2) Pearl Research Institute, Guangdong Ocean University, Zhanjiang 524088, China;
3) Guangdong Science and Innovation Center for Pearl Culture, Zhanjiang 524088, China;
4) Pearl Breeding and Processing Engineering Technology Research Center of Guangdong Province, Zhanjiang 524088, China;
5) Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, Zhanjiang 524088, China
Coloration patterns of animals are essential phenotypic characters related to their biological functions, including immunity (Clotfelter et al., 2007), thermoregulation (Smith et al., 2016), camouflage (Théry et al., 2005), selective mating (Houde and Endler, 1990), and speciation (Kocher, 2004). In addition, coloration can influence customers' choice, as they generally believe vivid and bright colors mean better quality (Shahidi and Brown, 1998; Wu and Sun, 2013). Species of phylum Mollusca exhibit different shell colors, and the shell color phenotype has been used as a breeding trait associated with survival, growth, and immunity (Brake et al., 2004; Wei et al., 2019). Therefore, shell color formation or pigmentation in molluscs, especially in pearl oyster, has attributed attention of researchers and were reported in multiple species. Genetic data analysis of noble scallop (Zheng et al., 2013) and oyster (Xing et al., 2018) has revealed that genes or genetic factors function in shell color, and environmental changes, including salinity (Sokolova and Berger, 2000) and diet (Liu et al., 2009), exert an influence on shell color. Transcriptome analysis provides a method to identify differentially expressed genes (DEGs) among different color phenotypes in aquatic animals. Transcriptome analysis of the clam Meretrix meretrix revealed that calcium signaling might participate in shell color patterning by activating the Notch pathway (Yue et al., 2015). In oysters with white shell, the Notch level was down-regulated by endocytosis (Feng et al., 2015). Betalain biosynthesis, tyrosine metabolism, and metalloporphyrins contribute to shell color formation in Yesso scallop Patinopecten yessoensis (Ding et al., 2015). Melanin synthesis and biomineralization associated genes may participate in the pigmentation of Yesso scallop (Sun et al., 2015) and Pinctada margaritifera (Lemer et al., 2015). Furthermore, color formation in Manila clam shell was associated with porphyrin and chlorophyll metabolism together with the calcium signaling pathway (Nie et al., 2020).
Pearl oysters exhibit multiple shell and pearl color phenotype variations, and their diversity is associated with color polymorphism of the mantle (Ky et al., 2019). The expression of biomineralization genes of the mantle leads to pearl production and shell formation (Joubert et al., 2010; Gardner et al., 2011; Wang et al., 2017). Shell color variation depends on genes involved in shell structure and pigment synthesis, and differential expression of genes involved in calcitic layer formation may also be involved in shell color variation (Lemer et al., 2015). In the biomineralization process of shell and pearl, the binding of aragonite to pigments also functions in nacre color formation (Snow et al., 2004). For example, changes in the expression of Pif-177, collagen alpha-1 XI chain precursor, and zinc metalloprotease, which participate in nacre formation and melanin biosynthesis, indicated that color variation or polymorphism occurred at different stages of shell formation (Lemer et al., 2015). Tyrosinase, a key enzyme that functions in biomineralization (Sun et al., 2015), is one of the potential factors involved in the coloration pigmentation of shell layers (Lemer et al., 2015).
Pearl oyster Pinctada fucata martensii, which mainly inhabits Southern China, Australia, Southeast Asia, Japan, and India (Li et al., 2018; Yang et al., 2020), is famous for producing high-quality pearls, and accounts for more than 90% of marine pearl production (He et al., 2020; Wu et al., 2022). The significant correlation between shell color and phenotypic traits can be used to select and cultivate varieties with characteristic shell color and growth traits. A three successive generation selection for yellow shell color in the base stock was conducted to produce the third-generation yellow-colored line. The base stock was developed by selecting yellow-colored individuals as parental breeders in the Liushagang stocks, Zhanjiang, Guangdong Province, China (Wang et al., 2012). The yellow-colored line of P. f. martensii displays yellow in the outer prismatic layer and white in the inner nacreous layer of shell (Figs.1A, B). The present study aimed to explore the molecular mechanism of pigmentation in these layers of P. f. martensii using mantle branch-specific ribonucleic acid (RNA) sequencing. The results can contribute to understand the underlying mechanism of shell color formation and promote the selection process for the desired color of shell and pearl.
|
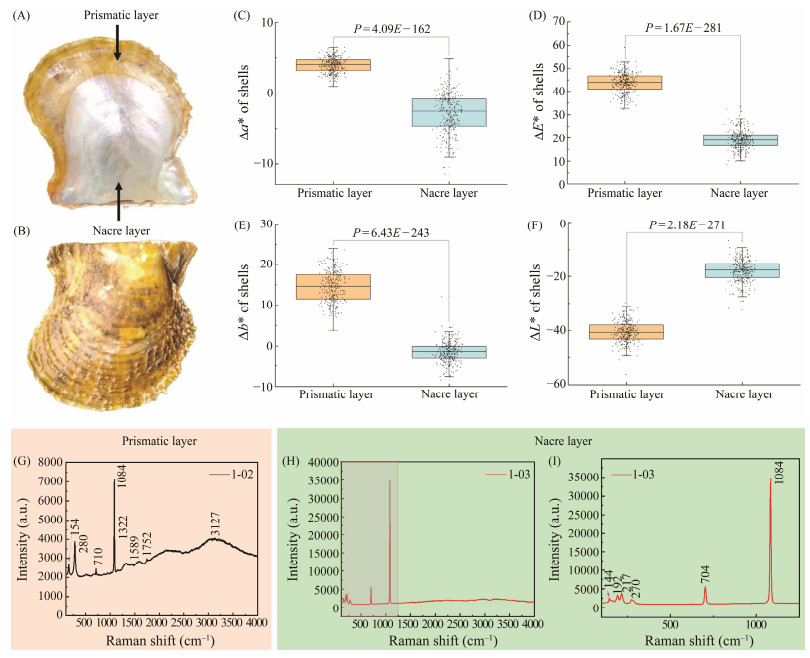
Fig. 1 Characteristics of yellow-colored line of P. f. martensii. (A) and (B) show the color phenotypes of yellow-colored line of P. f. martensii. (C), (D), (E), and (F) show the result of Δa*, ΔE, Δb*, and ΔL*, respectively. (G) shows microRaman spectroscopy analysis of prismatic layer. (H) shows micro-Raman spectroscopy analysis of nacreous layer. (I) is the enlargement of (H, left). |
A total of 284 pearl oysters with a mean shell length of 58.32 mm ± 4.96 mm were selected from our breed of the yellow-colored line of P. f. martensii (Deng et al., 2013) and cleaned to remove fouling animals. The yellow-colored line of P. f. martensii displays yellow in the outer prismatic layer and white in the inner nacreous layer of shell (Figs.1A, B). Regions of the nacreous and prismatic layers (three locations each) of shell were selected. Measurements were taken using a precision colorimeter (NR60CP; 3nh, Shenzhen, China) to obtain four parameters: L (lightness), a (red/green intensity), b (yellow/blue intensity), and ∆E (color difference). L > 0 indicates lighter shades, whereas L < 0 indicates darker shades. a > 0 indicates reddish tones, while a < 0 indicates greenish tones. b > 0 indicates yellowish hues, while b < 0 indicates bluish hues. ∆E represents the color difference, while higher ∆E values indicate darker colors. Marginal zone of mantle (ME) and central zone of mantle (MC) were collected from the P. f. martensii yellow-colored line, instantly preserved in liquid nitrogen, and stored at −80℃ for later analysis.
2.2 Micro-Raman Spectroscopy InvestigationThe shells were thoroughly cleaned and cut into small squares with a size of 0.5 cm × 0.5 cm. The nacreous and prismatic layers of the shells were dissected. High-resolution confocal Raman microscopy (laser: 532 nm; alpha-300R; WITec, Ulm, Germany) was used for detection. The testing conditions included a laser power of 40.0 mW, a grating of 300 g mm−1, a 20× objective lens (Olympus, Tokyo, Japan), an integration time of 20 s, and an accumulation of 8 times. Raman spectra were collected from three regions of each sample, and the data were processed using Origin 2019b software (OriginLab Corporation, Northampton, MA, USA).
2.3 Sequencing and Bioinformatics Analysis of MC and ME TranscriptomesTotal RNA from ME and MC was extracted separately with TRIzol (Invitrogen, Waltham, MA, USA) according to the manufacturer's instructions, and messenger RNA (mRNA) was enriched by Oligo (dT) magnetic beads. The enriched mRNA was fragmented and reverse-transcribed into complementary DNA (cDNA). Then, the prepared cDNA was sequenced on the NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA). Raw reads were filtered with fastp (v0.18.0) to obtain clean reads. HISAT2.2.4 was used to analyze the mapping of clean reads to the reference genome. The fragments per kilobase of transcript per million mapped reads (FPKM) of genes were analyzed by RSEM to evaluate gene expression.
2.4 DEG Identification and AnalysisDEGs were analyzed by DESeq2 (|log2 (FC)| > 1 (FC is fold change), and false discovery rate (FDR) < 0.05). They were further classified according to Gene Ontology (GO) terms (http://www.geneontology.org/) and GO enrichment (P ≤ 0.05). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs was also performed (P ≤ 0.05).
3 Results 3.1 Color Values of Shell ColorColor values of the prismatic and nacreous layers were detected using the International Commission on Illumination (CIE) model with the parameters of a, b, L, and ΔE. The values of Δa* and Δb* were calculated, and the higher values indicate greater red and yellow coloration, respectively. These values were significantly higher for the prismatic layer than for the nacreous layer (Figs.1C, D, E, F; P < 0.05). ΔL of the prismatic layer was significantly lower than that of the nacreous layer (Figs.1C, D, E, F; P < 0.05), indicating that the prismatic layer had a darker color.
3.2 Raman Spectra of ShellThe Raman spectra of the prismatic layer are shown in Fig.1G. A broad peak was observed at 3127 cm−1, which might be attributed to the stretching vibration of O-H or C-H in carotenoids present in the solution. The small peak at 1752 cm−1 was likely the sum frequency of the peaks at 710 and 1084 cm−1, indicating the vibration of carbonate ions. The small peak at 1589 cm−1 might have arisen from the stretching vibration of C=C in carotenoids. The small peak at 1322 cm−1 might be attributed to the out-of-plane bending vibration of C-H. These peaks suggest the presence of organic phase carotenoids in the prismatic layer. The strongest peak at 1084 cm−1 could be attributed to the symmetric stretching vibration of carbonate ions. Figs.1H and 1I show the Raman spectra of the nacre, which mainly concentrated between 100 and 1250 cm−1. The two characteristic peaks represent the stretching and bending vibrations of calcium carbonate at 1084 cm−1 and 704 cm−1, respectively (Zhang et al., 2001; Withnall et al., 2003).
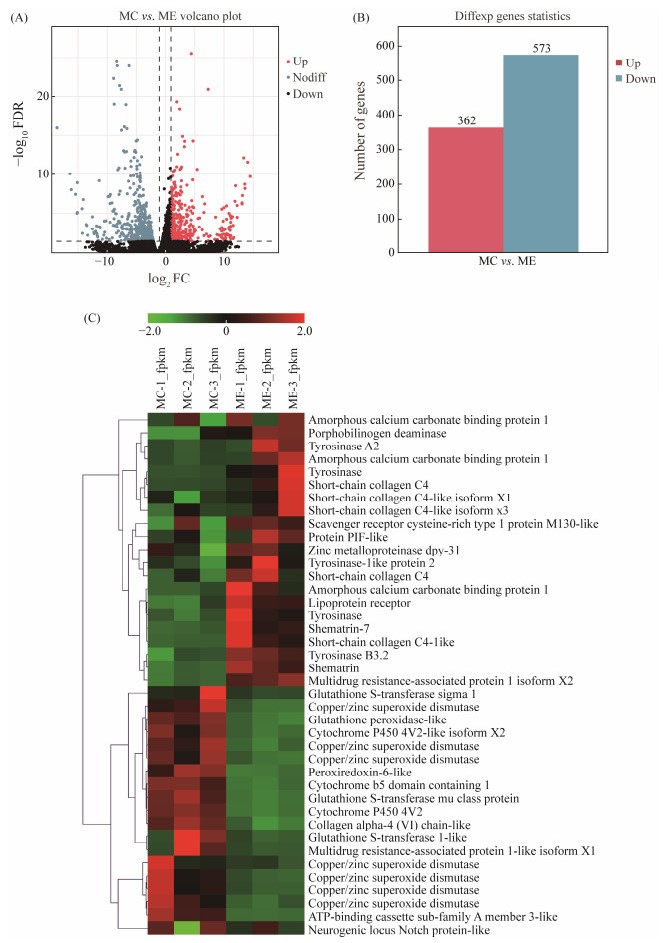
3.3 Transcriptome Sequencing and DEG AnalysisA total of 40944221 and 41112721 raw reads from ME and MC were sequenced respectively. From these reads, 40848235 and 41032457 clean reads were obtained, and the rate of clean reads mapped to the reference genome was 62.23% and 64.69%, respectively (Table 1). DEG identification and the transcriptome analysis results showed that 935 DEGs were obtained from MC and ME (FDR < 0.05 and |log2FC| > 1) (Fig.2A). Among these DEGs, the expressions of 573 genes were significantly downregulated, while the expressions of 362 genes were significantly upregulated in ME compared with MC (Fig.2B). DEGs were associated with melanoma/melanogenesis, such as tyrosinase, zinc metalloprotease, glutathione S-transferase (GST), and ATP-binding cassette sub-family; the carotenoid-related pathway, including scavenger receptors, cytochrome P450, and lipoprotein receptors; porphyrin metabolism, such as porphobilinogen deaminase and copper/zinc superoxide dismutase (SOD); and shell matrix proteins, including amorphous calcium carbonate binding protein (ACCBP), shematrin, Pif, and collagen (Fig.2C).
|
|
Table 1 Transcriptome analysis of ME and MC |
|
Fig. 2 Analysis results of DEGs. (A) Volcano plot of DEGs. Red and blue dots represent up-regulated and down-regulated genes in the ME compared with MC, respectively. (B) DEGs in ME and MC groups. Red and blue columns represent the number of up-regulated and down-regulated genes in the ME and MC, respectively. (C) Heatmap of the expression profiles of focus DEGs. Red and green colors represent high expression and low expression, respectively. |
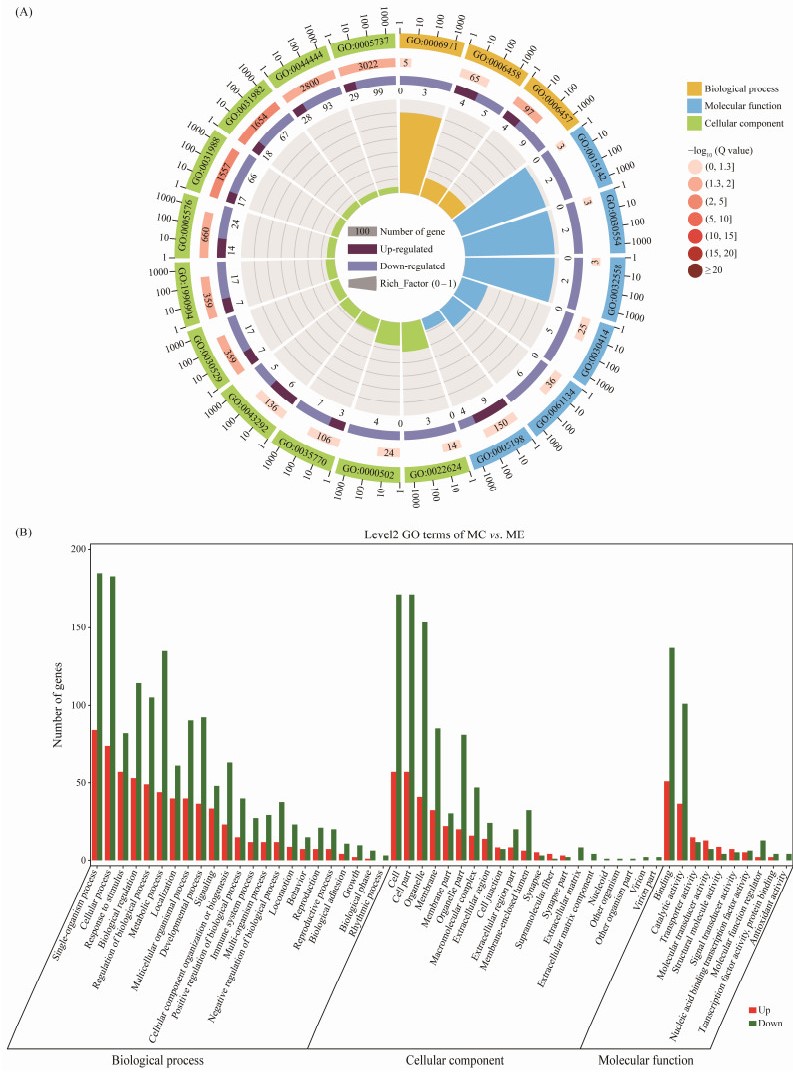
GO enrichment analysis of DEGs (Fig.3) showed that they were enriched in 2431 GO terms, including 206 GO terms that showed significant enrichment (P < 0.05). The significantly enriched GO terms involved 139 'biological process' terms, 44 'molecular function' terms, and 23 'cellular component' terms. GO terms with the most significant enrichment are shown in Fig.3, which include membrane-bounded vesicle (GO: 0031988), vesicle (GO: 0031 982), peptidase inhibitor activity (GO: 0030414), tricarboxylic acid transmembrane transporter activity (GO: 00151 42), and membrane transport-related terms.
|
Fig. 3 Enrichment analysis of DEGs. (A) The circle diagram of GO enrichment. Circle 1: the top 20 GO terms in the enrichment analysis, and the outside circle is the coordinate scale for the number of DEGs. Different colors represent different Ontologies. Circle 2: the DEG number and the Q value of the GO term in the background. The longer bars means more number of DEGs, and the redder color means lower Q value. Circle 3: the number of DEGs in the GO term. Circle 4: the Rich_Factor value of each GO term. (B) The histogram of GO enrichment classification. Red and green colors show the numbers of up-regulated and down-regulated genes in GO terms, respectively. |
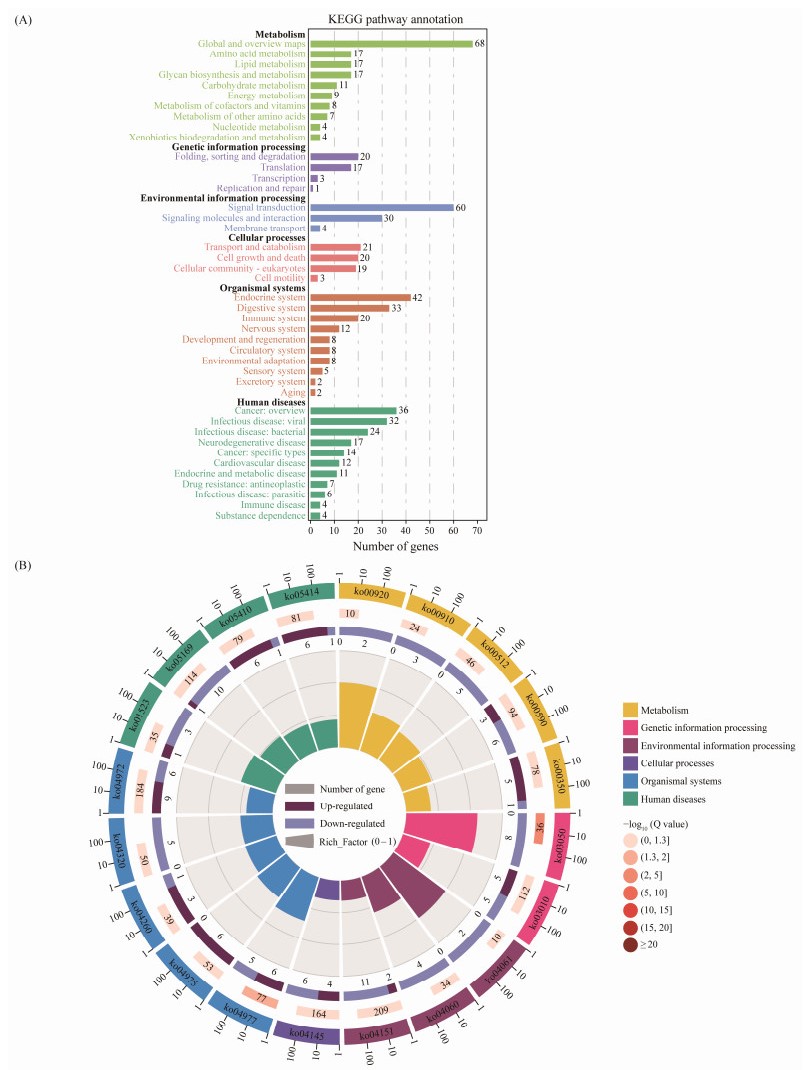
KEGG pathway enrichment analysis of DEGs showed that they were significantly enriched in 17 pathways (P < 0.05) (Fig.4), such as proteasome, vitamin digestion and absorption, and arachidonic acid metabolism. DEGs were also enriched in pigmentation or color-related signaling pathways, such as retinol metabolism, tyrosine metabolism, ABC transporter pathway, calcium signaling pathway, porphyrin metabolism, and melanogenesis.
|
Fig. 4 Pathway analysis of DEGs from ME and MC. (A) KEGG pathway annotation analysis of DEGs. Cyan, purple, blue, pink, red, and green represent metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, human diseases, respectively. (B) The circle diagram of KEGG enrichment. Circle 1: the top 20 pathways in the enrichment analysis, and the outside circle is the coordinate scale for the number of DEGs. Different colors represent different classes. Circle 2: the DEG number and the Q value of the pathway in the background. The longer bars mean higher number of DEGs, and the redder color means lower DEGs. Circle 3: the number of DEGs in the pathway. Circle 4: the Rich_Factor value of each pathway. |
The color of the prismatic and nacreous layers of the shell is crucial when selecting donor and recipient pearl oysters for producing pearls with desirable color (Wada and Komaru, 1996). The prismatic layer is formed by secretions from the ME area, whereas the nacreous layer is formed by secretions from the MC area (Marin et al., 2000). To understand the reasons causing the shell color variation, it is necessary to identify and quantify the substances responsible for shell coloration. Transcriptome profiling and functional analysis offer a suitable approach to gain a better understanding of how genetic and molecular factors contribute to differences in color-causing substances, ultimately influencing the coloration of both shell and pearl.
4.1 Melanin SynthesisMelanin, comprising two types of pigments eumelanin and pheomelanin, occurs in molluscs (Williams, 2017), and the colors of eumelanin and pheomelanin are brown to black and yellow to red, respectively (Ito and Wakamatsu, 2003). Enzymes and regulators are involved in melanin synthesis in molluscs. In the present study, melanin-related genes, including laccase and tyrosinase genes, were identified as DEGs. In melanogenesis, tyrosinase, a key rate-limiting enzyme, participated in the catalytic processes of hydroxyllation and oxidation (Oetting, 2000). Tyrosinases also contribute to mollusc tissue and shell formation, including native immune response (Asokan et al., 1997; Cong et al., 2005) and shell matrix formation (Feng et al., 2017). In P. persica, tyrosinases functioned in calcitic layer formation of shell and melanin synthesis between different color morphs (Stenger et al., 2021b), and similar results were also found in P. fucata (Nagai et al., 2007), Pteria penguin oyster (Yu et al., 2018), Pacific oyster Crassostrea gigas (Yu et al., 2014), Illex argentinus (Naraoka et al., 2003), and hard clam Mercenaria mercenaria (Hu et al., 2020). In P. fucata, zinc metalloprotease, a homolog of tyrosinase-related protein 1, participated in melanin biosynthesis of the periostracum layer (Zhang et al., 2006). Different from the results of Lemer et al. (2015) and Ky et al. (2019), tyrosinase (TYR) expression was significantly higher in ME than in MC. Orange phenotype may be associated with non-functional melanin protein composition, such as zinc metalloprotease (Parvizi et al., 2023). Laccase, tyrosinase, and zinc metalloprotease genes were more highly expressed in ME compared with MC in our study. Parvizi et al. (2023) also reported that TYR2A had higher expression in the orange phenotype of P. persica (Parvizi et al., 2023).
Melanin synthesis is regulated by transcriptional regulatory factors and signal transduction pathways (Walter et al., 2019). Calcium participates in the regulation of cellular processes (Petersen et al., 2005), and calcium ion binding genes were enriched in the present study. The Notch and calcium signaling pathways contributed to the development of pigment-producing cells in shell color formation in M. meretrix (Yue et al., 2015). Here, genes of the calcium signaling pathway were also enriched, and the neurogenic locus Notch protein-like gene showed higher expression in ME than in MC. ATP-binding cassette transporters (ABC transporters), one of the largest families of transporters, could hydrolyze ATP during the transport of substrates across membranes and showed an association with pigmentation in Arthropoda (Ito and Wakamatsu, 2003). Silkworm ABC gene (BmABCG6) showed low expression in the eyes and eggs of white mutants (Abraham et al., 2000; Xie et al., 2012). In oysters and hard clams, ABC transporter genes were associated with white coloration (Feng et al., 2015; Hu et al., 2020). White gene, as a homologous gene of ABC subfamily member were also researched and showed their potential function in pigmentation. In the compound eyes of Drosophila melanogaster, white gene could play a role in the transport of pigment precursors to pigment cells (Ewart and Howells, 1998; Xiao et al., 2017). Silkworm white gene (Bmwh3) inhibition led to white mutants in silkworm eggs (Quan et al., 2002). The knockdown of the Ar-white gene in Athalia rosae led to insufficience (nearly 60%) or complete absence (33%) of eye pigmentation in embryos (Sumitani et al., 2005). Therefore, ABC transporters, which were also enriched in the present study, might be responsible for shell pigmentation.
4.2 Carotenoid-Related PathwayCarotenoids have been identified in noble scallop Chlamys nobilis (Liu et al., 2015), pearl mussel Hyriopsis cumingii (Li et al., 2014), and Yesso scallop (Wade et al., 2009; Zhao et al., 2017) for their contribution to orange, red, and yellow shell color formation (Shahidi and Brown, 1998). Studies have shown that bivalves cannot synthesize carotenoids but can accumulate them from a microalgal diet (Petersen et al., 2005). Therefore, carotenoid pigmentation depends on carotenoid metabolism. Low-density lipoprotein and scavenger receptors, which are key receptors for carotenoid uptake (Walsh et al., 2012; Toews et al., 2017), showed higher expressions in ME in the present study. Low-density lipoprotein receptors participate in the pigmentation of Yesso scallop by accumulating carotenoids (Zhao et al., 2017). Scavenger receptors recognize lipoproteins with carotenoids and promote carotenoid transfer into cells (Kiefer et al., 2002; Sakudoh et al., 2013). They were found to participate in carotenoid absorption in P. f. martensii (Lei et al., 2017) and showed significant upregulated expression in yellow individuals compared with black individuals (Xu et al., 2019). Furthermore, the micro-Raman spectroscopy results indicated that the prismatic layer exhibited distinct characteristic peaks of carotenoids, whereas these peaks were not prominent in the nacreous layer. These findings indicate an important role of carotenoids in yellow shell pigmentation.
Stenger et al. (2023) identified cytochrome P450 genes that influence shell color of P. margaritifera. In our study, three genes from the cytochrome P450 family showed lower expression in ME than in MC. Cytochrome P450 3A4 (CYP3A4) could function in the metabolic processes of anthocyanidin and anthocyanin pigments, and showed lower expression in yellow shells than in darker shells of P. f. martensii (Srovnalova et al., 2014; Xu et al., 2019). CYP26A1, CYP2C8, and CYP3A29 could affect carotenoid concentrations by modifying the process of β-carotene degradation into all-trans-retinoic acids, resulting in β-carotene and pigmented carotenoid accumulation (Chichili et al., 2005; Hill and Johnson, 2012).
4.3 Porphyrin MetabolismTetrapyrroles, also known as porphyrins (either red or orange), comprise cyclic structures as shell pigments (Williams, 2017; Hu et al., 2020). In Clanculus pharaonius and C. margaritarius, uroporphyrin Ⅰ and Ⅲ were detected in shells (Williams et al., 2016). Porphyrin contributes to reddish-brown color formation or pigmentation of shells by combining with zinc and iron ions (Zheng et al., 2013; Ding et al., 2015). In this study, the porphobilinogen deaminase gene showed higher expression in ME than in MC. Pigmentation-related pathways generate reactive molecule porphyrins that can be oxidized and produce reactive oxygen species. Cu/Zn SOD catalyzes the dismutation of the superoxide radical into either hydrogen peroxide or ordinary molecular oxygen, and its expression is high in vitiligo patients because of the melanogenesis pathway perturbation (Wacewicz et al., 2018). In this study, seven copper/zinc SOD genes, three GST genes, and one glutathione peroxidase-like gene showed lower expressions in ME than in MC. Stenger et al. (2023) also reported that copper/zinc SOD influenced the shell color of P. margaritifera.
4.4 Shell Color Induced by Shell Matrix ProteinsIn addition to participating in nacre lamellae formation, restraining undesired growth of aragonite crystal faces, and stopping calcite nucleation and growth (Ma et al., 2007), ACCBP can also modify the shell color by physical regulation (Rousseau and Rollion-Bard, 2012; Stenger et al., 2021a). In this study, three ACCBP genes and one glutathione peroxidase-like gene were highly expressed in ME compared with MC. Stenger et al. (2023) also reported that ACCBP may impact the shell color of P. margaritifera. Shematrin proteins are important prismatic layer matrix proteins, which are synthesized in ME, and they provide a tenacious framework for calcification (Yano et al., 2006). In this study, two shematrin genes were highly expressed in ME compared with MC. Shem4 was also highly expressed in orange samples in the research of Ky et al. (2019).
In the present study, one Pif and six collagen genes were highly expressed in ME compared with MC. Pif-177 contributes to nacre growth in pearl oysters as organic matrix (Suzuki et al., 2009), which includes chitin-binding domain, protein-protein interaction domain, and aragonite binding domain (Feng et al., 2017). Pif plays an important role in shell color formation, and it shows lower expression in albino than in black individuals of P. margaritifera (Lemer et al., 2015). Pif expression was higher in orange individuals than in black individuals of P. persica (Parvizi et al., 2023), which was consistent with the results of the present study. Collagen VI-like gene is significantly expressed in the pallial mantle and responsible for nacre-layer formation in shells of pearl oyster (Zheng et al., 2017). Lemer et al. (2015) showed that both Pif and collagen functioned in the establishment of P. margaritifera shell color (Suzuki and Nagasawa, 2007). Thus, yellow pigmentation may be associated with special shell structure and biomineralization-related genes.
5 ConclusionsA total of 935 DEGs were identified in the transcriptome, with 362 genes upregulated and 573 genes downregulated in ME. Among these genes, those associated with melanoma/melanogenesis (tyrosinase, zinc metalloprotease, GST, and ATP-binding cassette sub-family), carotenoid-related pathway (scavenger receptors, cytochrome P450, and lipoprotein receptors), porphyrin metabolism (porphobilinogen deaminase and copper/zinc SOD), and shell matrix development (ACCBP, shematrin, Pif, and collagen) might result in the color difference between prismatic and nacreous layers in the yellow-colored line of pearl oyster P. f. martensii. These results provide insights into the complexity of shell color differentiation in shellfish and serve as a valuable resource for further research.
AcknowledgementsThis work was supported by the Science and Technology Program of Guangdong Province (No. 2022A1515010030), the National Natural Science Foundation of China (No. 32102817), the Program for Sientific Research Start-up Funds of Guangdong Ocean University (No. 060302022 304), the Department of Education of Guangdong Province (Nos. 2020ZDZX1045 and 2021KCXTD026), the Earmarked Fund for CARS-49, and the Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams (No. 2023KJ146).
Abraham, E. G., Sezutsu, H., Kanda, T., Sugasaki, T., Shimada, T., and Tamura, T., 2000. Identification and characterisation of a silkworm ABC transporter gene homologous to Drosophila white. Molecular and General Genetics, 21(1): 11-19. (  0) 0) |
Asokan, R., Arumugam, M., and Mullainadhan, P., 1997. Activation of prophenoloxidase in the plasma and haemocytes of the marine mussel Perna viridis Linnaeus. Developmental and Comparative Immunology, 21(1): 1-12. DOI:10.1016/S0145-305X(97)00004-9 (  0) 0) |
Bai, Z., Zheng, H., Lin, J., Wang, G., and Li, J., 2013. Comparative analysis of the transcriptome in tissues secreting purple and white nacre in the pearl mussel Hyriopsis cumingii. PLoS One, 8(1): e53617. DOI:10.1371/journal.pone.0053617 (  0) 0) |
Brake, J., Evans, F., and Langdon, C., 2004. Evidence for genetic control of pigmentation of shell and mantle edge in selected families of Pacific oysters Crassostrea gigas. Aquaculture, 229: 89-98. DOI:10.1016/S0044-8486(03)00325-9 (  0) 0) |
Chichili, G. R., Nohr, D., Schaffer, M., Von Lintig, J., and Biesalski, H. K., 2005. β-carotene conversion into vitamin A in human retinal pigment epithelial cells. Investigative Opthalmology and Visual Science, 46(10): 3562. DOI:10.1167/iovs.05-0089 (  0) 0) |
Clotfelter, E. D., Ardia, D. R., and McGraw, K. J., 2007. Red fish, blue fish: Trade-offs between pigmentation and immunity in Betta splendens. Behavioral Ecology, 18(6): 1139-1145. DOI:10.1093/beheco/arm090 (  0) 0) |
Cong, R., Sun, W., Liu, G., Fan, T., Meng, X., Yang, L., et al., 2005. Purification and characterization of phenoloxidase from clam Ruditapes philippinarum. Fish and Shellfish Immunology, 18(1): 61-70. DOI:10.1016/j.fsi.2004.06.001 (  0) 0) |
Deng, Y. W., Fu, S., Lu, Y. Z., Du, X. D., Wang, Q. H., Huang, H. L., et al., 2013. Fertilization, hatching, survival, and growth of third-generation colored pearl oyster (Pinctada martensii) stocks. Journal of Applied Aquaculture, 25: 113-120. DOI:10.1080/10454438.2013.788311 (  0) 0) |
Ding, J., Zhao, L., Chang, Y., Zhao, W., Du, Z., and Hao, Z., 2015. Transcriptome sequencing and characterization of Japanese scallop Patinopecten yessoensis from different shell color lines. PLoS One, 10(2): e0116406. DOI:10.1371/journal.pone.0116406 (  0) 0) |
Ewart, G. D., and Howells, A. J., 1998. ABC transporters involved in transport of eye pigment precursors in Drosophila melanogaster. Methods in Enzymology, 292: 213-224. (  0) 0) |
Feng, D., Li, Q., Yu, H., Kong, L., and Du, S., 2017. Identification of conserved proteins from diverse shell matrix proteome in Crassostrea gigas: Characterization of genetic bases regulating shell formation. Scientific Reports, 7(1): 45754. DOI:10.1038/srep45754 (  0) 0) |
Feng, D., Li, Q., Yu, H., Zhao, X., and Kong, L., 2015. Comparative transcriptome analysis of the Pacific oyster Crassostrea gigas characterized by shell colors: Identification of genetic bases potentially involved in pigmentation. PLoS One, 10(12): e0145257. DOI:10.1371/journal.pone.0145257 (  0) 0) |
Ferreira, B. S., De Almeida, C. G., Le Hyaric, M., De Oliveira, V. E., Edwards, H. G. M., and De Oliveira, L. F. C., 2013. Raman spectroscopic investigation of carotenoids in oils from amazonian products. Spectroscopy Letters, 46(2): 122-127. DOI:10.1080/00387010.2012.693569 (  0) 0) |
Gardner, L. D., Mills, D., Wiegand, A., Leavesley, D., and Elizur, A., 2011. Spatial analysis of biomineralization associated gene expression from the mantle organ of the pearl oyster Pinctada maxima. BMC Genomics, 12(1): 455. DOI:10.1186/1471-2164-12-455 (  0) 0) |
He, C. Z., Hao, R. J., Deng, Y. W., Yang, C. Y., and Du, X. D., 2020. Response of pearl oyster Pinctada fucata martensii to allograft-induced stress from lipid metabolism. Fish and Shellfish Immunology, 98(13): 1001-1007. (  0) 0) |
Hill, G. E., and Johnson, J. D., 2012. The vitamin A-redox hypothesis: A biochemical basis for honest signaling via carotenoid pigmentation. The American Naturalist, 180(5): E127-E150. DOI:10.1086/667861 (  0) 0) |
Houde, A. E., and Endler, J. A., 1990. Correlated evolution of female mating preferences and male color patterns in the guppy Poecilia reticulata. Science, 248(4961): 1405-1408. DOI:10.1126/science.248.4961.1405 (  0) 0) |
Hu, Z., Song, H., Zhou, C., Yu, Z. L., Yang, M. J., and Zhang, T., 2020. De novo assembly transcriptome analysis reveals the preliminary molecular mechanism of pigmentation in juveniles of the hard clam Mercenaria mercenaria. Genomics, 112(5): 3636-3647. DOI:10.1016/j.ygeno.2020.04.020 (  0) 0) |
Huang, R., Zheng, Z., Wang, Q., Zhao, X., Deng, Y., Jiao, Y., et al., 2015. Mantle branch-specific RNA sequences of moon scallop Amusium pleuronectes to identify shell color-associated genes. PLoS One, 10(10): e0141390. DOI:10.1371/journal.pone.0141390 (  0) 0) |
Ito, S., and Wakamatsu, K., 2003. Quantitative analysis of eumelanin and pheomelanin in humans, mice, and other animals: A comparative review. Pigment Cell Research, 16(5): 523-531. DOI:10.1034/j.1600-0749.2003.00072.x (  0) 0) |
Joubert, C., Piquemal, D., Marie, B., Manchon, L., Pierrat, F., Zanella-Cléon, I., et al., 2010. Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: Focus on biomineralization. BMC Genomics, 11(1): 613. DOI:10.1186/1471-2164-11-613 (  0) 0) |
Kiefer, C., Sumser, E., Wernet, M. F., and Von Lintig, J., 2002. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proceedings of the National Academy of Sciences, 99(16): 10581-10586. DOI:10.1073/pnas.162182899 (  0) 0) |
Kocher, T. D., 2004. Adaptive evolution and explosive speciation: The cichlid fish model. Nature Reviews Genetics, 5(4): 288-298. DOI:10.1038/nrg1316 (  0) 0) |
Ky, C. L., Blay, C., Broustal, F., Sham Koua, M., and Planes, S., 2019. Relationship of the orange tissue morphotype with shell and pearl colouration in the mollusc Pinctada margaritifera. Scientific Reports, 9(1): 5114. DOI:10.1038/s41598-019-41581-8 (  0) 0) |
Lei, C., Hao, R., Zheng, Z., Deng, Y., Wang, Q., and Li, J., 2017. Molecular cloning and characterisation of scavenger receptor class B in pearl oyster Pinctada fuctada martensii. Electronic Journal of Biotechnology, 30: 12-17. DOI:10.1016/j.ejbt.2017.08.003 (  0) 0) |
Lemer, S., Saulnier, D., Gueguen, Y., and Planes, S., 2015. Identification of genes associated with shell color in the blacklipped pearl oyster, Pinctada margaritifera. BMC Genomics, 16(1): 568. DOI:10.1186/s12864-015-1776-x (  0) 0) |
Li, J. H., Yang, C. Y., Wang, Q. H., Du, X. D., and Deng, Y. W., 2018. Growth and survival of host pearl oyster Pinctada fucata martensii (Dunker, 1880) treated by different biofouling-clean methods in China. Estuarine Coastal and Shelf Science, 207: 104-108. DOI:10.1016/j.ecss.2018.04.009 (  0) 0) |
Li, X., Bai, Z., Luo, H., Liu, Y., Wang, G., and Li, J., 2014. Cloning, differential tissue expression of a novel hcApo gene, and its correlation with total carotenoid content in purple and white inner-shell color pearl mussel Hyriopsis cumingii. Gene, 538(2): 258-265. DOI:10.1016/j.gene.2014.01.046 (  0) 0) |
Liu, H., Zheng, H., Zhang, H., Deng, L., Liu, W., Wang, S., et al., 2015. A de novo transcriptome of the noble scallop, Chlamys nobilis, focusing on mining transcripts for carotenoid-based coloration. BMC Genomics, 16(1): 44. DOI:10.1186/s12864-015-1241-x (  0) 0) |
Liu, X., Wu, F. C., Zhao, H. E., Zhang, G. F., and Guo, X. M., 2009. A novel shell color variant of the Pacific abalone Haliotis discus hannai Ino subject to genetic control and dietary influence. Journal of Shellfish Research, 28(2): 419-424. DOI:10.2983/035.028.0226 (  0) 0) |
Ma, Z., Huang, J., Sun, J., Wang, G., Li, C., Xie, L., et al., 2007. A novel extrapallial fluid protein controls the morphology of nacre lamellae in the pearl oyster, Pinctada fucata. Journal of Biological Chemistry, 282(32): 23253-23263. DOI:10.1074/jbc.M700001200 (  0) 0) |
Maoka, T., 2011. Carotenoids in marine animals. Marine Drugs, 9(2): 278-293. DOI:10.3390/md9020278 (  0) 0) |
Nagai, K., Yano, M., Morimoto, K., and Miyamoto, H., 2007. Tyrosinase localization in mollusc shells. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 146(2): 207-214. DOI:10.1016/j.cbpb.2006.10.105 (  0) 0) |
Naraoka, T., Uchisawa, H., Mori, H., Matsue, H., Chiba, S., and Kimura, A., 2003. Purification, characterization and molecular cloning of tyrosinase from the cephalopod mollusk, Illex argentinus. European Journal of Biochemistry, 270(19): 4026-4038. DOI:10.1046/j.1432-1033.2003.03795.x (  0) 0) |
Nie, H., Jiang, K., Jiang, L., Huo, Z., Ding, J., and Yan, X., 2020. Transcriptome analysis reveals the pigmentation related genes in four different shell color strains of the Manila clam Ruditapes philippinarum. Genomics, 112(2): 2011-2020. DOI:10.1016/j.ygeno.2019.11.013 (  0) 0) |
Oetting, W. S., 2000. The tyrosinase gene and oculocutaneous albinism type 1 (oca1): A model for understanding the molecular biology of melanin formation. Pigment Cell Research, 13(5): 320-325. DOI:10.1034/j.1600-0749.2000.130503.x (  0) 0) |
Oetting, W. S., and King, R. A., 1993. Molecular basis of type Ⅰ (tryrosinase-related) oculocutaneous albinism: Mutations and polymorphisms of the human tyrosinase gene. Human Mutation, 2(1): 1-6. DOI:10.1002/humu.1380020102 (  0) 0) |
Parvizi, F., Akbarzadeh, A., Farhadi, A., Arnaud-Haond, S., and Ranjbar, M. S., 2023. Expression pattern of genes involved in biomineralization in black and orange mantle tissues of pearl oyster, Pinctada persica. Frontiers in Marine Science, 9: 1038692. DOI:10.3389/fmars.2022.1038692 (  0) 0) |
Petersen, O. H., Michalak, M., and Verkhratsky, A., 2005. Calcium signalling: Past, present and future. Cell Calcium, 38: 161-169. DOI:10.1016/j.ceca.2005.06.023 (  0) 0) |
Quan, G. X., Kanda, T., and Tamura, T., 2002. Induction of the white egg 3 mutant phenotype by injection of the double-stranded RNA of the silkworm white gene. Insect Molecular Biology, 11(3): 217-222. DOI:10.1046/j.1365-2583.2002.00328.x (  0) 0) |
Rousseau, M., and Rollion-Bard, C., 2012. Influence of the depth on the shape and thickness of nacre tablets of Pinctada margaritifera pearl oyster, and on oxygen isotopic composition. Minerals, 2(1): 55-64. DOI:10.3390/min2010055 (  0) 0) |
Ruiz-Márvez, E., Ramírez, C. A., Rodríguez, E. R., Flórez, M. M., Delgado, G., Guzmán, F., et al., 2020. Molecular characterization of Tc964, a novel antigenic protein from Trypanosoma cruzi. International Journal of Molecular Sciences, 21(7): 2432. DOI:10.3390/ijms21072432 (  0) 0) |
Sakudoh, T., Kuwazaki, S., Iizuka, T., Narukawa, J., Yamamoto, K., Uchino, K., et al., 2013. CD36 homolog divergence is responsible for the selectivity of carotenoid species migration to the silk gland of the silkworm Bombyx mori. Journal of Lipid Research, 54(2): 482-495. DOI:10.1194/jlr.M032771 (  0) 0) |
Shahidi, F., and Brown, J. A., 1998. Carotenoid pigments in seafoods and aquaculture. Critical Reviews in Food Science and Nutrition, 38(1): 1-67. DOI:10.1080/10408699891274165 (  0) 0) |
Smith, K. R., Cadena, V., Endler, J. A., Porter, W. P., Kearney, M. R., and Stuart Fox, D., 2016. Colour change on different body regions provides thermal and signalling advantages in bearded dragon lizards. Proceedings of the Royal Society B: Biological Sciences, 283(1832): 20160626. DOI:10.1098/rspb.2016.0626 (  0) 0) |
Snow, M. R., Pring, A., Self, P., Losic, D., and Shapter, J., 2004. The origin of the color of pearls in iridescence from nano-composite structures of the nacre. American Mineralogist, 89(10): 1353-1358. DOI:10.2138/am-2004-1001 (  0) 0) |
Sokolova, I. M., and Berger, V. J., 2000. Physiological variation related to shell colour polymorphism in White Sea Littorina saxatilis. Journal of Experimental Marine Biology and Ecology, 245(1): 1-23. DOI:10.1016/S0022-0981(99)00132-X (  0) 0) |
Srovnalova, A., Svecarova, M., Kopecna Zapletalova, M., Anzenbacher, P., Bachleda, P., Anzenbacherova, E., et al., 2014. Effects of anthocyanidins and anthocyanins on the expression and catalytic activities of CYP2A6, CYP2B6, CYP2C9, and CYP3A4 in primary human hepatocytes and human liver microsomes. Journal of Agricultural and Food Chemistry, 62(3): 789-797. DOI:10.1021/jf404643w (  0) 0) |
Stenger, P., Ky, C., Vidal-Dupiol, J., Planes, S., and Reisser, C., 2023. Identifying genes associated with genetic control of color polymorphism in the pearl oyster Pinctada margaritifera var. cumingii (Linnaeus 1758) using a comparative whole genome pool-sequencing approach. Evolutionary Applications, 16(2): 408-427. DOI:10.1111/eva.13464 (  0) 0) |
Stenger, P. L., Ky, C. L., Reisser, C. M. O., Cosseau, C., Grunau, C., Mege, M., et al., 2021a. Environmentally driven color variation in the pearl oyster Pinctada margaritifera var. cumingii (Linnaeus, 1758) is associated with differential methylation of CpGs in pigment- and biomineralization-related genes. Frontiers in Genetics, 12: 630290. DOI:10.3389/fgene.2021.630290 (  0) 0) |
Stenger, P. L., Ky, C. L., Reisser, C. M. O., Duboisset, J., Dicko, H., Durand, P., et al., 2021b. Molecular pathways and pigments underlying the colors of the pearl oyster Pinctada margaritifera var. cumingii (Linnaeus 1758). Genes, 12(3): 421. (  0) 0) |
Sumitani, M., Yamamoto, D. S., Lee, J. M., and Hatakeyama, M., 2005. Isolation of white gene orthologue of the sawfly, Athalia rosae (Hymenoptera) and its functional analysis using RNA interference. Insect Biochemistry and Molecular Biology, 35(3): 231-240. DOI:10.1016/j.ibmb.2004.12.006 (  0) 0) |
Sun, X., Liu, Z., Zhou, L., Wu, B., Dong, Y., and Yang, A., 2016. Integration of next generation sequencing and EPR analysis to uncover molecular mechanism underlying shell color variation in scallops. PLoS One, 11(8): e0161876. DOI:10.1371/journal.pone.0161876 (  0) 0) |
Sun, X., Yang, A., Wu, B., Zhou, L., and Liu, Z., 2015. Characterization of the mantle transcriptome of yesso scallop (Patinopecten yessoensis): Identification of genes potentially involved in biomineralization and pigmentation. PLoS One, 10(8): e0122967. (  0) 0) |
Suzuki, M., and Nagasawa, H., 2007. The structure-function relationship analysis of Prismalin-14 from the prismatic layer of the Japanese pearl oyster, Pinctada fucata. FEBS Journal, 274(19): 5158-5166. DOI:10.1111/j.1742-4658.2007.06036.x (  0) 0) |
Suzuki, M., Saruwatari, K., Kogure, T., Yamamoto, Y., Nishimura, T., Kato, T., et al., 2009. An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science, 325(5946): 1388-1390. DOI:10.1126/science.1173793 (  0) 0) |
Théry, M., Debut, M., Gomez, D., and Casas, J., 2005. Specific color sensitivities of prey and predator explain camouflage in different visual systems. Behavioral Ecology, 16(1): 25-29. DOI:10.1093/beheco/arh130 (  0) 0) |
Toews, D. P. L., Hofmeister, N. R., and Taylor, S. A., 2017. The evolution and genetics of carotenoid processing in animals. Trends in Genetics, 33(3): 171-182. DOI:10.1016/j.tig.2017.01.002 (  0) 0) |
Wacewicz, M., Socha, K., Soroczyńska, J., Niczyporuk, M., Aleksiejczuk, P., Ostrowska, J., et al., 2018. Selenium, zinc, copper, Cu/Zn ratio and total antioxidant status in the serum of vitiligo patients treated by narrow-band ultraviolet-B phototherapy. Journal of Dermatological Treatment, 29(2): 190-195. DOI:10.1080/09546634.2017.1357797 (  0) 0) |
Wada, K. T., and Komaru, A., 1996. Color and weight of pearls produced by grafting the mantle tissue from a selected population for white shell color of the Japanese pearl oyster Pinctada fucata martensii (Dunker). Aquaculture, 142: 25-32. DOI:10.1016/0044-8486(95)01242-7 (  0) 0) |
Wade, N. M., Tollenaere, A., Hall, M. R., and Degnan, B. M., 2009. Evolution of a novel carotenoid-binding protein responsible for crustacean shell color. Molecular Biology and Evolution, 26(8): 1851-1864. DOI:10.1093/molbev/msp092 (  0) 0) |
Walsh, N., Dale, J., McGraw, K. J., Pointer, M. A., and Mundy, N. I., 2012. Candidate genes for carotenoid coloration in vertebrates and their expression profiles in the carotenoid-containing plumage and bill of a wild bird. Proceedings of the Royal Society B: Biological Sciences, 279(1726): 58-66. DOI:10.1098/rspb.2011.0765 (  0) 0) |
Walter, J., Selim, K. A., Leganés, F., Fernández-Piñas, F., Vothknecht, U. C., Forchhammer, K., et al., 2019. A novel Ca2+binding protein influences photosynthetic electron transport in Anabaena sp. PCC 7120. Biochimica et Biophysica Acta (BBA) – Bioenergetics, 1860(6): 519-532. (  0) 0) |
Wang, X., Liu, Z., and Wu, W., 2017. Transcriptome analysis of the freshwater pearl mussel (Cristaria plicata) mantle unravels genes involved in the formation of shell and pearl. Molecular Genetics and Genomics, 292(2): 343-352. DOI:10.1007/s00438-016-1278-9 (  0) 0) |
Wang, X. Y., Gao, Y. Z., Du, X. D., Deng, Y. W., and Lu, J., 2012. Genetic structure of the third generation yellow colored line and base stock of pearl oyster Pinctada martensii as revealed by SSR marker system. Marine Science Bulletin, 31(3): 324-328. DOI:10.3969/j.issn.1001-6392.2012.03.014 (  0) 0) |
Wei, L., Jiang, Q., Cai, Z., Yu, W., He, C., Guo, W., et al., 2019. Immune-related molecular and physiological differences between black-shelled and white-shelled Pacific oysters Crassostrea gigas. Fish and Shellfish Immunology, 92: 64-71. DOI:10.1016/j.fsi.2019.05.056 (  0) 0) |
Williams, S. T., 2017. Molluscan shell colour. Biological Reviews, 92(2): 1039-1058. DOI:10.1111/brv.12268 (  0) 0) |
Williams, S. T., Ito, S., Wakamatsu, K., Goral, T., Edwards, N. P., Wogelius, R. A., et al., 2016. Identification of shell colour pigments in marine snails Clanculus pharaonius and C. margaritarius (Trochoidea; Gastropoda). PLoS One, 11(7): e0156664. DOI:10.1371/journal.pone.0156664 (  0) 0) |
Withnall, R., Chowdhry, B. Z., and Silver, J., 2003. Raman spectra of carotenoids in natural products. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 59(10): 2207-2212. DOI:10.1016/S1386-1425(03)00064-7 (  0) 0) |
Wu, D., and Sun, D. W., 2013. Colour measurements by computer vision for food quality control – A review. Trends in Food Science and Technology, 29(1): 5-20. DOI:10.1016/j.tifs.2012.08.004 (  0) 0) |
Wu, H. L., Yang, C. Y., Hao, R. J., Liao, Y. S., Wang, Q. H., and Deng, Y. W., 2022. Lipidomic insights into the immune response and pearl formation in transplanted pearl oyster Pinctada fucata martensii. Frontiers in Immunology, 13: 1018423. DOI:10.3389/fimmu.2022.1018423 (  0) 0) |
Xiao, C., Qiu, S., and Robertson, R. M., 2017. The white gene controls copulation success in Drosophila melanogaster. Scientific Reports, 7(1): 7712. DOI:10.1038/s41598-017-08155-y (  0) 0) |
Xie, X., Cheng, T., Wang, G., Duan, J., Niu, W., and Xia, Q., 2012. Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in the silkworm, Bombyx mori. Molecular Biology Reports, 39(7): 7281-7291. DOI:10.1007/s11033-012-1558-3 (  0) 0) |
Xing, D., Li, Q., Kong, L., and Yu, H., 2018. Heritability estimate for mantle edge pigmentation and correlation with shell pigmentation in the white-shell strain of Pacific oyster, Crassostrea gigas. Aquaculture, 482: 73-77. DOI:10.1016/j.aquaculture.2017.09.026 (  0) 0) |
Xu, M., Huang, J., Shi, Y., Zhang, H., and He, M., 2019. Comparative transcriptomic and proteomic analysis of yellow shell and black shell pearl oysters, Pinctada fucata martensii. BMC Genomics, 20(1): 469. DOI:10.1186/s12864-019-5807-x (  0) 0) |
Yang, C. Y., Yang, J. M., Hao, R. J., Du, X. D., and Deng, Y. W., 2020. Molecular characterization of OSR1 in Pinctada fucata martensii and association of allelic variants with growth traits. Aquaculture, 516: 734617. DOI:10.1016/j.aquaculture.2019.734617 (  0) 0) |
Yano, M., Nagai, K., Morimoto, K., and Miyamoto, H., 2006. Shematrin: A family of glycine-rich structural proteins in the shell of the pearl oyster Pinctada fucata. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 144(2): 254-262. DOI:10.1016/j.cbpb.2006.03.004 (  0) 0) |
Yu, F., Pan, Z., Qu, B., Yu, X., Xu, K., Deng, Y., et al., 2018. Identification of a tyrosinase gene and its functional analysis in melanin synthesis of Pteria penguin. Gene, 656: 1-8. DOI:10.1016/j.gene.2018.02.060 (  0) 0) |
Yu, X., Yu, H., Kong, L., Guo, F., Zhu, G., and Li, Q., 2014. Molecular cloning and differential expression in tissues of a tyrosinase gene in the Pacific oyster Crassostrea gigas. Molecular Biology Reports, 41(8): 5403-5411. DOI:10.1007/s11033-014-3412-2 (  0) 0) |
Yue, X., Nie, Q., Xiao, G., and Liu, B., 2015. Transcriptome analysis of shell color-related genes in the clam Meretrix meretrix. Marine Biotechnology, 17(3): 364-374. DOI:10.1007/s10126-015-9625-0 (  0) 0) |
Zhang, C., Xie, L., Huang, J., Chen, L., and Zhang, R., 2006. A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata). Biochemical and Biophysical Research Communications, 342(2): 632-639. DOI:10.1016/j.bbrc.2006.01.182 (  0) 0) |
Zhang, G. S., Xie, X. D., and Wang, Y., 2001. Raman spectra of nacre from shells of main pearl-culturing mollusk in China. Spectroscopy and Spectral Analysis, 21(2): 193-196. DOI:10.3321/j.issn:1000-0593.2001.02.016 (  0) 0) |
Zhao, L., Li, Y. P., Li, Y. J., Yu, J., Liao, H., Wang, S., et al., 2017. A genome-wide association study identifies the genomic region associated with shell color in Yesso scallop, Patinopecten yessoensis. Marine Biotechnology, 19(3): 301-309. DOI:10.1007/s10126-017-9751-y (  0) 0) |
Zheng, H., Zhang, T., Sun, Z., Liu, W., and Liu, H., 2013. Inheritance of shell colours in the noble scallop Chlamys nobilis (Bivalve: Pectinidae). Aquaculture Research, 44(8): 1229-1235. DOI:10.1111/j.1365-2109.2012.03124.x (  0) 0) |
Zheng, Z., Du, X. D., Xiong, X. W., Jiao, Y., Deng, Y. W., Wang, Q. H., et al., 2017. PmRunt regulated by Pm-miR-183 participates in nacre formation possibly through promoting the expression of collagen VI-like and Nacrein in pearl oyster Pinctada martensii. PLoS One, 12(6): e0178561. DOI:10.1371/journal.pone.0178561 (  0) 0) |
 2024, Vol. 23
2024, Vol. 23


