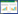文章信息
- 夏乾竣, 王飞, 李迅.
- XIA Qian-jun, WANG Fei, LI Xun.
- 解脂耶罗维亚酵母产油脂的研究进展
- Review of Yarrowia lipolytica for SCO Production
- 中国生物工程杂志, 2017, 37(3): 99-105
- China Biotechnology, 2017, 37(3): 99-105
- http://dx.doi.org/DOI:10.13523/j.cb.20170314
-
文章历史
- 收稿日期: 2016-10-17
- 修回日期: 2016-11-20
解脂耶罗维亚酵母是一种非常规酵母,因其体内有着复杂的机制能够代谢多种疏水碳源,所以在自然界中主要分布在富含酯类或者蛋白质的疏水环境中。解脂耶罗维亚酵母于1942年首次被分离得到,最初命名为Candida lipolytica,经多次更名最终定名为Yarrowia lipolytica [1-2]。野生型的解脂耶罗维亚酵母体内油脂含量能够达到菌体干重的36%,作为一种产油酵母有着很大的发展潜力[3]。在解脂耶罗维亚酵母之前,酿酒酵母已经作为微生物油脂代谢的模型被研究了多年。虽然人们对酿酒酵母的油脂代谢系统有了比较完备的认识[4],但是酿酒酵母并非产油酵母,其油脂含量最多只能达到菌体干重的15%。与酿酒酵母不同,解脂耶罗维亚酵母油脂含量高,并且其中90%以上为甘油三酯 (TAG),少部分以游离的脂肪酸形式存在[5]。甘油三酯是生物柴油生产的优良原料,随着石油资源的日益枯竭,以产油微生物的单细胞油脂生产生物柴油受到越来越多的关注[6]。解脂耶罗维亚酵母是目前为止唯一一个完成全基因测序的产油微生物,经过多年的研究用于解脂耶罗维亚酵母基因操作的手段已经相当完善[7-8]。在了解其遗传代谢的基础上,通过对油脂代谢途径的改造,多株油脂含量更高的工业化菌株被构建。本文就解脂耶罗维亚酵母的表达系统及油脂代谢途径的改造进行阐述。
1 产油微生物的介绍我们把油脂含量高于20%的微生物称为产油微生物[9]。目前主要的产油微生物有产油细菌、产油酵母、产油霉菌及产油微藻。产油细菌的油脂成分比较复杂,并且其体内油脂主要分布在细胞外膜上,提取困难。只有极少数的产油细菌能产生甘油三酯:在以乙酸为碳源时卡氏菌属油脂含量能达到菌体干重的49.7%;浑浊红球菌在以高浓度葡萄糖为碳源时,油脂含量达到菌体干重的38%;天蓝色链霉菌体内油脂的含量能超过菌体干重的80%[10-12]。产油微藻的优点是培养基中无需碳源能够直接吸收空气中的CO2作为碳源,缺点是生长缓慢,细胞密度低。其中绿藻和硅藻被认为是最具工业潜力的两类产油微藻。目前研究最多的是绿藻中的小球藻,通过基因手段构建的菌株含油量能达到51.8%[13]。产油酵母和产油霉菌具有发酵密度大、生长速率快等优点。现在研究较多的产油霉菌有深黄被孢霉、土曲霉、高山被孢霉等。产油酵母有斯达氏油脂酵母、圆红冬孢酵母、黏红酵母、解脂耶罗维亚酵母等[14-17]。其中解脂耶罗维亚酵母由于其产油量高,且能利用烷烃、甘油、废弃油脂等廉价的碳源来生产油脂,已经替代了酿酒酵母作为油脂代谢的模型微生物被研究。
2 解脂耶罗维亚酵母表达系统随着解脂耶罗维亚酵母全基因组测序的完成,其遗传背景日益清晰。自20世纪80年代起,解脂耶罗维亚酵母的表达系统被逐渐开发[18-21]。下面将从质粒和宿主菌两方面进行介绍。
2.1 质粒在解脂耶罗维亚酵母表达系统发展的初期,研究者开发了一系列的自主复制型质粒。此质粒主要由ARS和CEN两个原件构成[22]。不过该质粒在表达时需要外在选择性压力,表达的基因也有限,无法在工业上得到应用,为此又开发了整合型质粒。XPR2启动子在整合型质粒发展史上起着关键作用。XPR2在酵母内是起着启动碱性蛋白酶AEP的作用,但其启动外源蛋白的表达时也只在pH超过5.5时起作用,并且受到蛋白胨的诱导[23]。法国Catherine Madzak教授实验室受此启发,开发了pINA系列质粒,此表达质粒已用于商业化出售。pINA系列质粒采用的是hp4d启动子,hp4d启动子是由4个pXPR2上游的激活序列UAS1B串联,再加上最精简的LEU2启动子 (精简到只有TATA box) 杂交形成的组成型强启动子。hp4d启动子为生长依赖型启动子,无需诱导剂的诱导,受环境影响极小,在几乎所有的培养基中都能使用。外源蛋白将在菌体生长的稳定期的前期表达,一定程度上避免外源蛋白在菌体生长初期对细胞的毒害[24]。迄今为止已有近百种异源蛋白利用hp4d启动子在解脂耶罗维亚酵母中成功表达。
对于分泌到细胞外的蛋白质以及膜蛋白需要信号肽。异源信号肽难以在解脂耶罗维亚酵母中得到有效地应用,pINA系列质粒中主要有三种信号肽:XPR2 pre、XPR2 prepro、LIP2 prepro。这三种信号肽均来自于解脂耶罗维亚酵母本身,XPR2是碱性蛋白和酸性蛋白的分泌信号肽,LIP2 prepro是其脂肪酶的分泌信号肽。信号肽将在特定的XA、XP、KR位点进行切除从而表达出成熟完整的蛋白质[25]。常用pINA系列质粒如表 1所示[26-27]。
| 质粒 | 信号肽 | 选择标记 | 整合位点 |
| pBR为基础的单一整合型载体 | |||
| pINA1269 | 无 | LEU2 | pBR |
| pINA1296 | XPR2 pre | LEU2 | pBR |
| pINA1267 | XPR2 prepro | LEU2 | pBR |
| 单拷贝自复制载体 | |||
| pINA1311 | 无 | ura3d1 | Zeta |
| pINA1312 | 无 | ura3d1 | Zeta |
| pINA1313 | LIP2 prepro | ura3d1 | Zeta |
| pINA1317 | XPR2 prepro | ura3d1 | Zeta |
| pINA1314 | XPR2 pre | ura3d1 | Zeta |
| 多拷贝自复制载体 | |||
| pINA1291 | 无 | ura3d4 | Zeta |
| pINA1292 | 无 | ura3d4 | Zeta |
| pINA1293 | LIP2 prepro | ura3d4 | Zeta |
| pINA1294 | XPR2 prepro | ura3d4 | Zeta |
| pINA1297 | XPR2 pre | ura3d4 | Zeta |
为了增加质粒的整合效率,在质粒上增加了Zeta序列。该序列在酵母基因组DNA中为质粒提供了至少100个潜在的整合位点,极大提高了质粒的整合效率[28]。目标基因进入宿主染色体后,可以随宿主的DNA一起复制,遗传120代后无质粒丢失[29]。除了可使用Zeta序列外,pINA质粒还利用了pBR322序列作为整合位点,当使用pBR322序列为整合位点的表达质粒时,对应的宿主菌也应做相应的改造。
2.2 宿主菌解脂耶罗维亚酵母作为真核生物,氨苄青霉素、卡纳霉素等抗生素对其均不起作用,所以对酵母一般采用营养缺陷型作为筛选标记,通过基因改造可以得到所需要的宿主菌。Po1d (Po1d、Po1e、Po1f、Po1g、Po1h) 系列是在解脂耶罗维亚酵母的研究中应用最广泛的宿主系统。Po1d是由野生型解脂耶罗维亚酵母ATCC20460改造得到的,将菌体内对外源蛋白的表达有影响的碱性蛋白酶基因AEP敲除,减少对外源蛋白的降解,使其能够更高效地被表达[19]。并在Po1d中整合了Suc基因,使其能利用蔗糖作为底物,这一特点推动了其在工业上的应用。在Po1d的基础上Nicaud等[26]对其进行了改造,敲除了酸性蛋白基因AXP得到Po1f,彻底消除了胞外蛋白酶对外源蛋白表达时的不利影响。Po1e、Po1g被改造为适合以pBR322为基础的质粒整合菌系,在提高质粒的整合效率的同时使宿主菌具有不同的筛选标记。改造方式如图 1所示。
|
| 图 1 Po1d系列宿主菌改造图 Figure 1 Transformation diagram of Po1d series host bacteria |
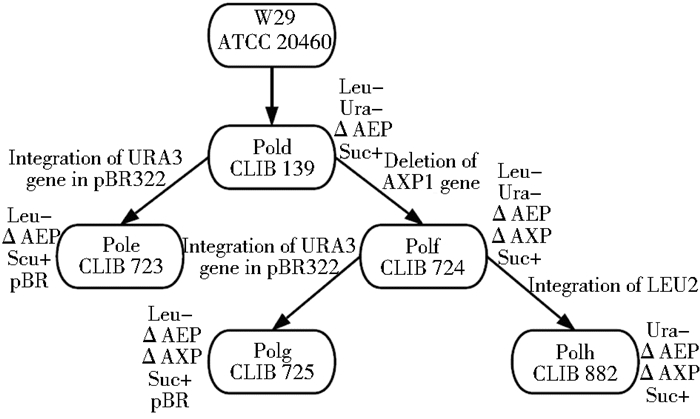
在产油微生物的生长过程中有多种因素会影响微生物细胞内油脂的累积。其中氮源的影响被认为是最关键的因素。一般认为在酵母培养过程中碳源被用来合成细胞内的糖类、酯类、核酸和蛋白质,而氮源仅仅用来合成核酸和蛋白质等细胞增殖必需的成分。所以在培养基中氮源缺乏时,微生物细胞的增殖速率迅速降低,过量的碳源通过油脂累积途径以甘油三酯的形式在细胞中累积。具体油脂累积途径如图 2所示。当微生物细胞内氮源缺乏时,腺嘌呤核糖核苷酸 (AMP) 脱氨酶活力增强,导致细胞内AMP含量下降,受AMP浓度影响的异柠檬酸脱氢酶 (ICDH) 活力下降,造成细胞内异柠檬酸 (isocitrate) 的累积[30-31]。异柠檬酸接着转化为柠檬酸 (citrate),由转运酶转运到细胞质中作为乙酰辅酶A (Acetyl-CoA) 合成的前体。当细胞内油脂累积达到极限时,细胞会分泌大量的柠檬酸,此时细胞油脂累积时期完毕进入柠檬酸生产时期[32]。控制培养基中氮源的含量,并将细胞生长控制在油脂的累积期是生产单细胞油脂的关键。目前的主要方法是改变氮源的添加方式,优化工艺条件来获得最高的油脂产率。Alexandre等[33]控制培养基中碳氮比为35:1得到油脂产率为12%。Aggelis和Komaitis[34]采用连续添加氮源的方式,当氮源添加量为0.032mol/h时,油脂产率达到了25%。
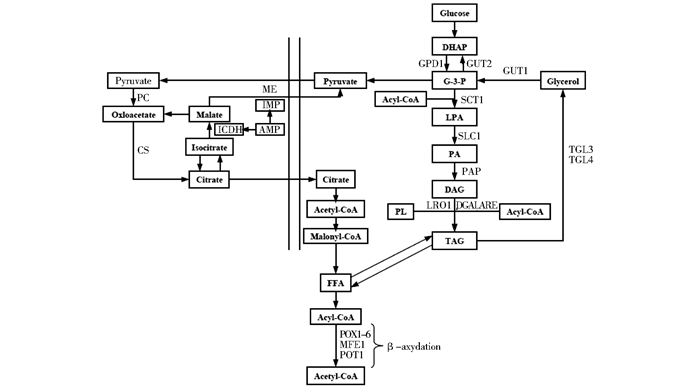
|
| 图 2 解脂耶罗维亚酵母油脂代谢途径 Figure 2 Lipid metabolism for Yarrowia lipolytica |
细胞内的柠檬酸在柠檬酸裂解酶 (ACL) 作用下生成乙酰辅酶A,乙酰辅酶A在油脂累积途径中参与了多步反应[35]。乙酰辅酶A在酰基辅酶A羧化酶 (ACC) 的作用下生成丙二酰辅酶A,丙二酰辅酶A作为前体,和多个乙酰辅酶A通过链增长反应生成脂肪酸。每一步的链增长反应需要两分子的NADPH,NADPH主要是由苹果酸脱氢酶 (ME) 产生。所以ACL、ACC、ME在油脂累积途径中起了极其重要的作用。具体反应如下[36]:
Citrate+HS-CoA+ATP→acetyl-CoA+oxaloacetate+ADP+Pi
Acetyl-CoA+HCO3-+ATP→malonyl-CoA+ADP+Pi
Malate+NADP+→pyruvate+NADPH
解脂耶罗维亚酵母合成的脂肪酸主要以甘油三酯形式存在于疏水小体-脂质体中,而没有进入细胞膜的磷脂双分子层中。甘油三酯的合成遵循Kennedy[37]途径,以由糖酵解途径生成的3-磷酸-甘油 (G-3-P) 为前体进行后续合成。3-磷酸-甘油在转酰酶 (SCT1) 和乙酰辅酶A的作用下生成溶血磷脂酸 (LPA),溶血磷脂酸接着在溶血磷脂酸转酰酶 (SLC1) 的作用下生成磷脂酸 (PA)。磷脂酸在磷酸水解酶 (PAP) 的脱磷酸作用下生成甘油二酯 (DAG)。甘油二酯通过酰基化反应生成最终产物甘油三酯。其中磷脂二酰基甘油酰基转移酶 (LRO1) 能将磷脂质 (LP) 中的酰基转移到甘油二酯中[38-39]。乙酰辅酶A作为酰基供体时,二酰基甘油酰基转移酶 (DGAT)、酰基辅酶A胆固醇酰基转移酶 (Are1、Are2) 起着催化作用,将乙酰辅酶A中的酰基转移至甘油二酯中生成甘油三酯。
同时细胞体内累积的TAG会发生水解,生成甘油和脂肪酸,为细胞膜的合成提供原料,也为细胞的生长提供能量[40]。在解脂耶罗维亚酵母中控制甘油三酯水解的关键酶为三酰甘油脂肪酶 (TGL3、TGL4)。游离的脂肪酸部分用来合成细胞膜,多余的会通过氧化反应多步降解成乙酰辅酶A。反应的第一步是在酰基辅酶氧化酶 (POX1-6) 的作用下,酰基辅酶A的2位置形成双键。多功能氧化酶 (MFE) 参与了后续的两步反应,这个蛋白质含有两个结构域,同时具有2-烯酰辅酶A水合酶和3-羟基辅酶A脱氢酶的作用。不饱和的酰基辅酶A在MFE的作用下先生成了3-羟烷基辅酶A,随后转化为3-酮酰基辅酶A。3-酮酰基辅酶A在裂解酶POT1的作用下变成了乙酰辅酶A[41]。油脂累积途径中各个基因的具体信息见表 2 [42]。
| 基因 | GB登陆号 | GeneID | 功能 |
| ME | YALI0E18634g | 50553401 | 苹果酸酶 |
| ACL1 | YALI0E34793g | 50554756 | ATP柠檬酸裂解酶,a族 |
| ACL2 | YALI0D24431g | 50551514 | ATP柠檬酸裂解酶,b族 |
| ACC1 | YALI0C11407g | 50548502 | 乙酰辅酶A羧化酶 |
| POX1 | YALI0E32835g | 50554588 | 酰基辅酶A氧化酶 |
| POX2 | YALI0F10857g | 50555711 | 酰基辅酶A氧化酶 |
| POX3 | YALI0D24750g | 50551538 | 酰基辅酶A氧化酶 |
| POX4 | YALI0E27654g | 50554132 | 酰基辅酶A氧化酶 |
| POX5 | YALI0C23859g | 50549456 | 酰基辅酶A氧化酶 |
| POX6 | YALI0E06567g | 50552443 | 酰基辅酶A氧化酶 |
| MFE1 | YALI0E15378g | 50553139 | 多功能氧化酶 |
| POT1 | YALI0E18568g | 50553395 | 过氧化物酶体酰基硫解酶 |
| GUT2 | YALI0B13970g | 50546784 | 3-磷酸甘油脱氢酶 |
| GPD1 | YALI0B02948g | 50545810 | 3-磷酸甘油脱氢酶 |
| SCT1 | YALI0C00209g | 50547610 | 3-磷酸甘油酰基转移酶 |
| SLC1 | YALI0E18964g | 50553431 | 3-磷酸甘油-1-酰基转移酶 |
| PAP | YALI0D27016g | 50551736 | 磷脂酸磷酸水解酶 |
| LRO1 | YALI0E16797g | 50553255 | 磷脂二酰基甘油酰基转移酶 |
| DGAT | YALI0E32769g | 50554582 | 二酰基甘油酰基转移酶 |
| ARE1 | YALI0F06578g | 50555354 | 酰基辅酶A胆固醇酰基转移酶 |
| ARE2 | YALI0D07986g | 50550168 | 酰基辅酶A胆固醇酰基转移酶 |
| TGL3 | YALI0D17534g | 50550940 | 三酰甘油脂肪酶 |
| TGL4 | YALI0F10010g | 50555643 | 三酰甘油脂肪酶 |
| GUT1 | YALI0F00484g | 50554824 | 甘油激酶 |
对表 2中的基因进行调控,可增强油脂累积途径向甘油三酯转化,目前已有多名学者对其中的一些关键基因进行了调控,得到了胞内油脂含量大幅度提高的菌株。张怀远等通过过表达来自小鼠的柠檬酸裂解酶ACL使酵母中油脂的累积量提高了50%~200%;Tai通过过表达耶氏酵母中的ACC1使油脂的含量提高了2倍,过表达DGAT使油脂含量提高了3倍;Beopoulos等通过对基因组中GUT2基因的敲除使酵母中油脂的含量提高了2倍。对解脂耶罗维亚酵母油脂累积途径具体基因的改造见表 3。
| 基因 | 改造方式 | 油脂变化 | 参考文献 |
| ME | 过表达 | 无变化 | [43] |
| ACL | 过表达 | 提高50%~200% | [44] |
| ACC1 | 过表达 | 提高2倍 | [45] |
| POX1-6 | 敲除 | 提高34% | [46] |
| POX1-6, MFE1, GUT2, GPD1 | POX1-6, MFE1, GUT2敲除GPD1过表达 | 含量增加 | [47] |
| GUT2 | 基因敲除 | 提高2倍 | [48] |
| GPD1 | 过表达 | 提高50% | [47] |
| GPD1, GUT2 | GPD1过表达,GUT2敲除 | 提高4.6倍 | [47] |
| LRO1 | 基因敲除 | 下降40% | [49] |
| DGAT | 过表达 | 提高3倍 | [45] |
| ACC1, DGAT | ACC1和DGAT共表达 | 提高4倍 | [45] |
| TGL3, TGL4 | TGL3和TGL4敲除 | 含量增加 | [47] |
| SLC1 | 过表达 | 提高99.91%~151% | [50] |
| AMPD, ACL1, ACL2, ME, DGAT | 过表达 | 油脂含量达到90% | [51] |
4 总结与展望
在化石燃料日益枯竭的今天,寻找化石燃料合适的替代品受到广泛的关注。其中以产油微生物的单细胞油脂作为生产生物柴油的原料最具发展潜力。解脂耶罗维亚酵母因其基因改造可操作性强,通过大量的改造使得单细胞油脂的产量逐步提高。现阶段对解脂耶罗维亚酵母产油脂的研究,主要集中在对其油脂累积过程的调控上。目前构建出的解脂耶罗维亚酵母,通过对其培养条件的优化油脂含量达到90%,产量达25g/L[51]。不过由于单细胞油脂的提取相对繁杂,以其为原料生产生物柴油相对于化石能源未建立明显优势。在未来通过代谢工程和生物工程等手段实现解脂耶罗维亚酵母胞内油脂的分泌,对单细胞油脂的生产具有重要意义。
由于解脂耶罗维亚酵母是美国食品药品管理局 (GRAS) 认定的最安全的微生物,所以可以将油脂的生产和食品加工相结合。在累积油脂的同时增加其中多不饱和脂肪酸,如亚油酸、共轭亚油酸、亚麻酸的含量。目前本实验室已构建出亚油酸含量上升50%的菌株,未来可以通过基因工程的进一步改造,使解脂耶罗维亚酵母细胞内的多不饱和脂肪酸含量进一步上升。这些菌株在食品饲料行业中有广泛的应用前景。
| [1] | Barth C, Gaillardin C. Yarrowia lipolytica Nonoconventional Yeasts in Biotechnology. Heidelberg: Springer-Verlag, 1996: 313-388. |
| [2] | Nicaud J M. Yarrowia lipolytica. Yeast, 2012, 29(10) : 409–418. DOI:10.1002/yea.v29.10 |
| [3] | Ratledge C, Wynn J P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Advances in Applied Microbiology, 2002, 51(1-44) : 1–51. |
| [4] | Czabany T, Athenstaedt K, Daum G. Synthesis, storage and degradation of neutral lipids in yeast. Biochimica Et Biophysica Acta, 2007, 1771(3) : 299–309. DOI:10.1016/j.bbalip.2006.07.001 |
| [5] | Beopoulos A, Cescut J, Haddouche R, et al. Yarrowia lipolytica as a model for bio-oil production. Progress in Lipid Research, 2009, 48(6) : 375–387. DOI:10.1016/j.plipres.2009.08.005 |
| [6] | Huang C, Chen X F, Xiong L, et al. Single cell oil production from low-cost substrates:The possibility and potential of its industrialization. Biotechnology Advances, 2013, 31(2) : 129–139. DOI:10.1016/j.biotechadv.2012.08.010 |
| [7] | 赵鹤云, 黄瑛, 杨江科, 等. 解脂耶氏酵母表达系统研究进展. 生物加工过程, 2008, 6(3) : 10–16. Zhao H Y, Huang Y, Yang J K, et al. Review of Yarrowia lipolytica expression system. Chinese Journal of Bioprocess Engineering, 2008, 6(3) : 10–16. |
| [8] | Kerscher S, Durstewitz G, Casaregola S, et al. The complete mitochondrial genome of Yarrowia lipolytica. Comparative & Functional Genomics,, 2001, 2(2) : 80–90. |
| [9] | Ratledge C, Wynn J P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Advances in Applied Microbiology, 2002, 51(1-44) : 1–51. |
| [10] | Alvarez H M, Souto M F, Viale A, et al. Biosynthesis of fatty acids and triacylglycerols by 2, 6, 10, 14-tetramethyl pentadecane-grown cells of Nocardia globerula 432. Fems Microbiology Letters, 2001, 200(2) : 195–200. DOI:10.1111/fml.2001.200.issue-2 |
| [11] | Kurosawa K, Boccazzi P, de Almeida N M, et al. High-cell-density batch fermentation of Rhodococcus opacus PD630 using a high glucose concentration for triacylglycerol production. Journal of Biotechnology, 2010, 147(3-4) : 212–218. DOI:10.1016/j.jbiotec.2010.04.003 |
| [12] | Arabolaza A, Rodriguez E, Altabe S, et al. Multiple pathways for triacylglycerol biosynthesis in Streptomyces coelicolor. Applied & Environmental Microbiology, 2008, 74(9) : 2573–2582. |
| [13] | Arora N, Patel A, Pruthi P A, et al. Synergistic dynamics of nitrogen and phosphorous influences lipid productivity in Chlorella minutissima for biodiesel production. Bioresource Technology, 2016, 213 : 79–87. DOI:10.1016/j.biortech.2016.02.112 |
| [14] | Mei L, Liu G L, Zhe C, et al. Single cell oil production from hydrolysate of cassava starch by marine-derived yeast Rhodotorula mucilaginosa TJY15a. Biomass & Bioenergy, 2010, 34(1) : 101–107. |
| [15] | Li Y, Zhao Z, Bai F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme & Microbial Technology, 2007, 41(3) : 312–317. |
| [16] | Wang Z P, Xu H M, Wang G Y, et al. Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2013, 1831(4) : 675–682. |
| [17] | Zhao X, Kong X, Hua Y, et al. Medium optimization for lipid production through co-fermentation of glucose and xylose by the oleaginous yeast Lipomyces starkeyi. European Journal of Lipid Science & Technology, 2008, 110(5) : 405–412. |
| [18] | Nicaud J M, Fabre E, Gaillardin C. Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Current Genetics, 1989, 16(16) : 253–260. |
| [19] | Nicaud J M, Fournier P, Bonnardière C L, et al. Use of ARS18 based vectors to increase protein production in Yarrowia lipolytica. Journal of Biotechnology, 1991, 19(2-3) : 259–270. DOI:10.1016/0168-1656(91)90063-2 |
| [20] | Hamsa P V, Chattoo B B. Cloning and growth-regulated expression of the gene encoding the hepatitis B virus middle surface antigen in Yarrowia lipolytica. Gene, 1994, 143(2) : 165–170. DOI:10.1016/0378-1119(94)90092-2 |
| [21] | Matsuoka M, Matsubara M, Daidoh H, et al. Analysis of regions essential for the function of chromosomal replicator sequences from Yarrowia lipolytica. Molecular & General Genetics:MGG, 1993, 237(3) : 327–333. |
| [22] | Müller S, Sandal T, Kamp-Hansen P, et al. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Yeast, 1998, 14(14) : 1267–1283. DOI:10.1002/(ISSN)1097-0061 |
| [23] | Madzak C, Gaillardin C, Beckerich J M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica:a review. Journal of Biotechnology, 2004, 109(1-2) : 63–81. DOI:10.1016/j.jbiotec.2003.10.027 |
| [24] | Matoba S, Ogrydziak D M. A novel location for dipeptidyl aminopeptidase processing sites in the alkaline extracellular protease of Yarrowia lipolytica. Journal of Biological Chemistry, 1989, 264(11) : 6037–6043. |
| [25] | Madzak C, Tréton B, Blanchin-Roland S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. Journal of Molecular Microbiology & Biotechnology, 2000, 2(2) : 207–216. |
| [26] | Nicaud J M, Madzak C, Broek P V D, et al. Protein expression and secretion in the yeast Yarrowia lipolytica. Fems Yeast Research, 2002, 2(3) : 371–379. |
| [27] | Juretzek T, Le Dall M, Mauersberger S, et al. Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast, 2001, 18(2) : 97–113. DOI:10.1002/(ISSN)1097-0061 |
| [28] | Pignède G, Wang H J, Fudalej F, et al. Autocloning and amplification of LIP2 in Yarrowia lipolytica. Applied & Environmental Microbiology, 2000, 66(8) : 3283–3289. |
| [29] | Nicaud J M, Fabre E, Gaillardin C. Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Current Genetics, 1989, 16(16) : 253–260. |
| [30] | Tang W, Zhang S, Wang Q, et al. The isocitrate dehydrogenase gene of oleaginous yeast Lipomyces starkeyi is linked to lipid accumulation. Canadian Journal of Microbiology, 2009, 55(9) : 1062–1069. DOI:10.1139/W09-063 |
| [31] | Li X, Wang P, Ge Y, et al. NADP (+)-specific isocitrate dehydrogenase from oleaginous yeast Yarrowia lipolytica CLIB122:biochemical characterization and coenzyme sites evaluation. Applied Biochemistry & Biotechnology, 2013, 171(2) : 403–416. |
| [32] | Kamzolova S V, Vinokurova N G, Lunina J N, et al. Production of technical-grade sodium citrate from glycerol-containing biodiesel waste by Yarrowia lipolytica. Bioresource Technology, 2015, 193 : 250–255. DOI:10.1016/j.biortech.2015.06.092 |
| [33] | Alexandre B, Tristan R, François K, et al. High-throughput fermentation screening for the yeast Yarrowia lipolytica with real-time monitoring of biomass and lipid production. Microbial Cell Factories, 2016, 15(1) : 1–12. DOI:10.1186/s12934-015-0402-6 |
| [34] | Aggelis G, Komaitis M. Enhancement of single cell oil production by Yarrowia lipolytica growing in the presence of Teucrium polium L. aqueous extract. Biotechnology Letters, 1999, 21(9) : 747–749. DOI:10.1023/A:1005591127592 |
| [35] | Ratledge C. Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie, 2004, 86(11) : 807–815. DOI:10.1016/j.biochi.2004.09.017 |
| [36] | Beopoulos A, Nicaud J M, Gaillardin C. An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Applied Microbiology & Biotechnology, 2011, 90(4) : 1193–1206. |
| [37] | Kennedy E P. Biosynthesis of complex lipids. Federation Proceedings, 1961, 20 : 934–940. |
| [38] | Dahlqvist A, Stahl U, Lenman M, et al. Phospholipid:diacylglycerol acyltransferase:an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proceedings of the National Academy of Sciences, 2000, 97(12) : 6487–6492. DOI:10.1073/pnas.120067297 |
| [39] | Sandager L, Gustavsson MH, Ståhl U, et al. Storage lipid synthesis is non-essential in yeast. Journal of Biological Chemistry, 2002, 277(277) : 6478–6482. |
| [40] | Athenstaedt K, Daum G. The life cycle of neutral lipids:synthesis, storage and degradation. Cellular & Molecular Life Sciences Cmls, 2006, 63(12) : 1355–1369. |
| [41] | Yamagami S, Iida T, Nagata Y, et al. Isolation and characterization of acetoacetyl-CoA thiolase gene essential for n-decane assimilation in yeast Yarrowia lipolytica. Biochemical & Biophysical Research Communications, 2001, 282(3) : 832–838. |
| [42] | Fickers P, Fudalej F, Dall M T L, et al. Identification and characterisation of LIP7, and LIP8, genes encoding two extracellular triacylglycerol lipases in the yeast Yarrowia lipolytica. Fungal Genetics & Biology, 2005, 42(3) : 264–274. |
| [43] | Zhang H, Zhang L, Chen H, et al. Regulatory properties of malic enzyme in the oleaginous yeast Yarrowia lipolytica and its non-involvement in lipid accumulation. Biotechnology Letters, 2013, 35(12) : 2091–2098. DOI:10.1007/s10529-013-1302-7 |
| [44] | Zhang H, Zhang L, Chen H, et al. Enhanced lipid accumulation in the yeast Yarrowia lipolytica by over-expression of ATP:citrate lyase from Mus musculus. Journal of Biotechnology, 2014, 192(1) : 78–84. |
| [45] | Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metabolic Engineering, 2013, 15(1) : 1–9. |
| [46] | Mlickova K, Luo Y, D'Andrea S, et al. Acyl-CoA oxidase a key step for lipid accumulation in the yeast Yarrowia lipolytica. Journal of Molecular Catalysis B Enzymatic, 2004, 28(2-3) : 81–85. DOI:10.1016/j.molcatb.2004.01.007 |
| [47] | Dulermo T, Nicaud J M. Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metabolic Engineering, 2011, 13(5) : 482–491. DOI:10.1016/j.ymben.2011.05.002 |
| [48] | Beopoulos A, Mrozova Z, Thevenieau F, et al. Control of lipid accumulation in the yeast Yarrowia lipolytica. Applied & Environmental Microbiology, 2008, 74(24) : 7779–7789. |
| [49] | Beopoulos A, Chardot T, Nicaud J M. Yarrowia lipolytica:a model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie, 2009, 91(6) : 692–696. DOI:10.1016/j.biochi.2009.02.004 |
| [50] | Silverman A M, Qiao K J, Xu P, et al. Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica. Applied Microbiology & Biotechnology, 2016, 100(8) : 3781–3798. |
| [51] | Blazeck J, Hill A, Liu L, et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nature Communications, 2014, 5(1) : 49–168. |
 2017, Vol. 37
2017, Vol. 37