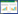文章信息
- 来亚鹏, 邓婷婷, 刘刚, 王娟.
- LAI Ya-peng, DENG Ting-ting, LIU Gang, WANG Juan.
- 同源过表达BglR对嗜热毁丝霉β-葡萄糖苷酶活性的影响
- The Influence of Homologous Overexpression of BglR on β-glucosidase Activities in Myceliophthora thermophila
- 中国生物工程杂志, 2017, 37(7): 64-71
- China Biotechnology, 2017, 37(7): 64-71
- http://dx.doi.org/DOI:10.13523/j.cb.20170712
-
文章历史
- 收稿日期: 2017-01-17
- 修回日期: 2017-03-11
2. 深圳市海洋生物资源与生态环境重点实验室 深圳 518060
2. Shenzhen Key Laboratory of Marine Bioresources and Ecology, Shenzhen 518060, China
β-葡萄糖苷酶(β-glucosidase,EC 3.2.1. 21) 属于糖苷水解酶。该酶对木质纤维素水解具有关键性作用,是纤维素酶代谢途径中的限速酶[1]。在纤维素酶系降解纤维素的过程中,内切葡聚糖酶(endoglucanase,EC 3.2.1.4) 与外切葡聚糖酶(cellobiohydrolase,EC 3.2.1.91) 协同产生纤维二糖,之后被β-葡萄糖苷酶水解生成葡萄糖,同时解除其对内切葡聚糖酶和外切葡聚糖酶的反馈抑制[2]。β-葡萄糖苷酶已被广泛应用于各种工业领域,如食品、饲料、制药和生物乙醇生产等[3-6]。此外,一些β-葡萄糖苷酶转糖基作用可用于生产高附加值的稀有寡糖[7]。
根据氨基酸序列相似性,β-葡萄糖苷酶属于糖苷水解酶家族(glycoside hydrolase families)GH第1、3、5、9、30和116家族(http://www.cazy.org/)[8],大多数真菌β-葡萄糖苷酶属于GH1和GH3家族。至今,许多GH3家族的β-葡萄糖苷酶从不同的真菌中被分离纯化并进行了功能鉴定,如黑曲霉(Aspergillus niger)[9]、烟曲霉(Aspergillus fumigatus)[10]、桧状青霉(Penicillium piceum)[11]、里氏木霉(Trichoderma reesei)[12]。最近,来自嗜热真菌的β-葡萄糖苷酶由于其具有良好的热稳定性而引起了广泛关注[13-17]。
嗜热毁丝霉(Myceliophthora thermophila)是一种嗜热丝状子囊真菌,生长在45~50℃条件下,是生产热稳定性纤维素水解酶的酶工厂[18]。Berka等[19]于2011年对其基因组和蛋白质组的测序结果表明,该菌具有纤维素分解所需的一套完整的酶,可产生大量植物细胞壁降解酶类,高效降解纤维素。近年来,在工业化生产纤维素酶方面,由于嗜热毁丝霉耐高温、纤维素降解酶含量丰富而引起了极大的兴趣[20]。
越来越多嗜热真菌来源的β-葡萄糖苷酶被分离纯化出来,并将其编码基因克隆表达于异源宿主菌中[21-27],然而关于β-葡萄糖苷酶调控因子方面的研究却鲜有报道。除通过基因重组技术高效表达β-葡萄糖苷酶基因之外,了解该酶基因的调控因子,开展其调控网络相关的研究也具有重要意义。Zn(Ⅱ)2Cys6型锌指蛋白BglR(β-glucosidase regulator)是通过单核苷酸多态性检测技术,新发现的一个特异性调控β-葡萄糖苷酶基因表达的转录因子,在里氏木霉中,BglR能上调β-葡萄糖苷酶基因的表达[28-29]。本实验拟使用丙酮酸脱羧酶启动子(MtPpdc)及终止子(MtTpdc)在嗜热毁丝霉中过表达Mtbglr基因,通过酶活测定、胞外蛋白质浓度测定、荧光定量PCR等方法确定该基因过表达对β-葡萄糖苷酶活性的影响。本研究将为嗜热真菌中β-葡萄糖苷酶的基因调控奠定理论基础。
1 材料与方法 1.1 菌株与质粒大肠杆菌JM107:由本学院苟德明教授课题组惠赠;嗜热毁丝霉(M. thermophila ATCC42464):购自美国菌种收藏中心(ATCC)。
pAN7-1质粒:带有真菌筛选标记潮霉素抗性基因hph及大肠杆菌筛选标记氨苄青霉素抗性基因Ampr;pUC19-MtPpdc-MtTpdc质粒:由本实验室构建[30]。
1.2 培养基及试剂购自TaKaRa公司的DNA限制性内切核酸酶Xba I、DNA Marker、RNA提取试剂(RNAiso Plus)、反转录试剂盒(PrimeScript reagent kit)、荧光定量试剂盒SYBR® Premix Ex Taq Ⅱ(Tli RNaseH Plus);购自Fermentas公司的DNA限制性内切核酸酶Not I;购自NEB公司的SLIC连接试剂T4 DNA polymerase、NEBuffer、超保真PCR试剂盒(Phusion® High-Fidelity PCR Kit);购自Omega公司的质粒提取试剂盒、PCR产物纯化试剂盒和DNA切胶回收试剂盒;购自Sigma公司的溶壁酶。其他生化试剂和试剂盒均购自上海生工生物工程股份有限公司(以下简称上海生工)。
孢子洗涤液:用于制备嗜热毁丝霉孢子悬液,为0.2%的吐温-80溶液。
PDA培养基:马铃薯200g/L,葡萄糖20g/L,琼脂15~20g/L。用于转化子筛选时加入潮霉素B,终浓度为50μg/ml。
Mandels营养盐浓缩液配制:(NH4)2SO4,14g/L;尿素,3g/L;KH2PO4,20g/L;CaCl2·2H2O,4g/L(或CaCl2,3g/L);MgSO4·7H2O,3g/L;加水至1L。
Mandels微量元素浓缩液配制:FeSO4·7H2O,5g/L;ZnSO4·7H2O,1.7g/L(或ZnCl2:0.7g/L);CoCl2·6H2O,3.7g/L(或CoCl2:2g/L);MnSO4·H2O,1.6g/L(或MnCl2:1.67g/L,或MnSO4·7H2O:2.6g/L);加水至1L。
液体基本培养基:含Mandels营养盐浓缩液100ml/L,Mandels微量元素浓缩液1.0ml/L,蛋白胨1.0g/L,无水葡萄糖20g/L,1mol/L的柠檬酸缓冲液(pH 4.5)50ml/L,吐温-80 1.0ml/L。
诱导产酶培养基:Mandels营养盐浓缩液200ml/L,Mandels微量元素浓缩液1.0ml/L, 蛋白胨10g/L,玉米芯粉(诱导碳源1%)10g/L。
1.3 嗜热毁丝霉基因组DNA、RNA的提取和cDNA的合成接种嗜热毁丝霉孢子于PDA平板上,于45℃恒温培养7d后,制备适量孢子悬液(约1×107个新鲜成熟孢子)接种于液体基本培养基中,于45℃、250r/min条件下培养2d后。取30ml菌液进行抽滤,刮取滤纸表面的菌体,用于液氮研磨。使用基因组DNA提取试剂盒进行基因组DNA的提取。使用RNAiso Plus试剂进行RNA的提取。将经过gDNA Eraser处理过的总RNA样品进行反转录,得到相应的cDNA模板,冻存于-20℃用于后续实验。
1.4 SLIC技术进行Mtbglr基因的扩增及过表达载体的构建通过NCBI获得里氏木霉(Trichoderma reesei)中的bglr基因(NW_006711177.1),通过Blast获得嗜热毁丝霉中同源Mtbglr基因(XM_003661910.1)。使用Primer 5.0引物设计软件获得一对有效扩增Mtbglr全长的引物Mtbglr-F和Mtbglr-R(引物序列见表 1)。使用超保真PCR试剂盒扩增出Mtbglr基因全序列。
| Name | Sequence(5′-3′) | Description |
| Mtbglr-F | CGCCAACAACAGACGCGGCCATGACGGCGACGGCAGTG | 扩增Mtbglr基因全长 |
| Mtbglr-R | CACCAAGCCACCATCTCTAGCTACCTGGAAAGACCGTTGAAGTC | |
| MtPpdc-F | CCCAAGCTTCCGAGTGTACTCCGTAAGGA | 载体完整性验证引物 |
| MtTpdc-R | CCGGAATTCGGATTACAGCGCAGTGCACG | |
| P-Mtbglr-T1-F | TATCGTGCCTGTGGTCTGA | 转化子验证引物 |
| P-Mtbglr-T1-R | TGAGCGTTTCCTTGGTGAG | |
| P-Mtbglr-T2-F | ATCGCCGACGGTCAACACTCT | |
| P-Mtbglr-T2-R | TCGTCAAAGCCACGGAATAGA | |
| Note:The 20bp homologous sequences of pUC19-MtPpdc-MtTpdc double digestion ends are underlined by double lines | ||
将本实验室的过表达载体pUC19-MtPpdc-MtTpdc经Not I和Xba I双酶切后,与带有载体末端20bp同源序列的Mtbglr基因利用不依赖基因序列和连接反应克隆(sequence and ligation-independent cloning,SLIC)技术连接(表 2)[31-32],转化大肠杆菌JM107。经氨苄抗性筛选,以MtPpdc-F和MtTpdc-R为上下游引物进行载体完整性PCR及测序验证,测序由上海生工完成。
| Reagent | Application amount |
| T4 DNA polymerase | 0.5μl |
| 10×NEBuffer 2.1 | 1μl |
| 带有载体双酶切末端20bp同源序列的Mtbglr基因 | DNA:Vector=3:1(摩尔比) |
| 双酶切的pUC19-MtPpdc-MtTpdc载体 RNase-Free H2O |
Up to 10μl |
1.5 嗜热毁丝霉原生质体制备及转化
嗜热毁丝霉的原生质体转化方法参照里氏木霉原生质体转化方法并进行了优化[33-34]。将转化子涂布于潮霉素抗性平板和普通PDA平板上纯化分离3次,直到得到单一的纯菌。将纯菌接种于PDA平板上,45℃倒置培养7天。
1.6 实时荧光定量PCR(RT-qPCR)使用SYBR Green I嵌合荧光定量PCR方法(RT-qPCR)分析目的基因的相对表达量(反应程序:预变性95℃ 30s;变性95℃ 5s,退火、延伸60℃ 31s,共40个循环;融解曲线95℃ 30s,60℃ 30s,95℃ 15s)反应结束后利用2-△△CT的方法计算样品中目的基因相对表达量[35]。基因表达水平以beta-tubulin基因(GenBank: AEO58945.1) 为内参对照。所用引物见表 3。
| Name | Sequence(5′-3′) |
| RTq-tub-F | AGGGTATGGATGAGATGGAG |
| RTq-tub-R | AGAAGCAAGCCCTGGAAC |
| RTq-bglr-F | ATCGCCGACGGTCAACACTCT |
| RTq-bglr-R | CGCTTGCTCCTCGTTCATCCTG |
| RTq-bgl1-F | AGCCAGGAGAATGGTAACGC |
| RTq-bgl1-R | TGACAGCCCAAACCCAAACT |
| RTq-bgl2-F | GGCGACCGTAAGAACCTGACTC |
| RTq-bgl2-R | CGGAAGACCAGCCCAGAGAATG |
| RTq-bgl3-F | CGGCGGCTATCTGAACAAGGAG |
| RTq-bgl3-R | CGGCTCGTTGAAGGTGATCCAG |
| RTq-egl1-F | TCGGGCAACGGTTACGAGG |
| RTq-egl1-R | AGCTCTTGACCTGGTTCTGG |
| RTq-egl3-F | TCCGCCACCAGCACCGCCCCTC |
| RTq-egl3-R | TTGCTGCCGAGCCACTTC |
1.7 酶活测定和蛋白质浓度测定
酶活测定:将嗜热毁丝霉接种于PDA平板上,45℃倒置培养7天,参考王宝林的实验用孢子洗涤液(0.2%吐温-80溶液)获得孢子悬液[36]。利用细胞计数板计数,取约1×107个孢子接入1%玉米芯粉为主要碳源的诱导产酶培养基(50ml),45℃,250r/min,摇床振荡培养。从第3天到第8天取发酵上清液,12 000r/min离心10min进行酶活测定。β-葡萄糖苷酶活(BG activity)、滤纸酶活(filter paper activity)、内切β-1.4-葡萄糖苷酶活(EG activity)参照国标QB 2583-2003及Eveleigh D E等的酶活测定方法并加以修改测定[37-38]。
酶活单位定义为1ml酶液在50℃、指定pH条件(酸性纤维素酶pH4.8),1h水解底物,产生出相当于1mg葡萄糖的还原糖量,为1个酶活力单位,以U/g(或U/ml)表示。以上酶活测定实验,每组均设置3个重复。
蛋白质浓度测定:使用改良型Bradford法蛋白质浓度测定试剂盒(上海生工,产品编号:C503041) 测定样品中蛋白质浓度,具体步骤参照该试剂盒说明书。
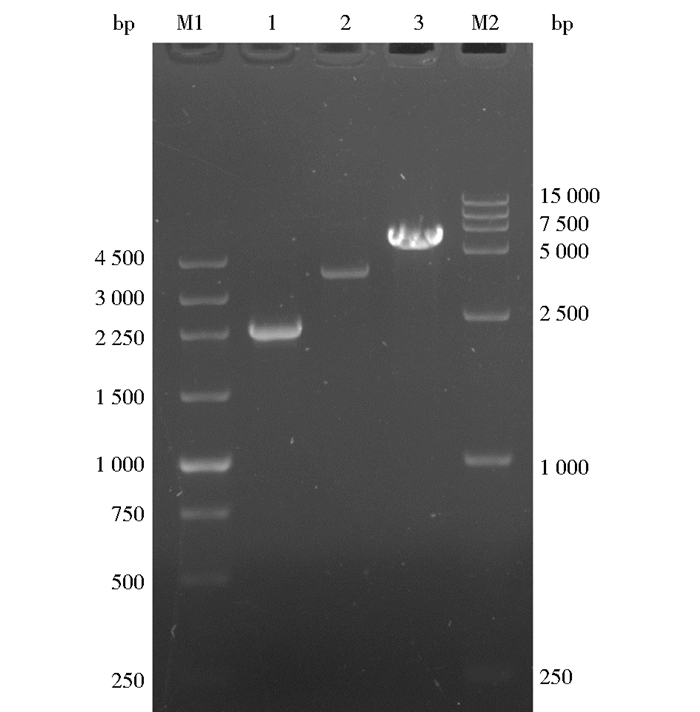
2 结果与分析 2.1 Mtbglr过表达载体的构建使用引物Mtbglr-F和Mtbglr-R从嗜热毁丝霉基因组中进行PCR扩增, 获得带有载体双酶切末端20bp同源序列的Mtbglr基因全长,(2 257bp),如图 1所示。pUC19-MtPpdc-MtTpdc载体经过Not I和Xba I双酶切后(3 534bp),与带有载体双酶切末端20bp同源序列的Mtbglr基因利用SLIC技术连接,转化E.coli。经过含100μg/ml氨苄青霉素抗性平板筛选、载体完整性PCR验证及测序,最终成功构建Mtbglr过表达载体(pUC19-MtPpdc-Mtbglr-MtTpdc)(5 831bp)。载体结构见图 2。
|
| 图 1 Mtbglr过表达载体的构建 Figure 1 Construction of overexpression vector of Mtbglr gene M1:250bp marker; 1:Mtbglr gene sequence; 2:pUC19-MtPpdc-MtTpdc sequence; 3:Mtbglr overexpression vector sequence; M2:DL 15 000TM marker |
|
| 图 2 Mtbglr过表达载体 Figure 2 Overexpression vector of Mtbglr gene |
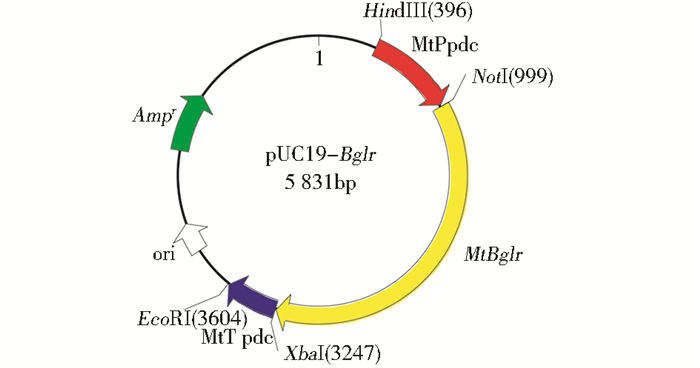
将Mtbglr过表达载体与pAN7-1质粒共同转化嗜热毁丝霉,经潮霉素抗性PDA平板筛选得到8株转化子菌株,于普通PDA平板上45℃倒置培养7天后,提取基因组,分别以P-Mtbglr-T1-F、P-Mtbglr-T1-R(518bp);MtPpdc-F、P-Mtbglr-T2-R(1 634bp);P-Mtbglr-T2-F,MtTpdc-R(2 026bp)这3对引物进行基因组PCR验证,结果如图 3所示。并且样品测序结果完全匹配,得到3株阳性转化子(或重组菌株)。依次命名为Mt3、Mt4、Mt8。
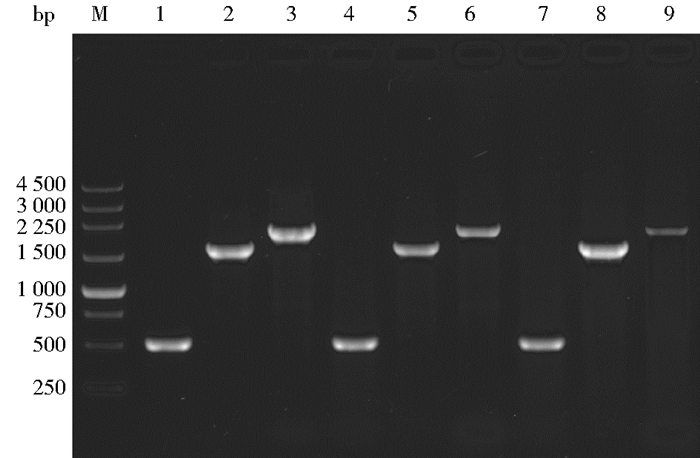
|
| 图 3 转化子PCR验证产物序列 Figure 3 Products of PCR amplification in transformants M:250bp marker; 1-3:Mt3 sequence; 4-6:Mt4 sequence; 7-9:Mt8 sequence; 1, 4, 7:518bp; 2, 5, 8:1634bp; 3, 6, 9:2026bp |
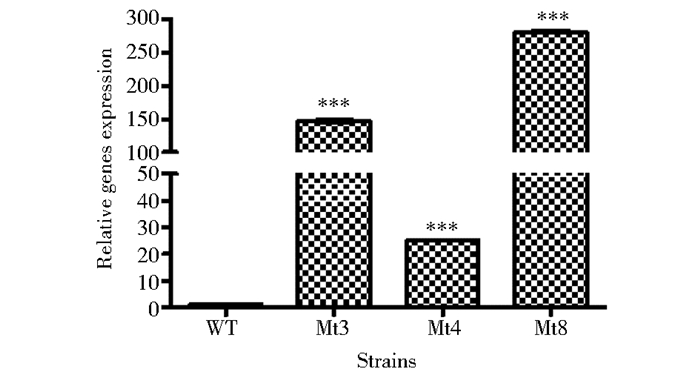
在液体基本培养基培养条件下,使用RT-qPCR分析了野生型WT和转化子Mt3、Mt4、Mt8的bglr mRNA表达水平。以野生型菌株WT的bglr mRNA表达量为1作为对照,转化子菌株Mt3、Mt4、Mt8该基因表达量分别为野生型菌株的147倍、25倍、279倍,见图 4。其中菌株Mt8的过表达效果最明显,故选转化子Mt8进行后续实验研究。
|
| 图 4 基因mRNA表达水平的荧光定量分析 Figure 4 RT-qPCR analysis of the bglr mRNA levels of wild type and transformants |
以1%玉米芯粉诱导产酶培养基,分别培养野生型菌株WT和转化子Mt8。从第3天到第8天,每天分别取两菌株的发酵上清液进行酶活和蛋白质含量测定。表 4显示的第5天和第6天菌株WT和转化子Mt8的总分泌蛋白质含量,结果表明Mt8的胞外蛋白质含量在第5天和第6天分别是WT的1.87倍和1.92倍。
| Strain | Total amount of secreted protein(μg/ml) | |
| 5 days | 6 days | |
| WT(野生型) | 103±1.5 | 106±3.0 |
| Mt8(转化子) | 193±2.5 | 203±5.0 |
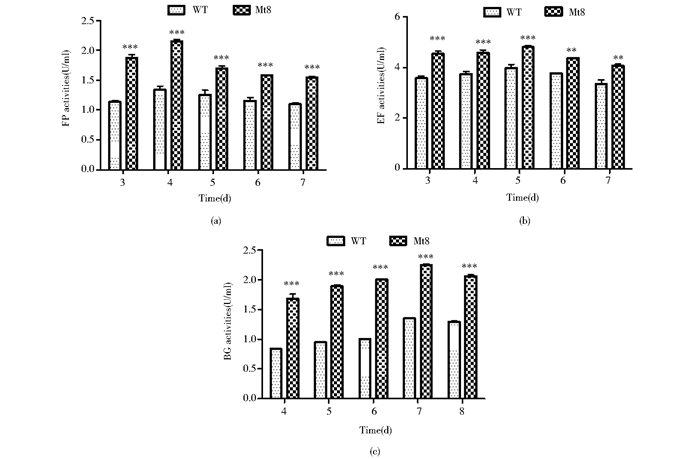
在1%玉米芯粉诱导产酶条件下培养野生型菌株与转化子菌株,分别测定了其从第3天到第8天发酵液的滤纸酶活、β-葡萄糖苷酶活、内切β-1.4-葡萄糖苷酶活。数据如图 5所示,滤纸酶活在第4天达到最高值,转化子Mt8的FPA是WT的1.6倍;内切β-1, 4-葡萄糖苷酶活在第5天达到最高值,Mt8是WT的1.2倍;β-葡萄糖苷酶活在第7天达到最高值,Mt8是WT的1.7倍。
|
| 图 5 诱导条件下野生型和转化子的纤维素酶活性 Figure 5 Cellulase activities of the wild strain and tranformant under the inducing condition (a) are the filter paper (FP) activities (b)β-1, 4-endoglucanase (EG) activities (c)β-glucosidase (BG) activities |
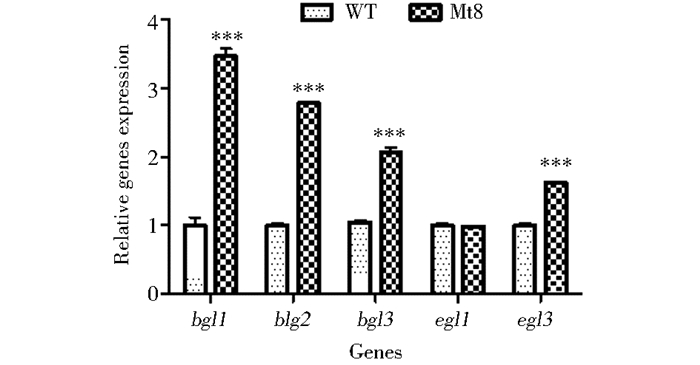
在玉米芯粉诱导培养条件下,使用RT-qPCR分析了野生型WT和转化子Mt8的几种主要纤维素酶表达量,包括β-葡萄糖苷酶基因bgl1、bgl2、bgl3和内切β-1, 4-葡萄糖苷酶基因egl1、egl3的表达水平。经内参基因beta-tubulin校正,设定野生型WT中各基因的表达量为1,计算出转化子Mt8中各基因的相对表达量。如图 6所示,Mt8中bgl1、bgl2、bgl3、egl3的表达量分别是WT中的3.5倍、2.8倍、2.1倍、1.6倍,egl1表达量没有明显差异。上述结果表明,过表达bglr对嗜热毁丝霉主要纤维素酶基因的表达具有一定的促进作用,该结果与转化子中EG、BG酶活的提高相吻合。
|
| 图 6 野生型和转化子主要纤维素酶基因表达水平 Figure 6 Expression of the main cellulase genes in the wild strain and tranformant |
在丝状真菌中,参与调节酶产量的调控因子众多。在碳代谢阻遏过程中,已知发挥重要作用的主要正调控因子有Ace2[39]、Hap2/3/5[40],Xyr1[41],负调控因子有Ace1[42]、Cre1[43]等。在里氏木霉中,bglr基因缺失抑制了诱导初期β-葡萄糖苷酶对纤维二糖的水解,在bglr缺失菌株中,bgl2、cel1b和cel3c等β-葡萄糖苷酶基因表达水平降低且表达时间出现不同程度的延迟,这说明BglR可以上调特定β-葡萄糖苷酶基因的表达[28],本研究在嗜热毁丝霉中过表达bglr基因,在酶活和基因表达量两方面确定,该基因可促进β-葡萄糖苷酶基因的表达,增强其基因表达量。获得的结果与里氏木霉中一致,表明在不同的丝状真菌中,该调控因子具有相似的功能。同时我们查询了嗜热真菌嗜热毁丝霉(XP_003661958.1)、太瑞斯梭孢壳霉(XP_003649523.1),以及常温真菌里氏木霉(XP_006969323.1)、粗糙脉孢菌(XP_958079.1) 的BglR氨基酸序列,通过多重序列比对,发现不同种属的BglR基因具有一个高度保守序列——典型的Zn(Ⅱ)2Cys6双核基因簇结构(-RACDACHRRKVKCDGINPCRNCSTAQLSCTYNAIPQK-)[28],这对BglR的人工改造和分子调控具有重要意义。然而目前关于BglR的研究刚刚起步,尚存在许多未知的问题。例如,BglR的作用位点、功能区域等,以及BglR与其他纤维素酶调控因子之间的关系如何,都有待于进一步的研究去揭示这些问题。
综上所述,本文在嗜热毁丝霉中过表达bglr基因,可有效提高诱导条件下菌株的滤纸酶活、内切纤维素酶活性及β-葡萄糖苷酶活性。结合本实验室以前的研究[30, 34, 44],不难发现针对纤维素酶基因表达调控的研究,是一种改造工业菌株的高效遗传操作方法。明确纤维素酶基因的调控机制,将有利于人们利用分子生物学方法改造工程菌,提高纤维素酶产量。
| [1] |
周庆新, 戴炳业, 陈蕾蕾, 等. 瑞氏木霉中β-葡萄糖苷酶基因功能研究进展. 中国农业科技导报, 2014, 16(2): 74-78. Zhou Q X, Dai B Y, Chen L L, et al. Progress on functional studies of β-glucosidase genes in Trichoderma reesei. Journal of Agricultural Science and Technology, 2014, 16(2): 74-78. |
| [2] |
Singhania R R, Patel A K, Sukumaran R K, et al. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresource Technology, 2013, 127: 500-507. DOI:10.1016/j.biortech.2012.09.012 |
| [3] |
Bhatia Y, Mishra S, Bisaria V S. Microbial β-glucosidases: cloning, properties, and applications. Critical Reviews in Biotechnology, 2002, 22(4): 375-407. DOI:10.1080/07388550290789568 |
| [4] |
Chandra M, Kalra A, Sangwan N S, et al. Biochemical and proteomic characterization of a novel extracellular β-glucosidase from Trichoderma citrinoviride. Molecular Biotechnology, 2013, 53(3): 289-299. DOI:10.1007/s12033-012-9526-7 |
| [5] |
Handa C L, Couto U R, Vicensoti A H, et al. Optimisation of soy flour fermentation parameters to produce β-glucosidase for bioconversion into aglycones. Food Chemistry, 2014, 152: 56-65. DOI:10.1016/j.foodchem.2013.11.101 |
| [6] |
Abedinifar S, Karimi K, Khanahmadi M, et al. Ethanol production by Mucor indicus and Rhizopus oryzae from rice straw by separate hydrolysis and fermentation. Biomass and Bioenergy, 2009, 33(5): 828-833. DOI:10.1016/j.biombioe.2009.01.003 |
| [7] |
Pal S, Banik S P, Ghorai S, et al. Purification and characterization of a thermostable intra-cellular β-glucosidase with transglycosylation properties from filamentous fungus Termitomyces clypeatus. Bioresource Technology, 2010, 101(7): 2412-2420. DOI:10.1016/j.biortech.2009.11.064 |
| [8] |
Cantarel B L, Coutinho P M, Rancurel C, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Research, 2009, 37(suppl 1): D233-D238. |
| [9] |
Dan S, Marton I, Dekel M, et al. Cloning, expression, characterization, and nucleophile identification of family 3, Aspergillus niger β-glucosidase. Journal of Biological Chemistry, 2000, 275(7): 4973-4980. DOI:10.1074/jbc.275.7.4973 |
| [10] |
Liu D, Zhang R, Yang X, et al. Characterization of a thermostable β-glucosidase from Aspergillus fumigatus Z5, and its functional expression in Pichia pastoris X33. Microbial Cell Factories, 2012, 11(25): 1-15. |
| [11] |
Gao L, Gao F, Zhang D, et al. Purification and characterization of a new β-glucosidase from Penicillium piceum and its application in enzymatic degradation of delignified corn stover. Bioresource Technology, 2013, 147: 658-661. DOI:10.1016/j.biortech.2013.08.089 |
| [12] |
Chen P, Fu X, Ng T B, et al. Expression of a secretory β-glucosidase from Trichoderma reesei in Pichia pastoris and its characterization. Biotechnology Letters, 2011, 33(12): 2475-2479. DOI:10.1007/s10529-011-0724-3 |
| [13] |
Haki G D, Rakshit S K. Developments in industrially important thermostable enzymes: a review. Bioresource Technology, 2003, 89(1): 17-34. DOI:10.1016/S0960-8524(03)00033-6 |
| [14] |
Morgenstern I, Powlowski J, Ishmael N, et al. A molecular phylogeny of thermophilic fungi. Fungal Biology, 2012, 116(4): 489-502. DOI:10.1016/j.funbio.2012.01.010 |
| [15] |
Basotra N, Kaur B, Di Falco M, et al. Mycothermus thermophilus (Syn. Scytalidium thermophilum): Repertoire of a diverse array of efficient cellulases and hemicellulases in the secretome revealed. Bioresource Technology, 2016, 222: 413-421. DOI:10.1016/j.biortech.2016.10.018 |
| [16] |
Xu X, Li J, Shi P, et al. The use of T-DNA insertional mutagenesis to improve cellulase production by the thermophilic fungus Humicola insolens Y1. Scientific Reports, 2016, 6: 31108. DOI:10.1038/srep31108 |
| [17] |
Mallek-Fakhfakh H, Belghith H. Physicochemical properties of thermotolerant extracellular β-glucosidase from Talaromyces thermophilus and enzymatic synthesis of cello-oligosaccharides. Carbohydrate Research, 2016, 419: 41-50. DOI:10.1016/j.carres.2015.10.014 |
| [18] |
Matsakas L, Antonopoulou I, Christakopoulos P. Evaluation of Myceliopthora thermophila as an enzyme factory for the production of thermophilic cellulolytic enzymes. Bio Resources, 2015, 10(3): 5140-5158. |
| [19] |
Berka R M, Grigoriev I V, Otillar R, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol, 2011, 29(10): 922-927. DOI:10.1038/nbt.1976 |
| [20] |
Visser H, Joosten V, Punt P J, et al. RESEARCH: Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1. Industrial Biotechnology, 2011, 7(3): 214-223. DOI:10.1089/ind.2011.7.214 |
| [21] |
Karnaouri A, Topakas E, Paschos T, et al. Cloning, expression and characterization of an ethanol tolerant GH3β-glucosidase from Myceliophthora thermophila. Peer J, 2013, 1: e46. DOI:10.7717/peerj.46 |
| [22] |
Zhao J, Guo C, Tian C, et al. Heterologous expression and characterization of a GH3β-glucosidase from thermophilic fungi Myceliophthora thermophila in Pichia pastoris. Applied Biochemistry and Biotechnology, 2015, 177(2): 511-527. DOI:10.1007/s12010-015-1759-z |
| [23] |
Murray P, Aro N, Collins C, et al. Expression in Trichoderma reesei and characterisation of a thermostable family 3β-glucosidase from the moderately thermophilic fungus Talaromyces emersonii. Protein Expression and Purification, 2004, 38(2): 248-257. DOI:10.1016/j.pep.2004.08.006 |
| [24] |
Hong J, Tamaki H, Kumagai H. Cloning and functional expression of thermostable β-glucosidase gene from Thermoascus aurantiacus. Applied Microbiology and Biotechnology, 2007, 73(6): 1331-1339. DOI:10.1007/s00253-006-0618-9 |
| [25] |
Yang X, Ma R, Shi P, et al. Molecular characterization of a highly-active thermophilic β-glucosidase from Neosartorya fischeri P1 and its application in the hydrolysis of soybean isoflavone glycosides. PLoS One, 2014, 9(9): e106785. DOI:10.1371/journal.pone.0106785 |
| [26] |
Guo Y, Yan Q, Yang Y, et al. Expression and characterization of a novel β-glucosidase, with transglycosylation and exo-β-1, 3-glucanase activities, from Rhizomucor miehei. Food Chemistry, 2015, 175: 431-438. DOI:10.1016/j.foodchem.2014.12.004 |
| [27] |
Xia W, Xu X, Qian L, et al. Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharification performance. Biotechnology for Biofuels, 2016, 9(1): 147. DOI:10.1186/s13068-016-0560-8 |
| [28] |
Nitta M, Furukawa T, Shida Y, et al. A new Zn (Ⅱ) 2 Cys 6-type transcription factor BglR r-egulates β-glucosidase expression in Trichoderma reesei. Fungal Genetics and Biology, 2012, 49(5): 388-397. DOI:10.1016/j.fgb.2012.02.009 |
| [29] |
Tani S, Kawaguchi T, Kobayashi T. Complex regulation of hydrolytic enzyme genes for cellulosic biomass degradation in filamentous fungi. Applied Microbiology and Biotechnology, 2014, 98(11): 4829-4837. DOI:10.1007/s00253-014-5707-6 |
| [30] |
Wang J, Wu Y, Gong Y, et al. Enhancing xylanase production in the thermophilic fungus Myceliophthora thermophila by homologous overexpression of Mtxyr1. Journal of Industrial Microbiology & Biotechnology, 2015, 42(9): 1233-1241. |
| [31] |
Li M Z, Elledge S J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nature Methods, 2007, 4(3): 251-256. DOI:10.1038/nmeth1010 |
| [32] |
Jeong J Y, Yim H S, Ryu J Y, et al. One-step sequence-and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Applied and Environmental Microbiology, 2012, 78(15): 5440-5443. DOI:10.1128/AEM.00844-12 |
| [33] |
Penttilä M, Nevalainen H, Rättö M, et al. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene, 1987, 61(2): 155-164. DOI:10.1016/0378-1119(87)90110-7 |
| [34] |
Yang F, Gong Y, Liu G, et al. Enhancing cellulase production in thermophilic fungus Myceliophthora thermophila ATCC42464 by RNA interference of cre1 gene expression. Journal of Microbiology and Biotechnology, 2015, 25(7): 1101-1107. DOI:10.4014/jmb.1501.01049 |
| [35] |
Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 2001, 25(4): 402-408. DOI:10.1006/meth.2001.1262 |
| [36] |
王宝林, 韩燕峰, 王玉荣, 等. 一株嗜热毁丝霉菌株产纤维素酶条件优化. 酿酒科技, 2012(10): 27-31. Wang B L, Han Y F, Wang Y R, et al. Optimization of cellulase-producing conditions of A Thermophilic myceliophthora sp. H127-1 strain. Liquor-Making Science & Tcchnology, 2012(10): 27-31. |
| [37] |
Eveleigh D E, Mandels M, Andreotti R, et al. Measurement of saccharifying cellulase. Biotechnology for Biofuels, 2009, 2(1): 21. DOI:10.1186/1754-6834-2-21 |
| [38] |
Mansour A A, Da Costa A, Arnaud T, et al. Review of lignocellulolytic enzyme activity analyses and scale-down to microplate-based assays. Talanta, 2016, 150: 629-637. DOI:10.1016/j.talanta.2015.12.073 |
| [39] |
Aro N, Saloheimo A, Ilmén M, et al. ACEⅡ, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. Journal of Biological Chemistry, 2001, 276(26): 24309-24314. DOI:10.1074/jbc.M003624200 |
| [40] |
Zeilinger S, Ebner A, Marosits T, et al. The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase Ⅱ-gene) activating element. Molecular Genetics and Genomics, 2001, 266(1): 56-63. DOI:10.1007/s004380100518 |
| [41] |
Stricker A R, Grosstessner-Hain K, Würleitner E, et al. Xyr1(xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryotic Cell, 2006, 5(12): 2128-2137. DOI:10.1128/EC.00211-06 |
| [42] |
Aro N, Ilmén M, Saloheimo A, et al. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Applied and Environmental Microbiology, 2003, 69(1): 56-65. DOI:10.1128/AEM.69.1.56-65.2003 |
| [43] |
Portnoy T, Margeot A, Linke R, et al. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genomics, 2011, 12(1): 269. DOI:10.1186/1471-2164-12-269 |
| [44] |
Wang S, Liu G, Yu J, et al. RNA interference with carbon catabolite repression in Trichoderma koningii for enhancing cellulase production. Enzyme and Microbial Technology, 2013, 53(2): 104-109. DOI:10.1016/j.enzmictec.2013.04.007 |
 2017, Vol. 37
2017, Vol. 37