘤是一种最常见的中枢神经系统恶性肿瘤[1],预后不良。主要原因是由于血肿瘤屏障(blood-tumor barrier,BTB)的存在严重限制了大分子化疗药物进入肿瘤组织,影响治疗效果[2]。因此,有效地增加BTB的通透性是切实提高脑胶质瘤疗效的关键。
人类基因组绝大部分转录为非编码RNAs(non-coding RNAs,ncRNAs),ncRNAs对生命活动具有重要的调控作用[3]。环状RNAs(circular RNAs,circRNAs)是近年新发现的一类ncRNAs,存在于多种细胞,具有调控基因表达的作用[4],与人类癌症的发生和发展密切相关[5]。研究[6]发现,在人脑胶质瘤组织中存在大量异常表达的circRNAs,这些异常表达的circRNAs可能参与胶质瘤的发生和发展。此外,缺氧可诱导内皮细胞中大量circRNAs表达水平的改变,提示circRNAs与血管功能密切相关。其中,circ-PTBP3(hsa_circ_0088072)在缺氧内皮细胞中高表达[7],但circ-PTBP3在BTB功能中的作用尚未见报道。因此,本研究拟探讨circ-PTBP3对BTB功能的调控作用,以期为切实提高胶质瘤的治疗提供新靶点。
1 材料与方法 1.1 材料 1.1.1 细胞人脑微血管内皮细胞(endothelial cells,ECs)系hCMEC/D3由Dr.Pierre-Olivier Couraud提供;人脑星形胶质母细胞瘤细胞系U87和人正常星形胶质细胞NHA购自中国科学院上海生命科学院细胞资源中心。
1.1.2 试剂EBM-2(美国Lonza公司);脂质浓缩物(美国Gibco公司);胎牛血清、HEPES(美国PAA公司);皮质醇(美国Sigma-Aldrich公司);抗坏血酸(美国Sigma-Aldrich公司);bFGF(美国Sigma-Aldrich公司);Cultrex Rat CollagenⅠ(美国R & D公司);Trizol(美国Thermo公司);one-step PrimeScript RT-PCR Kit(日本Takara公司);TaqMan Universal Master Mix Ⅱ(美国Thermo公司);TaqManGene Expression Assays(美国Thermo公司);RNase-R(美国Sigma-Aldrich公司);lipofectamine LTX和Plus试剂(美国Thermo公司);Lucifer yellow和FITC-Dextran(美国Sigma-Aldrich公司);兔抗ZO-1多克隆抗体(美国Thermo公司);小鼠抗occludin单克隆抗体(美国Thermo公司);小鼠抗claudin-5单克隆抗体(美国Thermo公司);小鼠抗GAPDH单克隆抗体(美国Santa Cruz公司);ECL试剂盒(美国Santa Cruz公司);Alexa Fluor 555标记的山羊抗小鼠和山羊抗兔荧光二抗(中国碧云天生物技术公司)。
1.2 方法 1.2.1 细胞培养hCMEC/D3细胞用含5%胎牛血清、1%青-链霉素、1.4 μmol/L皮质醇、1%化学定义脂质浓缩物、5 μg/mL抗坏血酸、10 mmol/L HEPES和1 ng/mL bFGF的EBM-2培养基培养于transwell小室上层(孔径0.4 μm,150 μg/mL Cultrex Rat CollagenⅠ处理)。人脑胶质母细胞瘤U87细胞培养于含有10%胎牛血清的DMEM高糖培养液中。正常星形胶质细胞NHA培养于含10%胎牛血清的RPMI-1640培养液中。用于本研究的ECs hCMEC/D3细胞在30~40代之间。所有细胞均置于37 ℃、5%CO2的恒温培养箱中培养。
1.2.2 体外血脑屏障(blood-brain barrier,BBB)与BTB模型的建立应用正常星形胶质细胞NHA和hCMEC/D3细胞共培养建立体外BBB模型,获得ECs;应用人脑胶质母细胞瘤细胞U87和hCMEC/D3细胞共培养建立体外BTB模型,获得脑胶质瘤微血管内皮细胞(glioma endothelial cells,GECs)。方法如下,用transwell小室(孔径0.4 μm)的下室,将U87或NHA细胞接种于6孔板中(2×104/孔),加入适量的培养液,培养48 h。然后,将hCMEC/D3细胞以2×105/孔的浓度接种于150 μg/mL Cultrex Rat CollagenⅠ处理的6孔transwell小室的上室,上下室均加入适量的EBM-2培养液,每2 d换液1次,共培养4 d[8]。
1.2.3 实时荧光定量PCR(qRT-PCR)Trizol法提取总RNA,用Nanodrop分光光度计测定RNA浓度及OD260/OD280比值。总RNA反转录后,应用one-step PrimeScript RT-PCR Kit检测circ-PTBP3和PTBP3 mRNA表达水平(RNase-R处理用于确认circ-PTBP3的存在并排除线性RNAs的干扰);应用TaqMan Universal Master MixⅡ和TaqMan Gene Expression Assays检测紧密连接相关蛋白[ZO-1(Hs01551861_m1)、occludin(Hs00170162_m1)、claudin-5(Hs00533949_s1)和GAPDH(Hs03929097_g1)]的表达。应用7500 Fast Real-Time PCR System测定Ct值,以2-ΔΔCt表示RNA的相对表达量。
1.2.4 细胞转染应用靶向circ-PTBP3的短发夹RNA序列连接到pGPU6/GFP/Neo载体上,构建circ-PTBP3表达沉默的质粒(sh-circ-PTBP3)。携带非靶向序列的质粒作为阴性对照(sh-NC)。以4×104/孔接种细胞于24孔板上,当细胞融合度达80%时,采用lipofectamine LTX和Plus试剂进行转染。4周后,G418抗性细胞克隆产生,用实时荧光定量PCR方法验证沉默效率。
1.2.5 跨内皮阻抗(transendothelial electric resistance,TEER)值的测量建立体外BBB和BTB模型,共培养第4天,将模型置于室温30 min后,应用millicell-ERS检测系统进行检测。每个样品的读数减去背景电阻值(包括transwell的微孔膜和培养液),并与transwell小室的表面积相乘,获得TEER值(Ω·cm2)。
1.2.6 通透性检测将transwell上下室培养液换为含有10 mmol/L HEPES和1 mmol/L丙酮酸钠的缓冲液,在上室缓冲液中加入50 mmol/L Lucifer yellow(LY,457)、50 mmol/L fluorescein isothiocyanate(FITC)-Dextran(4×103)或20 mmol/L FITC-Dextran(70×103)。使用Florescence multiwall plate reader分析,应用清除体积对时间作图,其曲线斜率为通透性系数。
1.2.7 Western blotting检测用含有1%PMSF的预冷RIPA蛋白裂解液提取总蛋白。SDS-PAGE电泳,转移至PVDF膜,封闭2 h后,分别加入抗ZO-1抗体(1︰500稀释)、抗occludin抗体(1︰250稀释)、抗claudin-5抗体(1︰250稀释)和抗GAPDH抗体(1︰1 000稀释),4 ℃孵育过夜。用辣根过氧化物酶标记的山羊抗兔或山羊抗小鼠IgG二抗孵育。ECL发光,用Microchemi仪器检测,ChemImager 5500 V2.03软件扫描,采用Fluorchem 2.0定量分析显色带的整合光密度值。
1.2.8 免疫荧光染色实验用冷丙酮固定细胞,室温封闭后,加入一抗4 ℃孵育过夜(1︰50稀释)。洗脱后,用Alexa Fluor 555标记的山羊抗小鼠或山羊抗兔荧光二抗(1︰500稀释)室温孵育2 h,洗脱后用0.5 μg/mL DAPI染核,甘油封片。应用荧光显微镜进行图像采集与分析。
1.3 统计学分析实验数据以x±s表示,采用SPSS 18.0统计软件进行分析。2组间比较采用Student’s t检验,3组及3组以上比较采用单因素方差分析和Dunnett’s post-test检验,P < 0.05为差异有统计学意义。
2 结果 2.1 circ-PTBP3在GECs中高表达应用人脑微血管ECs系hCMEC/D3与正常星形胶质细胞系NHA建立体外BBB模型,获得正常脑微血管ECs;应用人脑微血管内皮细胞系hCMEC/D3与胶质母细胞瘤细胞系U87建立体外BTB模型,获得GECs。应用qRT-PCR方法检测circ-PTBP3在ECs和GECs中的表达,如图 1所示,circ-PTBP3在GECs中的表达水平显著高于ECs(P < 0.01)。
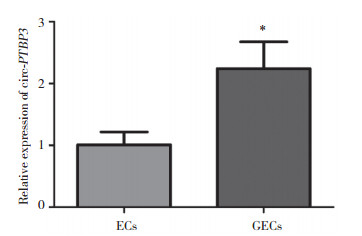
|
| Relative circ-PTBP3 expression in ECs and GECs. Data represent means ± SD (n = 5, each). *P < 0.01 vs ECs group. 图 1 circ-PTBP3在脑微血管内皮细胞中的表达水平 Fig.1 Expression of circ-PTBP3 in cultured brain endothelium |
2.2 RNase R处理后circ-PTBP3在GECs中高表达
为明确线性RNA linear-PTBP3是否参与BTB功能的调控,检测了PTBP3 mRNA在ECs和GECs中的表达水平。如图 2A所示,PTBP3 mRNA在ECs与GECs中的表达水平无统计学差异。RNase R可以降解线性RNA,却不能降解circRNA。用RNase R处理后检测circ-PTBP3在ECs和GECs中的表达水平,发现circ-PTBP3在GECs或RNase R处理后的GECs中高表达,且能抵抗RNase R的降解,其表达水平在GECs与RNase R处理后的GECs中无显著差异(图 2B)。
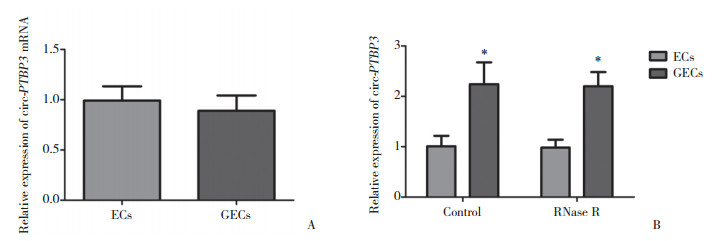
|
| A, expression of linear PTBP3 in ECs and GECs; B, expression of circ-PTBP3 in ECs and GECs with RNase R treatment. Data represent means ± SD (n = 5, each). **P < 0.01 vs ECs group. 图 2 线性PTBP3 RNA的表达及RNase R处理后circ-PTBP3的表达情况 Fig.2 Linear PTBP3 expression and circ-PTBP3 expression with RNase R treatment in cultured brain endothelium |
2.3 沉默circ-PTBP3破坏BTB的完整性并增加其通透性
应用TEER值和通透性检测法检测circ-PTBP3表达沉默对BTB的完整性和通透性的影响。通过转染sh-circ-PTBP3质粒建立稳定表达sh-circ-PTBP3的细胞系,circ-PTBP3的沉默效率如图 3A所示,sh-circ-PTBP3组的circ-PTBP3表达水平较sh-NC组显著降低(P < 0.01)。sh-circ-PTBP3组的TEER值较sh-NC组显著降低(P < 0.01)(图 3B)。进一步检测BTB对不同分子量物质的通透性。如图 3C所示,sh-circ-PTBP3组对LY和4×103 FITC-dextrans的通透性较sh-NC组均显著增加(均P < 0.01),对70×103 FITC-dextran的通透性与sh-NC组相比无统计学差异(P > 0.05)。提示沉默circ-PTBP3可显著破坏BTB的完整性并增加其通透性。
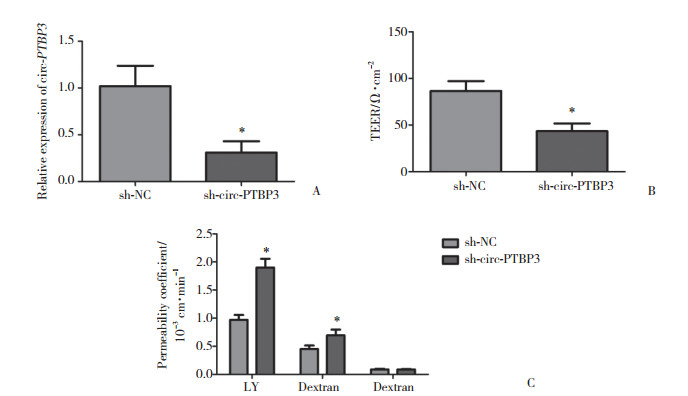
|
| A, relative circ-PTBP3 expression in sh-circ-PTBP3 stable expressing cells; B, the TEER values of BTB after circ-PTBP3 knockdown; C, the permeability coefficient values of LY (457), 4×103 FITC-dextrans and 70×103 FITC-dextran in BTB after circ-PTBP3 knockdown. Data represent means ± SD (n = 5, each). *P < 0.01 vs sh-NC group. 图 3 circ-PTBP3可调节BTB的完整性和通透性 Fig.3 circ-PTBP3 regulated the integrity and permeability of BTB |
2.4 沉默circ-PTBP3降低GECs中紧密连接相关蛋白的表达
应用Western blotting法检测ZO-1、occludin和claudin-5的蛋白表达变化。结果显示,转染sh-circ-PTBP3组的ZO-1、occludin和claudin-5的蛋白表达水平较sh-NC组显著降低(P < 0.05,P < 0.01,P < 0.05)(图 4A)。紧密连接相关蛋白的免疫荧光结果显示,转染sh-circ-PTBP3组的ZO-1、occludin和claudin-5的蛋白表达水平较sh-NC组显著降低。并且ZO-1和occludin位于GECs边缘,claudin-5位于细胞质。免疫荧光染色结果显示,sh-circ-PTBP3组ZO-1和occludin呈不连续性分布,claudin-5在细胞质中呈低表达状态(图 4B)。
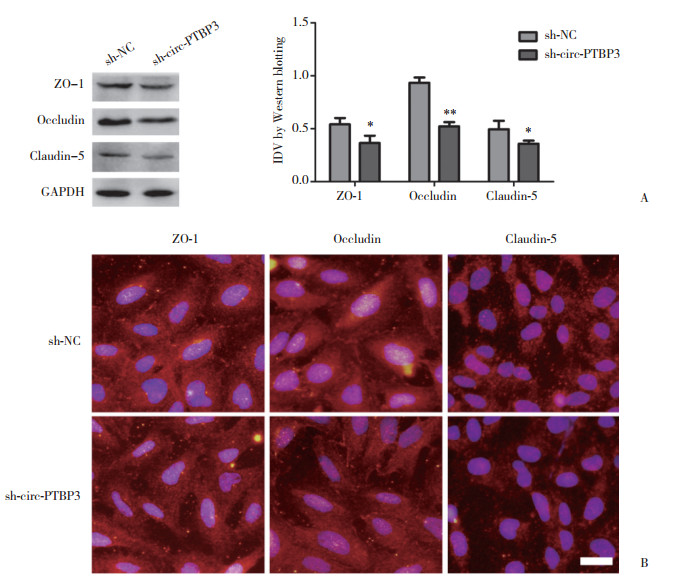
|
| A, Western blotting analysis of tight junction related proteins in GECs after circ-PTBP3 knockdown, using GAPDH as an endogenous control. Representative protein expression and their IDV are shown. Data represent means ± SD (n = 5, each). *P < 0.05 and **P < 0.01 vs sh-NC group. B, immunofluorescence staining of tight junction related proteins in GECs after circ-PTBP3 knockdown. Nuclei were labeled with DAPI. Images are representative of independent experiments (n = 5). Scale bar represents 20 μm. 图 4 circ-PTBP3可调节GECs紧密连接相关蛋白的表达 Fig.4 circ-PTBP3 regulated the expression of tight junction related proteins in GECs |
3 讨论
circRNAs普遍存在于真核细胞的基因组中,具有调控基因表达的作用。近年来发现circRNAs在肿瘤的发生和发展中发挥重要作用。越来越多的证据表明,circRNAs高度保守且稳定表达于细胞质[9],它在胶质瘤组织中存在大量的异常表达,可能在胶质瘤的发生、发展等方面发挥调控作用,有望成为胶质瘤诊断和治疗的潜在靶标[6]。
近期有研究报道了circRNAs对胶质瘤细胞的调节作用,发现circ-MMP9在胶质瘤组织中高表达并可促进胶质瘤细胞的增殖、迁移和侵袭能力[10],circ-U2AF1可通过增加NOVA2的表达促进胶质瘤细胞的恶性行为[11],circ-NFIX通过调节Notch1与Notch信号通路促进胶质瘤细胞的恶性行为[12]。此外,研究[7]发现缺氧可诱导ECs中大量circRNAs表达异常,提示circRNAs与血管功能密切相关。此外,还发现circ-SHKBP1在GECs中高表达,并可促进胶质瘤的血管新生[13]。以上提示circRNAs可能参与GECs功能的调控。研究[7]发现,circ-PTBP3(hsa_circ_0088072)在缺氧ECs中高表达。本研究结果证实,circ-PTBP3在GECs中高表达,提示,circ-PTBP3可能参与BTB微血管ECs功能的调控。
本研究证明,线性RNA linear-PTBP3在ECs和GECs中表达无差异。应用RNase R处理,降解了linear-PTBP3,未影响circ-PTBP3的表达。因此推测,circ-PTBP3与linear-PTBP3是2个相互独立的RNA,与circ-SHKBP1和circ-HIPK3相似[13-14]。有研究报道作为ncRNAs中的另一成员——lncRNAs可调控BTB屏障的通透性。沉默MIR17HG可降低紧密连接相关蛋白的表达,并增加BTB的通透性[15];过表达MEG3通过负性调控miR-330-5p增加BTB的通透性[16];沉默MALAT1可通过增加miR-140表达,增加BTB的通透性[17]。但目前尚未见circRNAs对BTB屏障通透性调控的报道。本研究沉默了circ-PTBP3,发现沉默circ-PTBP3可显著降低BTB的TEER值,增加BTB的通透性。BTB主要由肿瘤细胞和脑微血管ECs构成,相邻的脑微血管ECs之间的紧密连接是维持屏障完整性的重要结构和功能基础[18]。本研究组前期报道了紧密连接相关蛋白ZO-1、occludin和claudin-5表达的下调,在通过细胞旁途径增加BTB通透性的过程中起关键作用[8, 17, 19-20]。因此,本研究中检测了紧密连接相关蛋白ZO-1、occludin和claudin-5的表达,发现沉默circ-PTBP3可显著减少ZO-1、occludin与claudin-5的表达与分布。以上研究结果提示,沉默circ-PTBP3可降低BTB的完整性并增加其通透性。
综上所述,本研究证明了circ-PTBP3在GECs中高表达,沉默circ-PTBP3可破坏BTB的完整性并增加BTB的通透性,减少紧密连接相关蛋白的表达。此作用机制的阐明可为胶质瘤的治疗提供新的靶点。
| [1] |
OMURO A, DEANGELIS LM. Glioblastoma and other malignant gliomas:a clinical review[J]. JAMA, 2013, 310(17): 1842-1850. DOI:10.1001/jama.2013.280319 |
| [2] |
VAN TELLINGEN O, YETKIN-ARIK B, DE GOOIJER MC, et al. Overcoming the blood-brain tumor barrier for effective glioblastoma treatment[J]. Drug Resist Updat, 2015, 19: 1-12. DOI:10.1016/j.drup.2015.02.002 |
| [3] |
DJEBALI S, DAVIS CA, Merkel A, et al. Landscape of transcription in human cells[J]. Nature, 2012, 489(7414): 101-108. DOI:10.1038/nature11233 |
| [4] |
FANG Y. Circular RNAs as novel biomarkers with regulatory potency in human diseases[J]. Future Sci OA, 2018, 4(7): FSO314. DOI:10.4155/fsoa-2018-0036 |
| [5] |
LI J, YANG J, ZHOU P, et al. Circular RNAs in cancer:novel insights into origins, properties, functions and implications[J]. Am J Cancer Res, 2015, 5(2): 472-480. |
| [6] |
WANG D, YANG S, WANG H, et al. The progress of circular RNAs in various tumors[J]. Am J Transl Res, 2018, 10(6): 1571-1582. |
| [7] |
BOECKEL JN, JAE N, HEUMULLER AW, et al. Identification and characterization of hypoxia-regulated endothelial circular RNA[J]. Circ Res, 2015, 117(10): 884-890. DOI:10.1161/CIRCRESAHA.115.306319 |
| [8] |
MA J, WANG P, LIU Y, et al. Kruppel-like factor 4 regulates blood-tumor barrier permeability via ZO-1, occludin and claudin-5[J]. J Cell Physiol, 2014, 229(7): 916-926. DOI:10.1002/jcp.24523 |
| [9] |
MENG S, ZHOU H, FENG Z, et al. CircRNA:functions and properties of a novel potential biomarker for cancer[J]. Mol Cancer, 2017, 16(1): 94. DOI:10.1186/s12943-017-0663-2 |
| [10] |
WANG R, ZHANG S, CHEN X, et al. EIF4A3-induced circular RNA MMP9(circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis[J]. Mol Cancer, 2018, 17(1): 166. DOI:10.1186/s12943-018-0911-0 |
| [11] |
LI G, HUANG M, CAI Y, et al. Circ-U2AF1 promotes human glioma via derepressing neuro-oncological ventral antigen 2 by sponging hsa-miR-7-5p[J]. J Cell Physiol, 2019, 234(6): 9144-9155. DOI:10.1002/jcp.27591 |
| [12] |
XU HY, ZHANG Y, QI L, et al. NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via Notch signaling pathway[J]. Front Mol Neurosci, 2018, 11: 225. DOI:10.3389/fnmol.2018.00225 |
| [13] |
HE Q, ZHAO L, LIU Y, et al. circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways[J]. Mol Ther Nucleic Acids, 2018, 10: 331-348. DOI:10.1016/j.omtn.2017.12.014 |
| [14] |
MA XL, ZHU KP, ZHANG CL. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma[J]. J Cancer, 2018, 9(10): 1856-1862. DOI:10.7150/jca.24619 |
| [15] |
LENG X, MA J, LIU Y, et al. Mechanism of piR-DQ590027/MIR17HG regulating the permeability of glioma conditioned normal BBB[J]. J Exp Clin Cancer Res, 2018, 37(1): 246. DOI:10.1186/s13046-018-0886-0 |
| [16] |
SHEN S, YU H, LIU X, et al. PIWIL1/piRNA-DQ593109 regulates the permeability of the blood-tumor barrier via the MEG3/miR-330-5p/RUNX3 axis[J]. Mol Ther Nucleic Acids, 2018, 10: 412-425. DOI:10.1016/j.omtn.2017.12.020 |
| [17] |
MA J, WANG P, YAO Y, et al. Knockdown of long non-coding RNA MALAT1 increases the blood-tumor barrier permeability by up-regulating miR-140[J]. Biochim Biophys Acta, 2016, 1859(2): 324-338. DOI:10.1016/j.bbagrm.2015.11.008 |
| [18] |
BEGLEY DJ, BRIGHTMAN MW. Structural and functional aspects of the blood-brain barrier[J]. Prog Drug Res, 2003, 61: 39-78. |
| [19] |
MA J, YAO Y, WANG P, et al. MiR-181a regulates blood-tumor barrier permeability by targeting Kruppel-like factor 6[J]. J Cereb Blood Flow Metab, 2014, 34(11): 1826-1836. DOI:10.1038/jcbfm.2014.152 |
| [20] |
ZHAO W, WANG P, MA J, et al. MiR-34a regulates blood-tumor barrier function by targeting protein kinase Cepsilon[J]. Mol Biol Cell, 2015, 26(10): 1786-1796. DOI:10.1091/mbc.E14-10-1474 |
 2019, Vol. 48
2019, Vol. 48




