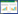文章信息
- 吴红丽, 薛勇, 刘健, 甘礼惠, 龙敏南
- WU Hong-li, XUE Yong, LIU Jian, GAN Li-hui, LONG Min-nan
- 乙酰木聚糖酯酶研究进展
- Research Progress of Acetyl Xylan Esterase
- 中国生物工程杂志, 2016, 36(3): 102-110
- China Biotechnology, 2016, 36(3): 102-110
- http://dx.doi.org/DOI:10.13523/j.cb.20160315
-
文章历史
- 收稿日期: 2015-10-12
- 修回日期: 2015-11-02
2. 福建生物工程职业技术学院 福州 350002
2. Fujian Vocational College of Bioengineering, Fuzhou 350002, China
半纤维素是植物细胞壁中仅次于纤维素的第二大可再生生物质资源[1]。它的利用与转化对解决能源危机、温室效应等一系列全球问题有很大的促进作用。木聚糖是半纤维素的主要组成成分,它是一种杂合多聚糖,其主链由多个吡喃木糖基通过β-1,4木糖苷键相连,而大约80%的木聚糖含有侧链,如O-乙酰基、L-阿拉伯糖残基、阿魏酸和4-O-甲基-D-葡萄糖醛酸残基等[2]。由于木聚糖的来源及其结构的复杂性,其完全降解需要多种酶的协同作用来切开主链骨架及侧链,包括β-1,4内切木聚糖酶、β-D-木糖苷酶、α-L-阿拉伯呋喃糖酶、α-D-葡萄糖醛酸酶、阿魏酸酯酶和乙酰木聚糖酯酶等[3]。在阔叶材中,被乙酸所酯化的木糖残基大约占50%[4],乙酰基的存在限制了木聚糖降解酶的作用[5]。因此在木聚糖的生物降解过程中,对乙酰基的降解显得尤为重要[6]。
乙酰木聚糖酯酶(acetyl xylan esterase,AXE,EC3.1.1 .6 )主要作用于乙酰化木聚糖中木糖残基的2位和3位的O-乙酰基[7, 8, 9]。该酶在1985年由Biely[5]首次发现之后,越来越多的不同类型的乙酰木聚糖酯酶被发现和研究。
1 乙酰木聚糖酯酶的分类与微生物来源根据氨基酸序列的相似性,来自不同微生物的乙酰木聚糖酯酶分别归属于CE(carbohydrate esterase)家族第1~7家族和12家族(表 1)。Aspergillus、Penicillium、Schizophyllum、Trichoderma等真菌来源的乙酰木聚糖酯酶及细菌来源的乙酰木聚糖酯酶属于CE家族1和5,来源于厌氧菌Neocallimastix patriciarum[10]的乙酰木聚糖酯酶属于CE家族2、3和6,来源于Streptomyces的乙酰木聚糖酯酶属于CE家族4,它们与根瘤菌和一些酵母的几丁质脱乙酰酯酶具有同源性。
| Family | Microorganism |
| CE1 | Aspergillus niger[11],Myceliophthora thermophile C1[12],Schizophyllum commune[13],Talaromyces emersonii[14],Aspergillus oryzae[15] |
| CE2 | Cellvibrio japonicas[16],Neocallimastix patriciarum[17],Butyrivibrio proteoclasticus[16] |
| CE3 | Candidatus methanosphaerula palustris*,Actinoplanes friuliensis* |
| CE4 | Streptomycces lividans[18],Clostridium thermocellum[19],Anoxybacillus flavithermus[20] |
| CE5 | Trichoderma reesie[21],Chrysosporium lucknowense C1[13],Penicillium purpurogenum[15] |
| CE6 | Orpinomyces sp.[22] |
| CE7 | Thermotoga maritime[23],Bacillus pumilus[24] |
| CE12 | Bacillus subtilis[25] |
| *The gene has not been cloned | |
不同家族的乙酰木聚糖酯酶在作用于乙酰化木聚糖时显示出不同的底物特异性。图 1所示为阔叶材中乙酰化木聚糖上4种乙酰化类型,以及来自CE家族第1家族、第4家族、第5家族、第6家族的乙酰木聚糖酯酶对不同位置乙酰基的特异性。第1家族、第4家族、第5家族和第6家族的乙酰木聚糖酯酶均能够脱除吡喃木糖残基上的2位或3位上的乙酰基,而第4家族与其他三个家族的不同之处在于它不能脱除2位和3位同时乙酰化的基团[26, 27]。第2家族的乙酰木聚糖酯酶对吡喃木糖残基的4位乙酰基有很强的特异性。第3家族和第7家族的乙酰木聚糖酯酶对乙酰化木聚糖的作用范围较广,并没有很强的特异性。
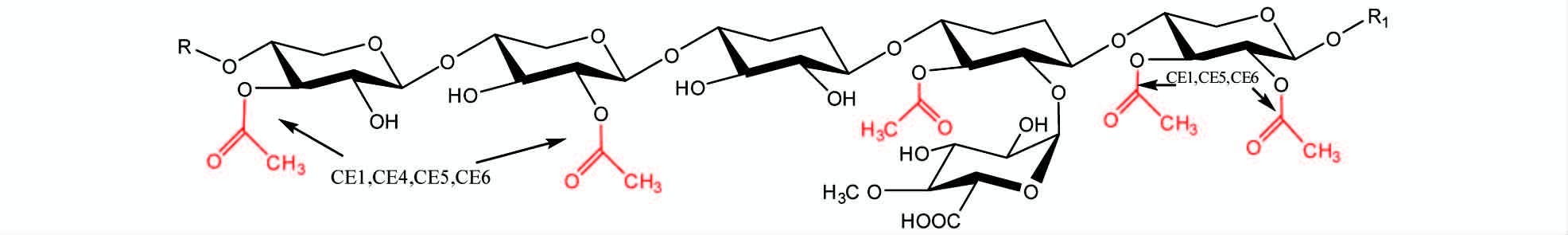 |
| 图 1 不同家族乙酰木聚糖酯酶作用于阔叶材乙酰化木聚糖时的底物特异性 Fig. 1 The substrate specificity of acetyl xylan esterase CE1,CE4,CE5 and CE6 |
一般来说,作为单体酶存在的乙酰木聚糖酯酶的分子质量为2 348kDa,大多数真菌和一些细菌来源的乙酰木聚糖酯酶是典型的单体酶,然而一些嗜热厌氧菌来源的乙酰木聚糖酯酶以低聚物形式存在。不同微生物来源的乙酰木聚糖酯酶的等电点、分子质量、温度和pH作用范围会有一定差异。几种不同来源的乙酰木聚糖酯酶的酶学特性见表 2。
| Organism | MW(kDa) | pI | pH-optimum | Temperature-optimum(C) | Reference |
| Aspergillus ficuum | 29.5 | n.d | 7.0 | 37 | [28] |
| Aspergillus oryzae | 30~31 | n.d | 6 | 45 | [15] |
| Chrysosporium lucknowense Axe2 | 23.6 | 3.7 | 7.0 | 40 | [13] |
| Chrysosporium lucknowense Axe3 | 33.6 | 5.4 | 7.0 | 34~45 | [13] |
| Caldanaerobacter subterraneus | 34 | n.d | 8.0 | 60 | [29] |
| Fibrobacter succinogenes | 55 | 4.0 | 7.0 | 45 | [30] |
| Penicillium purpurogenum AcXEI | 48 | 7.5 | 5.3 | 50 | [31] |
| Penicillium purpurogenum AcXEII | 23 | 7.8 | 6.0 | 60 | [31] |
| Penicillium purpurogenum AcXEIII | 85.7 | 6.3 | 5.3 | 20 | [32] |
| Streptomyces griseus | 37 | 4.95 | 8.0 | 50 | [33] |
| Streptomyces lividans | 34 | 9.0 | 7.5 | 70 | [18] |
| Talaromyces emersonii | 32 | 3.94 | 7.0 | 60 | [14] |
| Thermobifida fusca NTU22 | 28 | 6.34 | 7.5 | 60 | [34] |
| Note:n.d=Not determined | |||||
热稳定性是衡量酶工业用途的重要参数。不同来源的乙酰木聚糖酯酶的最适反应温度多数为45~60℃(表 2)。一些从嗜热细菌和某些真菌中提取的乙酰木聚糖酯酶的最适反应温度较高,如从Thermobifida fusca NTU22和Caldanaerobacter subterraneus subsp.Tengcongensis(表 2)中提纯得到的乙酰木聚糖酯酶的最适反应温度都达到60℃,且后者在65℃下保温100min酶活仅损失10%左右,说明有较好的热稳定性。从Streptomyces lividans(表 2)中提纯得到的乙酰木聚糖酯酶最适反应温度更是高达70℃。较高的最适反应温度和耐热性会使酶有很大的工业利用价值,所以更多的嗜热性乙酰木聚糖酯酶基因还有待我们发掘。
2.2 乙酰木聚糖酶的最适pH真菌来源的乙酰木聚糖酯酶最适pH大多呈弱酸性或中性,而细菌来源的乙酰木聚糖酯酶最适pH比真菌来源的pH偏碱性(表 2)。Moriyoshi等[29]将从细菌C.subterraneus中克隆得到的乙酰木聚糖酯酶基因在大肠杆菌中表达,其表达产生的乙酰木聚糖酯酶最适pH为8.0。刘伟娜等[33]从灰色链霉菌Streptomyces griseus中克隆得到了乙酰木聚糖酯酶基因SgrAxe,其最适pH为8.0,pH为8.0~9.5时仍保持较高的O-乙酰基团水解活性,pH作用范围较广。
2.3 金属离子对乙酰木聚糖酯酶活性的影响除温度和pH之外,金属离子对酶活性也会产生较大的影响。在工业应用中某些金属离子对酶的激活或抑制作用也是衡量该酶工业用途的重要参数。研究表明,金属离子对不同来源的乙酰木聚糖酯酶有着不同的影响。来自于C.subterraneus[29]和Aspergillus usamii E001[35]的乙酰木聚糖酯酶几乎不受Cu2+、Ca2+、Mg2+、Fe2+、Mn2+、Co2+、Zn2+及EDTA的影响(表 3),而来自于Streptomyces griseus[33]的乙酰木聚糖酯酶SgrAxe则受Mn2+、Co2+和Mg2+的影响较大,加入Al3+、Ba2+、Ca2+、Zn2+ 4种金属离子后,抑制的重组酶SgrAxe相对活力超过了90%。值得注意的是,来自于S.lividans和C.thermocellum的乙酰木聚糖酯酶属于CE4家族,该家族的酶属于金属依赖性酶,EDTA会完全抑制该酶的活性[36]。
| Organism | EDTA | Ca2+ | Mg2+ | Zn2+ | Co2+ | Mn2+ | Cu2+ | Cd2+ | Reference |
| Aspergillus usamii E001 | + | ++ | + | + | + | n.d. | [35] | ||
| Bacillus pumilus | n.d. | ++++ | n.d. | +++++ | ++++ | ++++ | +++++ | n.d. | [37] |
| Clostridium thermocellum | ++++ | n.d. | +++++ | +++++ | +++ | n.d. | +++++ | [36] | |
| Phanerochaete chrysosporium | n.d. | ++++ | ++++ | ++ | ++ | n.d. | [38] | ||
| Streptomyces albus | +++ | ++ | n.d. | [39] | |||||
| Streptomyces griseus | +++++ | ++++ | +++++ | ++++ | ++++ | n.d. | [33] | ||
| Streptomyces lividans | ++++ | n.d. | +++++ | +++ | +++ | n.d. | +++ | [36] | |
| Volvariella volvacea | n.d. | +++ | ++ | n.d. | [40] | ||||
| Note:n.d=Not determined;“+”Represent the degree of inhibition; “+++++”Completely inhibited | |||||||||
许多编码乙酰木聚糖酯酶的基因已被克隆。被克隆的真菌来源有Aspergillus、Penicillium、Schizophyllum、Trichoderma、Neocallimastix、Rhodotorula、Orpinomyces和Talaromyces等。Blum等[22]克隆了Orpinomyces sp. PC-2乙酰木聚糖酯酶基因(axeA)在大肠杆菌中表达,并纯化得到了重组体蛋白。axeA基因包含一个939bp的开放阅读框,编码313个氨基酸。Koseki等[15]从Aspergillus oryzae中克隆得到了乙酰木聚糖酯酶基因Aoaxe,该基因编码276个氨基酸,通过蛋白氨基酸序列比对发现其与A.niger、A.ficuum和A.awamori等来源的乙酰木聚糖酯酶分别有70%、70%、71%的同源性,并在毕赤酵母中实现了高效表达。Waters等[14]从嗜热真菌Talaromyces emersonii中克隆了乙酰木聚糖酯酶基因(TeAXE)并在大肠杆菌中成功表达,对其酶学特性也进行了研究。研究发现,TeAXE的催化特性属于丝氨酸型催化。Huy等[38]从Phanerochaete chrysosporium中克隆得到了乙酰木聚糖酯酶基因PcAxe2,该基因编码367个氨基酸,包含一个碳水化合物结合组件(CBM)。在国内,曹杰[41]也从草菇(Volvariella volvacea)中克隆得到了乙酰木聚糖酯酶基因,并成功在毕赤酵母中表达。目前已被克隆的细菌来源有Cellvibrio、Clostridium、Pseudomonas、 Streptomyces、 Thermobifida、Bacillus和Caldanaerobacter等。Dupont等[18]在Streptomyces lividans中实现了乙酰木聚糖酯酶的过表达。Huang等[34]从Thermobifida fusca NTU22中克隆得到了乙酰木聚糖酯酶axe基因,该基因片段长786bp,编码262个氨基酸,包含一个Ser-Asp-His的催化三联体。汪文静等[39]从白色链霉菌(Streptomyces albus)中克隆得到了乙酰木聚糖酯酶基因,其核酸序列含有741bp的开放阅读框,编码247个氨基酸。Moriyoshi等[29]从嗜热细菌Caldanaerobacter subterraneus中克隆得到了乙酰木聚糖酯酶基因TTE0866并在大肠杆菌中成功表达,结果显示该酶具有较好的热稳定性。Tian等[42] 从Bacillus subtilis中克隆得到了乙酰木聚糖酯酶基因,并在大肠杆菌中成功表达,该基因由957bp的开放阅读框组成,编码318个氨基酸,通过蛋白氨基酸序列比对发现,其与Bacillus sp.916、B.subtilis 168和Bacillus pumilus Cect5072具有较高的同源性。Eminolu等[20]从Anoxybacillus flavithermus DSM2641T中克隆得到了CE4家族乙酰木聚糖酯酶基因,该基因片段长762bp,编码253个氨基酸。
4 乙酰木聚糖酯酶结构和催化特性乙酰木聚糖酯酶是目前了解最少的植物细胞壁降解酶系之一。目前关于乙酰木聚糖酯酶的催化机制最普遍的是Ser-His-Asp催化三联体联合起到催化作用,这种机制包含两种主要元素:一个丝氨酸提供亲核的羟基和广义的酸碱催化基团。早在1996年,Peter等[43, 44, 45]提出当单取代的乙酰基邻位有羟基存在时,会促使一个五元过渡状态酶催化产物的形成。说明乙酰木聚糖酯酶的催化过程中有中间产物的产生。Hakulinen等[21]通过分子置换法得出了来自Trichoderma reesei的乙酰木聚糖酯酶催化中心的晶体结构为三层的α/β /α三明治结构,中心的β束包含6个平行的 折叠片,折叠片的周围为4个α螺旋线,分布在折叠片两侧。该酶的结构见图 2。同时提出木聚糖上乙酰基的去除与催化三联体有密切联系,乙酰木聚糖酯酶活性中心存在Ser90-His187-Asp175催化三联体,其中His187从Ser90位的羟基夺去一个质子,亲核氧攻击乙酰基上的羰基氧来形成四面体的中间产物。同样,Ghosh等[46]通过定点突变及X射线结晶体学研究发现,来自Penicillium purpurogenum的乙酰木聚糖酯酶AXEⅡ三维结构含有两个α/β三明治结构,中心有6个β折叠束,折叠束两侧分别有一个α螺旋线。在β折叠片的末端,即C端存在Ser90-His187-Asp175催化三联体。同时发现Tyr57、Gln91和Phe152与底物结合有直接联系。随着对功能性氨基酸残基研究的深入,乙酰木聚糖酯酶的催化机制得到进一步揭示。Correia等[47]也通过X射线衍射发现,来自Clostridium thermocellum的CE3家族酯酶CtCes3 1为典型的α/β折叠结构,定点突变表明,CtCes3-1拥有Ser44-Asp205-His208催化三联体,以及能够使氢键成为过渡状态的氧阴离子穴Ser44、Gly95和Asn124。丝氨酸作为亲核体与底物结合,组氨酸通过吸收一个质子来活化亲核体Ser44,天冬氨酸通过带有咪唑环的氢键使组氨酸上的正电荷稳固。来自于Streptomyces lividans的CE4家族的乙酰木聚糖酯酶为(α/β)8桶状结构。该酶以二聚体的形式实现其催化功能。其中每个单体都包含有一个金属活性中心、两个组氨酸残基(His62和His66)、Asp13、一个单一的水分子和乙酸盐分子 [36, 48]。该酶属于金属依赖性酶,带有一个对金属离子具有特殊偏好的金属中心。其中二价金属起着路易斯酸的作用,它和His及Asp残基一起作为催化的基础来活化水分子成为亲核试剂。由于酯键的裂解,会形成不稳定的四面体中间物,这与丝氨酸型酯酶具有相似的作用机制。Huy等[38]从Phanerochaete chrysosporium中克隆得到了一种含有双功能团的乙酰木聚糖酯酶,该酶包含一个碳水化合物结合组件(CBM),对木聚糖有催化活性,并且通过研究发现,该酶包含两个不同的催化结构域。Tong等[49]通过序列比对发现,来自Chaetomium thermophilum的两个乙酰木聚糖酯酶CtAxeA和CtAxeB分别含有Ser119-Asp200-His257和Ser97-Asp184-His196催化三联体结构。进一步研究发现,Pro146和Gly144对CtAxeA酶与底物的结合中起着重要作用;而在CtAxeB中发现,底物结合位点的凹槽中存在Thr20、Tyr64、Tyr186、Pro141和Phe159等氨基酸残基,说明这些氨基酸对酶与底物的结合有一定作用,但具体作用还不清楚。而在国内,对于乙酰木聚糖酯酶的催化机制及晶体结构的研究并不多见,但近年来已陆续有文献对乙酰木聚糖酯酶进行一些深入研究。汪文静等[39]对白色链霉菌(Streptomyces albus)来源的乙酰木聚糖酯酶进行同源建模时发现其三维结构中的一个多肽片段与已知晶体结构乙酰木聚糖酯酶的C端极为相似,该片段由1个β折叠和9个α螺旋组成,其中9个α螺旋环绕于β折叠的周围。田斌[50]在研究草菇来源的乙酰木聚糖酯酶AXE1时发现,N-糖基化对乙酰木聚糖酯酶AXE1起着类似“蛋白伴侣”的作用,在体内帮助初生AXE1的折叠。表 4展现了来自于不同CE家族的乙酰木聚糖酯酶的酯键的水解机制及蛋白质结构。除了CE4家族,其他CE家族的酶都是典型的α/β水解酶折叠方式。来自于CE1、CE5、CE6、CE7家族的乙酰木聚糖酯酶都是典型的丝氨酸型酯酶,包含Ser-His-Asp催化三联体结构,CE4家族则属于天冬氨酸金属依赖型酶。
由于半纤维素结构的复杂性,需要各种酶的协同降解。早在1986年,就有研究[53]表明,乙酰木聚糖酯酶与其他类型的木聚糖水解酶同时作用于半纤维素可以大大提高乙酰化木聚糖的水解率。之后陆续有文献[54, 55]指出其他类型的木聚糖酶和乙酰木聚糖酯酶在水解乙酰木聚糖时存在协同作用,乙酰木聚糖酯酶在水解乙酰化木聚糖时可以在木聚糖主链上产生新的木聚糖酶作用位点。同样的,主链的断裂可以为乙酰木聚糖酯酶提供新的作用位点。Puls等[56]通过对木聚糖酯酶和乙酰木聚糖酯酶添加顺序的研究发现,不同的添加顺序不仅会影响水解的程度,也会影响水解产物的组成。Raweesri等[57]通过乙酰木聚糖酯酶和木聚糖酶的协同作用分析发现乙酰木聚糖酯酶及阿拉伯呋喃糖苷酶等侧链酶对木聚糖的降解起着很大的促进作用。Moriyoshi等[29]通过乙酰木聚糖酯酶和纤维素酶协同降解醋酸纤维素,结果显示,当两种酶同时作用时,无论是脱乙酰的比率还是降解所产生还原糖的得率都得到了大幅度的提升,均提高了60%以上,说明乙酰木聚糖酯酶的存在能够提高醋酸纤维素分子中β-1,4-糖苷键的水解效率,醋酸纤维素中乙酰基的脱除能够为纤维素酶提供新的作用位点。Huy等[38]也通过研究发现,乙酰木聚糖酯酶rPcAxe2与木聚糖酶rPcXynC协同作用于山毛榉木聚糖、桦木木聚糖及小麦阿拉伯木聚糖能够大大提高还原糖的得率。Tong等[49]通过半纤维素酶水解小麦阿拉伯木聚糖发现,当木聚糖水解酶和乙酰木聚糖酯酶协同作用于小麦阿拉伯木聚糖时,总糖得率由原来木聚糖酶单独作用时的8%提高至二者协同作用时的34%。
6 乙酰木聚糖酯酶的应用前景及展望工业上降解木聚糖经常采用酸法或碱法,但酸、碱水解过程往往伴有副反应,产生较多的有毒物质,对于后期微生物发酵有抑制作用,且产生的酸、碱废液会造成较大的环保压力,因此,酶法降解木聚糖以其温和环保的优势将会成为未来木聚糖降解的主要方式。乙酰木聚糖酯酶作为半纤维素酶系成员之一具有良好的应用前景。在工程菌构建方面,乙酰木聚糖酯酶的研究对于构建产量高、协同效果优异的半纤维素酶工程菌具有重要意义。在工业应用方面,乙酰木聚糖酯酶能够协同内切木聚糖酶和β -木糖苷酶将半纤维素彻底降解为木糖,进而转化为木糖醇等。另外,一些乙酰木聚糖酯酶能够作用于甲壳素的乙酰基[58, 59],因此可以应用酶法制备壳聚糖,在药物、纺织、造纸、化妆品等方面具有广阔应用前景。
目前,相对于其他木质纤维素降解酶系,对乙酰木聚糖酯酶的研究还远远不够,乙酰木聚糖酯酶与其他木质纤维素降解酶系的协同作用已经得到认可,其潜在应用价值还有待开发。
致谢 感谢厦门大学校长基金(20720150090)对本文的资助。
| [1] | Prade R A. Xylanases: from biology to biotechnology. Biotechnology and Genetic Engineering Reviews, 1996, 13(1): 101-132. |
| [2] | Tang C, Guo J, Wu M, et al. Cloning and bioinformatics analysis of a novel acidophilic β-mannanase gene, Auman5A, from Aspergillus usamii YL-01-78. World Journal of Microbiology and Biotechnology, 2011, 27(12): 2921-2929. |
| [3] | Wang J, Zhang H, Wu M, et al. Cloning and sequence analysis of a novel xylanase gene, Auxyn10A, from Aspergillus usamii. Biotechnology Letters, 2011, 33(5): 1029-1038. |
| [4] | Koutaniemi S, Van Gool M, Juvonen M, et al. Distinct roles of carbohydrate esterase family CE16 acetyl esterases and polymer-acting acetyl xylan esterases in xylan deacetylation. Journal of Biotechnology, 2013, 168(4): 684-692. |
| [5] | Biely P. Microbial xylanolytic systems. Trends in Biotechnology, 1985, 3(11): 286-290. |
| [6] | Bouveng H, Garegg P, Lindberg B. Position of the O-acetyl groups in Birch Xylan. London:SOC Chemical Industry 14 Belgrave Souare, London SW1X 8PS, England, 1958: 1727-1727. |
| [7] | Biely P, Mastihubová M, Tenkanen M, et al. Action of xylan deacetylating enzymes on monoacetyl derivatives of 4-nitrophenyl glycosides of β-D-xylopyranose and α-L-arabinofuranose. Journal of Biotechnology, 2011, 151(1): 137-142. |
| [8] | Christov L P, Prior B A. Esterases of xylan-degrading microorganisms: production, properties, and significance. Enzyme and Microbial Technology, 1993, 15(6): 460-475. |
| [9] | Poutanen K, Sundberg M, Korte H, et al. Deacetylation of xylans by acetyl esterases of Trichoderma reesei. Applied Microbiology and Biotechnology, 1990, 33(5): 506-510. |
| [10] | Dalrymple B P, Cybinski D H, Layton I, et al. Three Neocallimastix patriciarum esterases associated with the degradation of complex polysaccharides are members of a new family of hydrolases. Microbiology, 1997, 143(8): 2605-2614. |
| [11] | Kormelink F, Lefebvre B, Strozyk F, et al. Purification and characterization of an acetyl xylan esterase from Aspergillus niger. Journal of Biotechnology, 1993, 27(3): 267-282. |
| [12] | Kool M M, Schols H A, Wagenknecht M, et al. Characterization of an acetyl esterase from Myceliophthora thermophila C1 able to deacetylate xanthan. Carbohydrate Polymers, 2014, 111: 222-229. |
| [13] | Pouvreau L, Jonathan M, Kabel M, et al. Characterization and mode of action of two acetyl xylan esterases from Chrysosporium lucknowense C1 active towards acetylated xylans. Enzyme and Microbial Technology, 2011, 49(3): 312-320. |
| [14] | Waters D M, Murray P G, Miki Y, et al. Cloning, overexpression in Escherichia coli, and characterization of a thermostable fungal acetylxylan esterase from Talaromyces emersonii. Applied and Environmental Microbiology, 2012, 78(10): 3759-3762. |
| [15] | Koseki T, Miwa Y, Akao T, et al. An Aspergillus oryzae acetyl xylan esterase: molecular cloning and characteristics of recombinant enzyme expressed in Pichia pastoris. Journal of Biotechnology, 2006, 121(3): 381-389. |
| [16] | Till M, Goldstone D C, Attwood G T, et al. Structure and function of an acetyl xylan esterase (Est2A) from the rumen bacterium Butyrivibrio proteoclasticus. Proteins: Structure, Function, and Bioinformatics, 2013, 81(5): 911-917. |
| [17] | Topakas E, Kyriakopoulos S, Biely P, et al. Carbohydrate esterases of family 2 are 6-O-deacetylases. FEBS Letters, 2010, 584(3): 543-548. |
| [18] | Dupont C, Daigneault N, Shareck F, et al. Purification and characterization of an acetyl xylan esterase produced by Streptomyces lividans. Biochem J, 1996, 319(3): 881-886. |
| [19] | Biely P, Mastihubová M, Puchart V. The vicinal hydroxyl group is prerequisite for metal activation of Clostridium thermocellum acetylxylan esterase. Biochimica et Biophysica Acta (BBA)-General Subjects, 2007, 1770(4): 565-570. |
| [20] | Emino lu A, lker S, Sandalli C. Cloning, purification and characterization of acetyl xylane esterase from Anoxybacillus flavithermus DSM 2641T with activity on low molecular-weight acetates. The Protein Journal, 2015, 34(4): 237-242. |
| [21] | Hakulinen N, Tenkanen M, Rouvinen J. Three-dimensional structure of the catalytic core of acetylxylan esterase from Trichoderma reesei: insights into the deacetylation mechanism. Journal of Structural Biology, 2000, 132(3): 180-190. |
| [22] | Blum D L, Li X L, Chen H, et al. Characterization of an acetyl xylan esterase from the anaerobic fungus Orpinomyces sp. strain PC-2. Applied and Environmental Microbiology, 1999, 65(9): 3990-3995. |
| [23] | Hedge M K, Gehring A M, Adkins C T, et al. The structural basis for the narrow substrate specificity of an acetyl esterase from Thermotoga maritima. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2012, 1824(9): 1024-1030. |
| [24] | Krastanova I, Guarnaccia C, Zahariev S, et al. Heterologous expression, purification, crystallization, X-ray analysis and phasing of the acetyl xylan esterase from Bacillus pumilus. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2005, 1748(2): 222-230. |
| [25] | Navarro-Fernández J, Martínez-Martínez I, Montoro-García S, et al. Characterization of a new rhamnogalacturonan acetyl esterase from Bacillus halodurans C-125 with a new putative carbohydrate binding domain. Journal of Bacteriology, 2008, 190(4): 1375-1382. |
| [26] | Uhliariková I, Vráanská M, Mccleary B V, et al. Positional specifity of acetylxylan esterases on natural polysaccharide: An NMR study. Biochimica et Biophysica Acta (BBA)-General Subjects, 2013, 1830(6): 3365-3372. |
| [27] | Biely P, Cziszárová M, Uhliariková I, et al. Mode of action of acetylxylan esterases on acetyl glucuronoxylan and acetylated oligosaccharides generated by a GH10 endoxylanase. Biochimica et Biophysica Acta (BBA)-General Subjects, 2013, 1830(11): 5075-5086. |
| [28] | Chung H J, Park S M, Kim H R, et al. Cloning the gene encoding acetyl xylan esterase from Aspergillus ficuum and its expression in Pichia pastoris. Enzyme and Microbial Technology, 2002, 31(4): 384-391. |
| [29] | Moriyoshi K, Koma D, Yamanaka H, et al. Expression and characterization of a thermostable acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis involved in the degradation of insoluble cellulose acetate. Bioscience, Biotechnology, and Biochemistry, 2013, 77(12): 2495-2498. |
| [30] | Mcdermid K P, Mackenzie C R, Forsberg C W. Esterase activities of Fibrobacter succinogenes subsp. succinogenes S85. Applied and Environmental Microbiology, 1990, 56(1): 127-132. |
| [31] | Egana L, Gutierrez R, Caputo V, et al. Purification and characterization of two acetyl xylan esterases from Penicillium purpurogenum. Biotechnology and Applied Biochemistry, 1996, 24(1): 33-99. |
| [32] | Chávez R, Bull P, Eyzaguirre J. The xylanolytic enzyme system from the genus Penicillium. Journal of Biotechnology, 2006, 123(4): 413-433. |
| [33] | 刘伟娜,梁迪,王晓宇,等. 灰色链霉菌中乙酰木聚糖酯酶基因的克隆, 表达及酶学性质研究. 生物技术通报, 2015, 31(2): 153-159. Liu W N, Liang D, Wang X Y, et al. Cloning, expression and characterization of acetyl xylan esterase from Streptomyces griseus.Biotechnology Bulletin, 2015, 31(2): 153-159. |
| [34] | Huang Y C, Chen G H, Chen Y F, et al. Heterologous expression of thermostable acetylxylan esterase gene from Thermobifida fusca and its synergistic action with xylanase for the production of xylooligosaccharides. Biochemical and Biophysical Research Communications, 2010, 400(4): 718-723. |
| [35] | 朱天地, 殷欣, 邬敏辰, 等. 乙酰木聚糖酯酶基因的克隆, 表达及酶学性质研究. 食品与生物技术学报, 2014, 33(10): 1084-1089. Zhu T D, Yin X, Wu M C, et al. Gene cloning, expression and enzymatic characterization of acetyl xylan esterase. Journal of Food Science and Biotechnology, 2014, 33(10): 1084-1089. |
| [36] | Taylor E J, Gloster T M, Turkenburg J P, et al. Structure and activity of two metal ion-dependent acetylxylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases. Journal of Biological Chemistry, 2006, 281(16): 10968-10975. |
| [37] | Degrassi G, Okeke B C, Bruschi C V, et al. Purification and characterization of an acetyl xylan esterase from Bacillus pumilus. Applied and Environmental Microbiology, 1998, 64(2): 789-792. |
| [38] | Huy N D, Thiyagarajan S, Kim D H, et al. Cloning and characterization of a novel bifunctional acetyl xylan esterase with carbohydrate binding module from Phanerochaete chrysosporium. Journal of Bioscience and Bioengineering, 2013, 115(5): 507-513. |
| [39] | 汪文静. 白色链霉菌乙酰木聚糖酯酶基因的克隆、表达及酶学性质的研究. 北京: 北京林业大学, 生命科学与技术学院,2012. Wang W J. Gene Cloning, Expression and Characterization of Acetyl xylan Esterase from Streptomyces albus. Beijing: Beijing Forestry University, College of Biological Sciences and Biotechnology, 2012. |
| [40] | 陈艳. 乙酰木聚糖酯酶的优化表达及催化特性研究. 南京: 南京林业大学, 化学工程学院,2010. Chen Y. Fermentation Optimization and Catalytic Properties of Acetyl Xylan Esterase. Nanjing: Nanjing Forestry University, College of Chemical Engineerig, 2012. |
| [41] | 曹杰. 草菇乙酰木聚糖酯酶的重组表达及其酶学特性. 南京: 南京林业大学, 化学工程学院,2007. Cao J. Recombination Expression and Enzymatic Characterization of Acetate Xylan Esterase in Olvariella volvacea. Nanjing: Nanjing Forestry University, College of Chemical Engineerig, 2007. |
| [42] | Tian Q, Song P, Jiang L, et al. A novel cephalosporin deacetylating acetyl xylan esterase from Bacillus subtilis with high activity toward cephalosporin C and 7-aminocephalosporanic acid. Applied Microbiology and Biotechnology, 2014, 98(5): 2081-2089. |
| [43] | Biely P, Côté G L, Kremnick ý L, et al. Substrate specificity of acetylxylan esterase from Schizophyllum commune: mode of action on acetylated carbohydrates. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology, 1996, 1298(2): 209-222. |
| [44] | Biely P, Heinrichová K, Kru M. Induction and inducers of the pectolytic system in Aureobasidium pullulans. Current Microbiology, 1996, 33(1): 6-10. |
| [45] | Kremnick ý L, Biely P. β-Mannanolytic system of Aureobasidium pullulans. Archives of Microbiology, 1997, 167(6): 350-355. |
| [46] | Ghosh D, Sawicki M, Lala P, et al. Multiple conformations of catalytic serine and histidine in acetylxylan esterase at 0.90Å. Journal of Biological Chemistry, 2001, 276(14): 11159-11166. |
| [47] | Correia M A, Prates J A, Brás J, et al. Crystal structure of a cellulosomal family 3 carbohydrate esterase from Clostridium thermocellum provides insights into the mechanism of substrate recognition. Journal of Molecular Biology, 2008, 379(1): 64-72. |
| [48] | Puchart V, Gariépy M C, Shareck F, et al. Identification of catalytically important amino acid residues of Streptomyces lividans acetylxylan esterase a from carbohydrate esterase family 4. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2006, 1764(2): 263-274. |
| [49] | Tong X, Lange L, Grell M N, et al. Hydrolysis of wheat arabinoxylan by two acetyl xylan esterases from Chaetomium thermophilum. Applied Biochemistry and Biotechnology, 2015, 175(2): 1139-1152. |
| [50] | 田斌. N糖基化和CBM对乙酰木聚糖酯酶分泌尧活力及稳定性影响. 南京:南京林业大学, 化学工程学院,2012. Tian B. Effect of N-glycosylaton and CBM on Secretion, Activity and Stability of AXE1 Expressed in Pichia pastoris. Nanjing: Nanjing Forestry University, College of Chemical Engineerig,2012. |
| [51] | Levisson M, Sun L, Hendriks S, et al. Crystal structure and biochemical properties of a novel thermostable esterase containing an immunoglobulin-like domain. Journal of Molecular Biology, 2009, 385(3): 949-962. |
| [52] | Levisson M, Han G W, Deller M C, et al. Functional and structural characterization of a thermostable acetyl esterase from Thermotoga maritima. Proteins: Structure, Function, and Bioinformatics, 2012, 80(6): 1545-1559. |
| [53] | Biely P, Mackenzie C, Puls J, et al. Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Nature Biotechnology, 1986, 4(8): 731-733. |
| [54] | Selig M J, Knoshaug E P, Adney W S, et al. Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresource Technology, 2008, 99(11): 4997-5005. |
| [55] | Zhang J, Siika-Aho M, Tenkanen M, et al. The role of acetyl xylan esterase in the solubilization of xylan and enzymatic hydrolysis of wheat straw and giant reed. Biotechnology for Biofuels, 2011, 4(1): 1-10. |
| [56] | Puls J, Tenkanen M, Korte H E, et al. Products of hydrolysis of beechwood acetyl-4-O-methylglucuronoxylan by a xylanase and an acetyl xylan esterase. Enzyme and Microbial Technology, 1991, 13(6): 483-486. |
| [57] | Raweesri P, Riangrungrojana P, Pinphanichakarn P. α-L-arabinofuranosidase from Streptomyces sp. PC22: Purification, characterization and its synergistic action with xylanolytic enzymes in the degradation of xylan and agricultural residues. Bioresource Technology, 2008, 99(18): 8981-8986. |
| [58] | Liu X, Ding S. Molecular characterization of a new acetyl xylan esterase (AXEII) from edible straw mushroom Volvariella volvacea with both de-O-acetylation and de-N-acetylation activity. FEMS Microbiology Letters, 2009, 295(1): 50-56. |
| [59] | Morley K L, Chauve G, Kazlauskas R, et al. Acetyl xylan esterase-catalyzed deacetylation of chitin and chitosan. Carbohydrate Polymers, 2006, 63(3): 310-315. |
 2016, Vol. 36
2016, Vol. 36