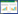文章信息
- 刘红霞, 施琼, 周一青, 安利钦, 严树涓, 张汝益, 翁亚光.
- LIU Hong-xia, SHI Qiong, ZHOU Yi-qing, AN Li-qin, YAN Shu-juan, ZHANG Ru-yi, WENG Ya-guang.
- 过表达miR-155抑制BMP9诱导间充质干细胞C3H10T1/2成骨分化
- Overexpression of miR-155 Inhibits the Osteogenic Differentiation of Mesenchymal Stem Cells C3H10T1/2 Induced by BMP9
- 中国生物工程杂志, 2017, 37(5): 9-18
- China Biotechnology, 2017, 37(5): 9-18
- http://dx.doi.org/DOI:10.13523/j.cb.20170502
-
文章历史
- 收稿日期: 2016-11-08
- 修回日期: 2017-01-23
间充质干细胞(mesenchymal stem cell, MSC)是一种具有自我复制能力和多向分化潜能的成体干细胞,可以分化为成骨细胞、软骨细胞、脂肪细胞和成肌细胞等多种结缔组织细胞,已成为骨应用组织工程学研究中成骨细胞的重要来源。BMP9属于TGF-β超家族成员,已有研究证实其可以诱导间充质干细胞成骨分化[1]。但目前BMP9尚未进入临床使用,对成骨诱导的调控机制仍然研究的不够完整,这就需要我们对其中包含的具体机制进行深入的研究。microRNA(miRNA)是一类长度约为22个核苷酸的非编码单链小分子RNA,通过抑制靶基因mRNA的翻译过程或降解靶基因的mRNA分子,介导转录后基因表达的调控,其在细胞的增值、分化以及很多疾病的发生发展过程中发挥巨大作用[2-8]。
miR-155是一个多功能miRNA[9],目前对它的研究主要集中在炎症[10]、免疫性疾病[11-13]、血液肿瘤[14-16]、实体肿瘤及心血管疾病[17-21]中,但对成骨分化的作用了解的并不多。有研究表明,在人类肺表皮细胞系A549中,miR-155可以通过抑制Smad1和Smad5来抑制BMP信号通路[17]。但miR-155在BMP9诱导间充质干细胞C3H10T1/2成骨分化中的作用尚未见报道,文章旨在研究BMP9诱导成骨分化过程中miR-155的作用,并对其相关的作用机制进行初步探究。
1 材料与方法 1.1 材料 1.1.1 细胞和重组腺病毒来源小鼠间充质干细胞系C3H10T1/2细胞购于ATCC公司;重组腺病毒BMP9由本实验室前期构建保存。本实验所保存的BMP9携带绿色荧光标签GFP,因此可用荧光显微镜来观察其感染效率。
1.1.2 试剂来源DMEM高糖培养基购于Hyclone公司;胎牛血清购于美国Gibco公司,链霉素、青霉素、茜素红S染料、维生素C和β-磷酸甘油购于Sigma公司;转染试剂EntransterTM-R4000购于北京英格恩生物科技有限公司;碱性磷酸酶(ALP)染色试剂盒购于碧云天生物科技有限公司;碱性磷酸酶(ALP)活性检测试剂盒购于BD公司;RNA提取试剂Trizol及Lipofectamine 2000购于Invitrogen公司;M-MLV逆转录酶、RT-PCR试剂盒购于TaKaRa公司;内切酶和T4连接酶、qRCR试剂SYBR Green购于TaKaRa公司;pMIR-REPORT microRNA(miRNA)expression reporter购于Ambion公司;荧光素酶检测试剂盒购自Promega公司;PCR引物由南京金斯瑞生物科技有限公司合成。p-Smad1/5/8、Smad1/5/8抗体购于Santa Cruz公司,HIF1α和VEGF抗体购于沈阳万类生物科技有限公司。β-actin抗体和二抗购于北京中杉金桥生物技术有限公司,其他试剂均为进口分装或国产分析纯。
1.1.3 转染所用寡核苷酸序列miR-155模拟物(miR-155) 及miR-155抑制剂(anti-miR-155) 阴性对照物(NC)由上海吉玛制药技术有限公司合成。与miR-155结合的靶基因的野生和突变序列由南京金斯瑞生物科技有限公司按照表 1序列信息合成。
| Name | Sequence(5′→3′) |
| mmu-miR-155 mimic | (F)UUAAUGCUAAUUGUGAUAGGGGU |
| (R)CCCUAUCACAAUUAGCAUUAAUU | |
| mmu-miR-155 inhibitor | (F)ACCCCUAUCACAAUUAGCAUUAA |
| Negative control (NC) | (F)UUCUCCGAACGUGUCACGUTT |
| (R)ACGUGACACGUUCGGAGAATT | |
| WT-HIF1α | (F)CTAGGAGTAATTTTAGAAGCATTATTTTAGGAATATATAGTTGTCACAG |
| (R)CTGTGACAACTATATATTCCTAAAATAATGCTTCTAAAATTACTC | |
| MT-HIF1α | (F)CTAGGAGTAATTTTAGAACGTAAATTTTAGGAATATATAGTTGTCACAG |
| (R)AGCTCTGTGACAACTATATATTCCTAAAATTTACGTTCTAAAATTACTC | |
| Note: WT-HIF1α, HIF1α-3′-UTR wild type; MT-HIF1α, HIF1α-3′-UTR mutant type; F. Forward; R. Reverse | |
1.2 方法 1.2.1 细胞培养
C3H10T1/2细胞系和HEK 293细胞系均培养于含有10%胎牛血清的DMEM高糖培养基中,其中含有100U/ml青霉素和100μg/ml链霉素,在37℃、5% CO2的饱和湿度培养箱中培养。
1.2.2 细胞转染用胰酶消化处于对数生长期的细胞,接种于6孔板中,待细胞密度长至70%左右时,按照EntransterTM-R4000试剂说明书进行操作,把miR-155、anti-miR-155或NC转染到C3H10T1/2细胞中,6h后换液,再转染BMP9诱导其成骨分化,6h后换液。
1.2.3 ALP染色及活性测定将C3H10T1/2细胞接种于24孔板后,待细胞密度达到70%时进行不同的处理,6h后换液,培养7d后,弃去培养基用PBS冲洗2次,4℃ 100%乙醇固定1h,每孔加入NBT/BCIP溶液200μl进行ALP染色,避光30min后观察染色结果。根据试剂盒说明书进行ALP活性测定。
1.2.4 茜素红S染色C3H10T1/2细胞接种于24孔板,细胞融合度达到70%时给予不同的处理,6h后换液,同时加入工作浓度50μg/ml的维生素C、10mmol/L的β-磷酸甘油,继续培养14d进行茜素红S染色:弃去孔板中的培养基,PBS冲洗3次后用4℃ 0.05%戊二醛固定10min,去离子水洗3次后加入0.4%茜素红S染液,染色5min,弃去染液,去离子水终止反应并洗涤,显微镜下观察并成像。
1.2.5 PCR6孔板培养C3H10T1/2细胞,细胞覆盖达到70%时给予不同处理,分别在适当时间点提取RNA,根据试剂盒说明书逆转录成cDNA进行RT-PCR或qPCR(miRNA检测以U6为内参,一般基因检测以β-actin为内参)。qPCR数据分析采用比较CT法(ΔΔCT),目的基因相对表达量=2-[(CT处理-CT内参)-(CT对照-CT内参)],所有数值取三次重复的平均值。实验所用引物序列见表 2。
| Genes | Sequence(5′→3′) | |
| mmu-miR-155 | (RT) GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCCCT | |
| U6 | (RT) AAAATATGGAACGCTTCACGAATTTG | |
| mmu-miR-155 | (F) GGCGTTAATGCTAATTGTGAT | (R) GTGCAGGGTCCGAGGT |
| U6 | (F) CTCGCTTCGGCAGCACATATACT | (R) ACGCTTCACGAATTTGCGTGTC |
| Runx2 | (F) TCTGACAAAGCCTTCATGTCC | (R) AAATAGTGATACCGTAGATGCG |
| OSX | (F) GAAGTCCAATGGGGATCTGA | (R) GAATCCCTTTCCCTCTCCAG |
| COL1A1 | (F) CGGCTCCTGCTCCTCTTA | (R) TTCATTGCATTGCACGTCAT |
| ALP | (F) TGACCTTCTCTCCTCCATCC | (R) CTTCCTGGGAGTCTCATCCT |
| OCN | (F) CTGCTTGTGACGAGCTATCAG | (R) TGATACCGTAGATGCGTTTGT |
| OPN | (F) GAGGAAACCAGCCAAGGTAAG | (R) AAAGCAAATCACTGCCAATCTC |
| β-actin | (F) ATGAAGGCGTGGCAACAT | (R) GCCATTGGCTCTGTCCTG |
| HIF1α | (F) TCAGCATACAGTGGCACTCA | (R) GGTTAAGGCTCCTTGGATGA |
| VEGF | (F) AGCACAGCAGATGTGAATGC | (R) AATGCTTTCTCCGCTCTGAA |
| Note: RT. Reverse transcription; F. Forward; R. Reverse | ||
1.2.6 Western blot
C3H10T1/2细胞经不同处理因素处理至需要的天数后,用RIPA裂解液于冰上裂解细胞,离心取上清液,BCA法检测上清液总蛋白浓度,加入适量load buffer煮沸10min,取总量50μg的蛋白质经过SDS-PAGE电泳、转膜、封闭、孵一抗、洗膜、孵二抗、洗膜显影后,最后成像保存。
1.2.7 荧光素酶报告基因实验TargetScan和PicTar靶基因预测软件预测到的miR-155靶基因位点加上HindⅢ和SpeⅠ酶切位点后用T4连接酶连接到报告载体上。用Lipofectamine 2000将阳性质粒和对照质粒转染到HEK 293细胞中,48h后根据试剂盒说明书检测荧光素酶活性。
1.3 统计学分析数据采用均数±标准差(x±s)表示,组间比较采用单因素方差分析,两组间比较采用t检验,统计所用软件为SPSS17.0,P < 0.05认为差异有统计学意义。
2 结果 2.1 在BMP9诱导间充质干细胞C3H10T1/2细胞成骨分化过程中miR-155的表达变化用腺病毒BMP9诱导C3H10T1/2细胞成骨分化的不同天数里,miR-155的表达先升高,3d时降低,5d升到最高(P < 0.01),7d又下降[图 1(a)]。荧光显微镜下观察BMP9成功感染细胞[图 1(b)]。在BMP9成骨分化的不同天数里,Runx2和ALP的表达也升高,说明BMP9诱导成骨分化成功[图 1(c)]。

|
| 图 1 miR-155在BMP9诱导C3H10T1/2细胞成骨分化不同天数中的表达 Figure 1 The expression of miR-155 in different osteogenic differentiation days of C3H10T1/2 cells induced by BMP9 (a) Tested the expression of miR-155 in different differentiation days of C3H10T1/2 cells induced by BMP9 used qPCR **P < 0.01 compared with 0d (b) BMP9 infected C3H10T1/2 cells successful (×100) (c) RT-PCR detected the expression of Runx2 and ALP in different differentiation days of C3H10T1/2 cells that induced by BMP9 |
在BMP9诱导C3H10T1/2细胞成骨分化过程中,转染miR-155后7d, qPCR检测miR-155表达明显升高(P < 0.001);而anti-miR-155转染则可有效抑制内源性miR-155的表达(P < 0.001)[图 2(a)]。与阴性对照组BMP9+NC组相比,过表达miR-155可显著减少细胞的ALP活性(P < 0.01),减弱ALP染色;而抑制miR-155则能逆转其对ALP活性以及ALP染色的抑制作用[图 2(b)、(c)]。

|
| 图 2 过表达miR-155抑制BMP9诱导的C3H10T1/2细胞早期成骨分化 Figure 2 Overexpressed miR-155 inhibits the osteogenic differentiation of C3H10T1/2 cells induced by BMP9 in early stage (a) Tested the expression of miR-155 after overexpressing or inhibiting miR-155 in C3H10T1/2 cells osteogenic differentiation induced by BMP9 used qPCR **P < 0.001 compared with control group (b) Detected ALP activity of C3H10T1/2 cells in 7d **P < 0.01 (c) ALP staining of C3H10T1/2 cells in the 7d of differentiation (×100) |
过表达miR-155并用BMP9诱导C3H10T1/2细胞成骨分化14d,茜素红S染色检测发现与对照组相比,过表达miR-155明显减少了细胞的钙盐沉积,而抑制miR-155则促进了细胞的钙盐沉积(图 3)。

|
| 图 3 过表达miR-155抑制BMP9诱导的C3H10T1/2细胞晚期成骨分化 Figure 3 Overexpressed miR-155 inhibits the later stage of osteogenic differentiation of C3H10T1/2 cells induced by BMP9 |
在用BMP9诱导C3H10T1/2细胞成骨分化过程中,过表达miR-155抑制成骨分化相关因子的表达。利用qPCR检测转染miR-155并成骨诱导3d的细胞,发现与对照组相比Runx2、OSX和COL1A1的表达受到明显的抑制(P < 0.001、P < 0.05、P < 0.01)[图 4(a)~(c)]。在成骨分化7d时检测ALP、OCN和OPN的表达,结果与前面的三个基因一致,过表达miR-155显著降低了它们的表达(P < 0.05、P < 0.001、P < 0.01),而降低内源性miR-155的表达后,miR-155对以上成骨分化相关基因的抑制作用消失[图 4(d)~(f)]。
|
| 图 4 过表达miR-155抑制BMP9诱导的成骨分化相关因子的表达 Figure 4 Overexpressed miR-155 decreased the expression of osteogenesis related genes induced by BMP9 Tested the expression of Runx2(a), OSX(b) and COL1A1(c) used qPCR *P < 0.05, **P < 0.01, ***P < 0.001 compared with BMP9+NC group. Tested the expression of ALP(d), OCN(e) and OPN(f) by qPCR *P < 0.05, **P < 0.01, ***P < 0.001 compared with BMP9 +NC group |
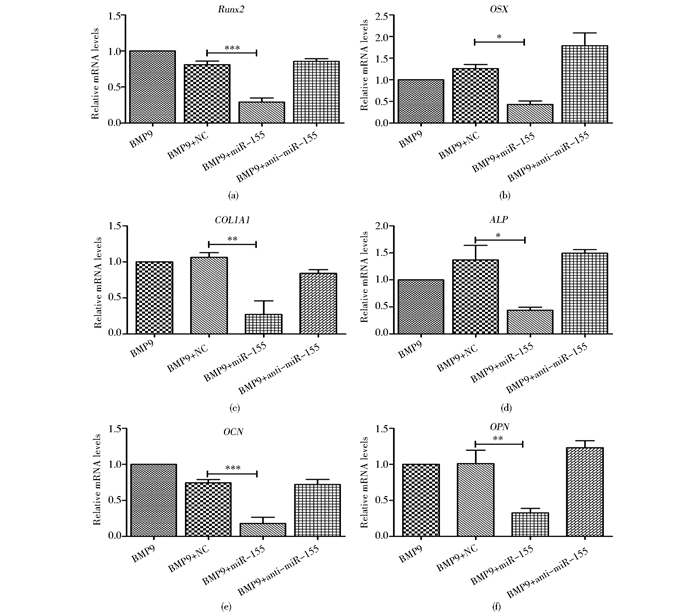
在BMP9诱导C3H10T1/2细胞成骨分化3d时,Western blot检测蛋白的表达水平,结果显示,与对照组BMP9+NC相比,过表达miR-155显著抑制Runx2和p-Smad1/5/8蛋白的表达水平(P < 0.01、P < 0.001);相反,抑制miR-155的表达则能促进Runx2的表达(P < 0.001),而对p-Smad1/5/8的表达无影响[图 5(a)]。在成骨诱导的7d时检测OCN和OPN的表达,结果显示,过表达miR-155减少了OCN和OPN蛋白的表达水平(P < 0.05、P < 0.001);而抑制miR-155的表达则增加了OCN和OPN蛋白的表达水平(P < 0.001、P < 0.05)[图 5(b)]。
|
| 图 5 过表达miR-155抑制p-Smad1/5/8以及OCN和OPN的表达 Figure 5 Overexpressed miR-155 decreased the expression of p-Smad1/5/8, OCN and OPN |
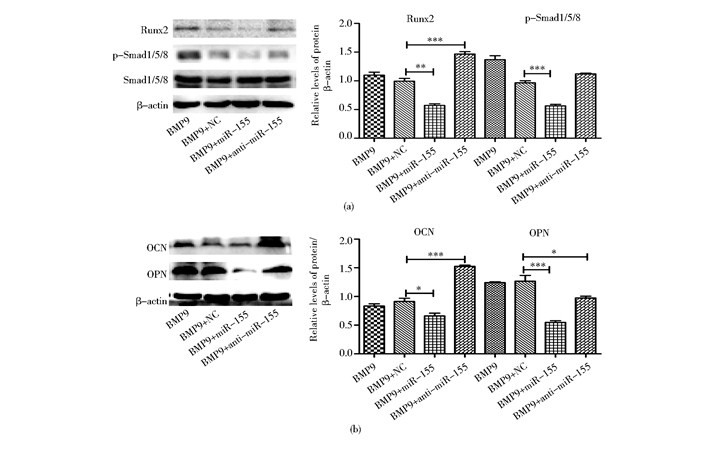
qPCR检测HIF1α mRNA水平的表达,结果显示,在成骨分化过程中,过表达miR-155明显抑制了HIF1α的表达(P < 0.01),而下调miR-155的表达则能逆转其抑制作用[图 6(a)]。qPCR检测HIF1α下游靶基因VEGF mRNA的表达水平,结果同HIF1α的结果一致(P < 0.001)[图 6(b)]。Western blot检测结果与qPCR结果一致,过表达miR-155能够显著下调HIF1α和VEGF蛋白的表达水平(P < 0.001),同时,抑制内源性miR-155的表达则可以使其抑制作用消失[图 6(c)]。
|
| 图 6 过表达miR-155抑制HIF1α和VEGF mRNA蛋白的表达水平 Figure 6 Overexpressed miR-155 repressed the expression of HIF1α and VEGF on mRNA and protein level |
转染miR-155进入C3H10T1/2细胞,qPCR检测其转染效率,miR-155表达显著升高(P < 0.001),证明转染成功[图 7(a)]。qPCR检测HIF1α mRNA的表达水平,结果显示与对照组NC相比,过表达miR-155组和抑制miR-155组HIF1α的表达均无明显差异[图 7(b)]。Western blot检测其蛋白质表达水平,结果显示与对照组相比,miR-155抑制了HIF1α蛋白的表达水平(P < 0.001),而抑制miR-155的表达则对HIF1α蛋白的表达水平无影响[图 7(c)]。靶基因预测软件找到miR-155与HIF1α的结合位点,并对其位点进行突变[图 7(d)],得到野生型和突变型结合序列。荧光素酶活性检测结果显示,miR-155显著抑制HIF1α 3′-UTR荧光素酶活性(P < 0.001),而突变了结合位点后,其抑制作用消失[图 7(e)]。

|
| 图 7 miR-155直接靶向HIF1α Figure 7 miR-155 targets HIF1α directly |
骨骼的重塑是伴随个体一生的连续的过程,这个重塑过程需要骨的重吸收和骨生成相互协调,从而取代损伤的骨或者满足机体代谢的需要[22]。骨重建,或者骨转换是由成骨细胞和破骨细胞在数量和活性上保持微妙的平衡来调控的。很多重要的信号分子参与调控间充质干细胞分化为成骨细胞,BMP信号通路在促进成骨细胞分化以及骨生成中有很显著的作用[23]。研究发现,除了传统的BMP2、BMP7之外,BMP9诱导MSC成骨分化能力最强,但其具体的调控机制还不是特别清楚,仍需要对其进行深入研究以促进其临床使用。目前,一些研究集中在BMP信号通路能够调控miRNA,并以此作为研究miRNA在成骨细胞中的作用方式[24-25]。从这些研究中可以看出,这类BMP调控的miRNA在调节成骨细胞的分化中有很重要的作用。
前期课题组用BMP9诱导小鼠间充质干细胞C3H10T1/2细胞成骨分化过程中,用micro芯片筛选出三个差异表达的miRNA,又用qPCR对芯片结果进行验证,其中就包括miR-155这个差异表达的miRNA。在本实验前期结果显示,过表达miR-155抑制BMP9诱导的C3H10T1/2细胞的ALP活性、ALP染色及钙盐沉积,且过表达miR-155降低了成骨分化相关基因如Runx2、OSX、COL1A1、ALP、OCN和OPN mRNA水平的表达,并且从蛋白质表达水平检测到其对早期成骨标志物Runx2有显著的抑制作用,对晚期成骨标志物OCN和OPN也有明显的抑制作用。为进一步探讨miR-155抑制BMP9所诱导的成骨分化的作用机制,我们通过实验发现过表达miR-155能够抑制p-Smad1/5/8蛋白的表达,表明miR-155能抑制Smad/BMP信号的活化。已有研究证明HIF1α能够增强BMP9诱导间充质干细胞成骨分化的能力,结合生物信息学分析,我们进一步探讨在BMP9诱导C3H10T1/2细胞成骨分化过程中,miR-155是否对HIF1α的表达有影响。本实验研究证明,在BMP9诱导C3H10T1/2细胞成骨分化过程中,过表达miR-155明显抑制HIF1α和其下游靶基因VEGF mRNA和蛋白质水平的表达,并通过荧光素酶报告基因实验证实了HIF1α是miR-155的一个靶基因。结合此部分实验与相关文献显示,miR-155也可能是通过抑制了HIF1α的表达而减弱了BMP9诱导间充质干细胞成骨分化的能力来发挥其在整个过程中的抑制作用。总而言之,在BMP9所诱导的C3H10T1/2细胞成骨分化过程中,miR-155作为一个负调节因子,可能是通过抑制Smad/BMP信号通路发挥作用,也可能是通过直接抑制其靶基因HIF1α的表达,而使HIF1α促进BMP9诱导成骨分化的能力减弱来发挥作用。而在后期的研究中,我们可以研究miR-155这两个抑制间充质干细胞成骨分化机制的相互关系,它们是简单的相加关系还是具有协同作用。本课题研究将为探讨BMP调控的miRNA在调节成骨细胞的分化中的重要作用提供基础,并为BMP9诱导的间充质干细胞向成骨分化的全面调控机制研究提供理论和实验依据。
| [1] | Brown M A, Zhao Q, Baker K A, et al. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. The Journal of Biological Chemistry, 2005, 280(26) : 25111–25118. DOI:10.1074/jbc.M503328200 |
| [2] | Zuo B, Zhu J, Li J, et al. microRNA-103a functions as a mechanosensitive microRNA to inhibit boneformation through targeting Runx2. Journal of Bone and Mineral Research:The Official Journal of the American Society for Bone and Mineral Research, 2015, 30(2) : 330–345. DOI:10.1002/jbmr.2352 |
| [3] | Zeng Y, Qu X, Li H, et al. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Letters, 2012, 586(16) : 2375–2381. DOI:10.1016/j.febslet.2012.05.049 |
| [4] | Taipaleenmaki H, Bjerre Hokland L, Chen L, et al. Mechanisms in endocrinology:micro-RNAs:targets for enhancing osteoblast differentiation and bone formation. European Journal of Endocrinology/European Federation of Endocrine Societies, 2012, 166(3) : 359–371. DOI:10.1530/EJE-11-0646 |
| [5] | Li Y, Kowdley K V. MicroRNAs in common human diseases. Genomics, Proteomics & Bioinformatics, 2012, 10(5) : 246–253. |
| [6] | Ivey K N, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell, 2010, 7(1) : 36–41. DOI:10.1016/j.stem.2010.06.012 |
| [7] | Itoh T, Takeda S, Akao Y. MicroRNA-208 modulates BMP-2-stimulated mouse preosteoblast differentiation by directly targeting V-ets erythroblastosis virus E26 oncogene homolog 1. The Journal of Biological Chemistry, 2010, 285(36) : 27745–27752. DOI:10.1074/jbc.M110.105080 |
| [8] | Hwang H W, Mendell J T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. British Journal of Cancer, 2006, 94(6) : 776–780. DOI:10.1038/sj.bjc.6603023 |
| [9] | Faraoni I, Antonetti F R, Cardone J, et al. miR-155 gene:a typical multifunctional microRNA. Biochimica et Biophysica Acta, 2009, 1792(6) : 497–505. DOI:10.1016/j.bbadis.2009.02.013 |
| [10] | Du F, Yu F, Wang Y, et al. MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice. Arteriosclerosis, Thrombosis, and Vascular Biology, 2014, 34(4) : 759–767. DOI:10.1161/ATVBAHA.113.302701 |
| [11] | Vigorito E, Perks K L, Abreu-Goodger C, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity, 2007, 27(6) : 847–859. DOI:10.1016/j.immuni.2007.10.009 |
| [12] | O'Connell R M, Kahn D, Gibson W S, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity, 2010, 33(4) : 607–619. DOI:10.1016/j.immuni.2010.09.009 |
| [13] | Nazari-Jahantigh M, Wei Y, Noels H, et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. The Journal of Clinical Investigation, 2012, 122(11) : 4190–4202. DOI:10.1172/JCI61716 |
| [14] | Wang M, Tan L P, Dijkstra M K, et al. miRNA analysis in B-cell chronic lymphocytic leukaemia:proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. The Journal of Pathology, 2008, 215(1) : 13–20. DOI:10.1002/path.v215:1 |
| [15] | Lawrie C H. MicroRNAs and lymphomagenesis:a functional review. British Journal of Haematology, 2013, 160(5) : 571–581. DOI:10.1111/bjh.2013.160.issue-5 |
| [16] | Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(18) : 7024–7029. DOI:10.1073/pnas.0602266103 |
| [17] | Yin Q, Wang X, Fewell C, et al. MicroRNA miR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation. Journal of Virology, 2010, 84(13) : 6318–6327. DOI:10.1128/JVI.00635-10 |
| [18] | Yanaihara N, Caplen N, Bowman E, et al. microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell, 2006, 9(3) : 189–198. DOI:10.1016/j.ccr.2006.01.025 |
| [19] | Volinia S, Galasso M, Costinean S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Research, 2010, 20(5) : 589–599. DOI:10.1101/gr.098046.109 |
| [20] | Jiang S, Zhang H W, Lu M H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Research, 2010, 70(8) : 3119–3127. DOI:10.1158/0008-5472.CAN-09-4250 |
| [21] | Omrane I, Benammar-Elgaaied A. The immune microenvironment of the colorectal tumor:Involvement of immunity genes and microRNAs belonging to the TH17 pathway. Biochimica et Biophysica Acta, 2015, 1856(1) : 28–38. |
| [22] | Kristina K, Anne M D. MicroRNA biogenesis and regulation of bone. Arthritis Research & Therapy, 2011, 13(3) : 220. |
| [23] | Wang R N, Green J, Wang Z, et al. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes & diseases, 2014, 1(1) : 87–105. |
| [24] | Inose H, Ochi H, Kimura A, et al. microRNA regulatory mechanism of osteoblast differentiation. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(49) : 20794–20799. DOI:10.1073/pnas.0909311106 |
| [25] | Li Z, Hassan M Q, Volinia S, et al. microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(37) : 13906–13911. DOI:10.1073/pnas.0804438105 |
 2017, Vol. 37
2017, Vol. 37