2) College of Fisheries and Life, Shanghai Ocean University, Shanghai 201306, China;
3) College of Marine Life Sciences, Ocean University of China, Qingdao 266237, China
Interferons (IFNs) are enormous family of cytokines that play vital roles in defense, immune activation, and regulation of cell growth in vertebrates (Samuel, 2001). The IFN type Ⅰ and Ⅱ systems are pivotal in both innate and adaptive immunity against viral infection in teleost (Zou and Secombes, 2011). Viral dsRNA triggers IFNs to bind their receptors in virus-infected cells and subsequently initiate the JAK/STAT signaling pathway, which induces the transcription of interferon-stimulated genes (ISGs) (Robertsen, 2006). ISGs form the molecular mainstay of the innate immune system and limit the spread of virus as well as intra- and inter-cellular viral replication. Therefore, ISGs network is a critical therapeutic target against viral infections (Hubel et al., 2019).
Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1), also called IFN-induced protein 56 kDa (IFI56) or Interferon-stimulated protein 56 (ISG56), was initially cloned from human fibroblastoid cells treated with IFN-beta (Chebath et al., 1983; Wathelet et al., 1986). IFIT1/ ISG56 and other three IFIT family members including IFIT2/ISG54, IFIT3/ISG60, and IFIT5/ISG58 have been characterized in humans and found to be localized in a cluster on chromosome 10q23 (Lafage et al., 1992; Fensterl and Sen, 2011). Different IFIT family members contain distinct numbers and arrangements of tetratricopeptide repeats (TPR) motif which mediate protein-protein interactions, thus substantiate their multifunctional role in cell proliferation and migration, translation initiation, double-stranded RNA signal recognition, and inhibition of virus replication (D'Andrea and Regan, 2003; Fensterl and Sen, 2011; Diamond, 2014). Recent reports have demonstrated that IFIT1 can detect viral RNA as a sensor molecular and subsequently inhibit viral translation or sequester them for active replication function as effector molecules (Daffis et al., 2010; Kimura et al., 2013; Diamond, 2014). Meanwhile, human immunodeficiency virus type 1 (HIV-1) viral protein R (Vpr) upregulates IFIT1 gene expression in human monocyte-derived macrophages and dendritic cells, indicating its function in fighting HIV (Zahoor et al., 2014; Zahoor et al., 2015). IFIT proteins have broad-spectrum antiviral functions. They can fight against certain viruses by binding to the subunits of eukaryotic initiation factor (eIF3) translation initiation complex and inhibiting protein translation (Hui et al., 2003). Human IFIT1 and IFIT2 bind to eIF3E, while human IFIT2, mouse IFIT1, and mouse IFIT2 bind to eIF3C (Hui et al., 2005).
IFIT1 gene is conservatively found in mammalian species, birds, and amphibians. According to gene scan and sequence analysis, only a few IFIT1 homologs have been reported in fishes including large yellow croaker (Larimich-thyscrocea), zebrafish (Denio rerio), crucian carp (Carassius auratus L.), and olive flounder (Paralichthys olivaceus). From the previous results, IFIT1 gene expression is upregulated by fish IFNs, Poly (I: C) or viral hemorrhagic septicemia virus (VHSV) in zebrafish or olive flounder, indicating the important role of IFIT1 in response against viral infection (Zhang and Gui, 2004; Wan and Chen, 2008; Liu et al., 2013; Varela et al., 2014; Hwang et al., 2017). Giant grouper (E. lanceolatus) is a commercially important marine fish, and is the largest grouper type in the world (Williams, 2009). Because of its fast growth performance, E. lanceolatus is preferably selected as the male parent in order to produce the hybrid grouper with Epinephelus coicoides, Epinephelus fuscoguttatus, and Epinephelus moara and so on (Kiriyakit et al., 2011; Chen and Long, 2018; Chen et al., 2018). The chromosome-level genome assembly of giant grouper has been well-documented in 2019 and it provides the characteristics of innate immune response and rapid growth (Zhou et al., 2019). Many grouper farms have suffered from financial losses induced by viral infection. In the present study, we cloned the full length of IFIT1 gene in giant grouper and characterized its expression pattern in different tissues. Furthermore, we studied the expression level of ELIFIT1 after stimulation with SKIV. Our results will contribute to the knowledge of ELIFIT1 in grouper and its essential role in antiviral progress in giant grouper.
2 Materials and Methods 2.1 Ethical StatementExaminations and investigations were performed according to the guidelines and ethical standards of Regulations for the Administration of Affairs Concerning Experimental Animals of the State Science and Technology Commission of Shandong Province. This study was approved by the Ethics Committee of Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences.
2.2 Fish, Virus Infection, and Sample CollectionThirty-five healthy giant grouper (body weight: 700 g ± 25 g; body length: 30 cm ± 3 cm) were purchased from Chen-hai Aquaculture Company (Hainan, China) and cultured in circulating seawater at 25℃. Ten tissues (liver, spleen, kidney, intestine, gill, skin, muscle, brain, heart, and blood) were collected from five normal fish and transferred immediately into liquid nitrogen and stored at −80℃. Twenty-five fish were intraperitoneally injected with 106 TCID50/ fish spotted knifejaw iridovirus (SKIV) and five control fish were injected with sterilized phosphate-buffered saline (PBS, pH7.4) at a dose of 0.5 mL per 200 g fish. Spleen and kidney were collected from each group (5 fish each) at 0 h, 12 h, 24 h, 72 h, and 96 h after the infection.
2.3 Cloning of ELIFIT1 cDNA Sequence and Multiple AlignmentBased on the partial sequence of ELIFIT1 gene from the genome database (Zhou et al., 2019), we cloned the full length open reading frame (ORF) of ELIFIT1 using the primers (ELIFIT1-F1 and ELIFIT1-R1) listed in Table 1 through polymerase chain reaction (PCR) assay. Afterward, we obtained the 5' and 3' untranslated regions (UTR) of ELIFIT1 by rapid amplification of cDNA ends (RACE) PCR using SMART RACE 5'/3' Kit (Clontech) with the primers listed in Table 1. The cDNA sequence of ELIFIT1 was linked by DNAstar software and conserved domains of the predicted protein were analyzed using SMART program (http://smart.embl-heidelberg.de/). ProtScale (http://web.expasy.org/protscale/) was used for hydropathicity/ hydrophobicity analysis of the protein and TMHMM v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict the transmembrane domain.
|
|
Table 1 Primers used in this study |
The predicted protein was analyzed by BLAST program (http://www.ncbi.nlm.nih.gov/blast). Putative TPR motifs was identified using TPRpred (http://toolkit.tuebingen.mpg.de/tools/tprpred/) (Karpenahalli and Soding, 2007) by comparing it with Homo sapiens IFIT1 (HsIFIT1), Mus musculus IFIT1 (MmIFIT1) and Epinephelus coioides IFIT1 (EcIFIT1), and the E-value inclusion TRP & SEL is 1e-2. Putative amino acidsequence alignment was performed using ClustalX and ESPript 3.0 (http://espript.ibcp.fr/ES-Pript/cgi-bin/ESPript.cgi) (Robert and Gouet, 2014). A phylogenetic tree was constructed with the sequences listed in Table 2 using neighbor-joining algorithm. Reliability of the branching was tested through bootstrap resampling with 1000 replications.
|
|
Table 2 Information of IFIT protein family in various species |
Total RNA was extracted using Trizol Reagent (Takara, Dalian, China) from various tissues, respectively. The first strand cDNA was synthesized using PrimeScript RT Reverse kit (Takara, Dalian, China). Real-time quantitative PCR (RT-qPCR) using ELIFIT1-RT-F and ELIFIT1-RT-R was carried out according to the instructions for SYBR® Green I Premix Ex Taq™ (Takara, Japan) in a 7500 fast real-time PCR system (Applied Biosystems, USA). The as-say condition entails initial denaturation at 95℃ for 5 min, followed by 40 cycles of 95℃ for 10 s and 60℃ for 34 s. Gene β-actin was used as an internal reference and the relative abundance of transcripts was analyzed by 2ΔΔCt method. The primers are listed in Table 1.
2.6 Expression of ELIFIT1 Gene in Immune Tissues of E. lanceolatus Under Infection with SKIVThree immunity-related tissues including spleen, kidney, and liver were collected from the experimental groups and PBS group at 0, 12, 24, 72, and 96 h after infection and five fish were dissected at each time point. RNA extraction and RT-qPCR were performed in triplicate as described previously.
2.7 Statistical AnalysisData was expressed as mean ± standard error (SE) of five repetitions for each experimental group. Data were analyzed by one-way ANOVA and t test using SPSS version 19.0 software package (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered as significant and P < 0.01 was considered as extremely significant.
3 Results 3.1 ELIFIT1 cDNA and AA Sequence AnalysisThe full length sequence of ELIFIT1 cDNA was obtained from the spleen of E. lanceolatus by normal PCR and RACE. Complete ELIFIT1 gene was 2921 bp, with an ORF of 1314 bp, which encodes a 437-AA peptide. The 5' UTR is 99 bp and the 3' UTR is 1508 bp, with a poly (A) tail (Fig.1). Molecular weight of the ELIFIT1 protein is 50.64 kDa, and the isoelectric point is 6.353. The ELIFIT1 protein is a tetratricopeptide repeat (TPR) protein, which contains multiple tetratricopeptide repeat domains (domain architecture ID 12138572), according to the conserved domain analysis. As shown in Fig.1, 9 TPR motifs are located in position 52−85, 95-128, 140-173, 179-213, 214-247, 250-283, 293-326, 330-363, and 369- 402. In architecture analysis, a low complexity region (266IDEAI-DLAEEALE278) was found in the ELIFIT1 protein (Fig.1). There are 147 hydrophobic AAs and 290 hydrophilic AAs in the ELIFIT1 protein and most of the AAs have values below zero in the hydrophilic/hydrophobicity analysis (Fig.2). ELIFIT1 protein is a secreted protein and has no transmembrane domain with TMHMM analysis.

|
Fig. 1 Nucleotides and deduced amino acid sequences of ELIFIT1 cDNA. The TAG stop codon is indicated with an asterisk. Nine TPR motifs located in position 52-85, 95-128, 140-173, 179-213, 214-247, 250-283, 293-326, 330-363, and 369-402 are single-underlined and low complexity regions are double-underlined. |

|
Fig. 2 Hydrophilic/hydrophobic analysis of ELIFIT1 protein. Hydrophobic segments have values above zero in the Y-axis, while hydrophilic segments have values below zero in the Y-axis. |
A Blast-protein search revealed that ELIFIT1 protein is significantly homologous with IFIT1 members in bony fish, which contain multiple TPR motifs. The TPR motif numbers of HsIFIT1 and MmIFIT1 are 10 and 11 in TPR-pred, and the TPR motif numbers of EcIFIT1 and ELIFIT1 are 6 and 4 (Fig.3). The first TPR motif in mouse was identified in 13 aa-51 aa. Some TPR motifs were identified to SEL-1 motif in EcIFIT1 and ELIFIT1, while the probability is only 0.09% and 0.37%, separately. Based the results of alignment, the number of TPR motifs were confirmed to be 9 in ELIFIT1. The entire amino acid sequences of fish IFIT1s and mammal IFIT1s were aligned using the ClustalX program and the TPRs were annotation in Fig.4.

|
Fig. 3 TPR predicted results of HsIFIT1, MmIFIT1, EcIFIT1 and ELIFIT1 in TPRpred. |

|
Fig. 4 Multiple sequence alignment of IFIT1 proteins from 11 fishes and two mammals. The amino acid numbers are shown on the top of the figure. Conservative amino acids have a red background and similar amino acids have a yellow background. TRP motifs are marked in blank wireframe and numbers of TRPs are covered with blue shadow. |
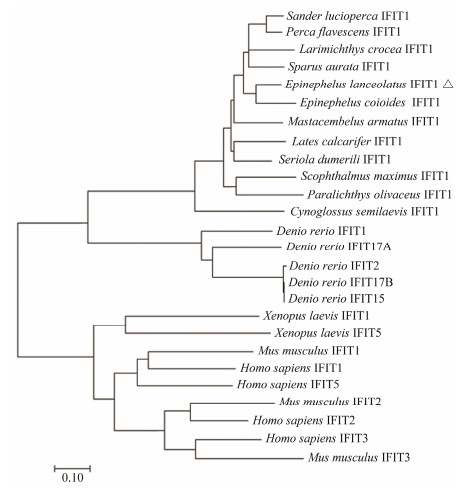
The deduced ELIFIT1 protein shares > 80% identity with the sequences found in orange-spotted grouper (E. coioides) and two kinds of perch (Perca flavescens, Sander luci-operca). The identity between ELIFIT1 and three flatfish IFIT1 (Scophthalmus maximus, Paralichthys olivaceus, Cynoglossus semilaevis) are 67.4%, 62.4%, and 59.9%, respectively. The identity between ELIFIT1 and two mammals (Homo sapiens, Mus musculus) are 24.7% and 22.8%, respectively. The phylogenic analysis showed that IFIT1 members from 12 marine fishes form an independent clad and five IFIT1 members of Denio rerio form another independent clad. The IFIT1 family of Xenopus laevis (African clawed frog) and mammals form a unitary clad, which reflect that the distance of the evolutionary relationship was relatively far away between fish and mammalian IFITs (Fig.5).
|
Fig. 5 Phylogenetic relationship of IFIT protein family. The tree was constructed using Mega 7.0 by neighbor-joining method. The bootstrap confidence values are based on 1000 replications. |
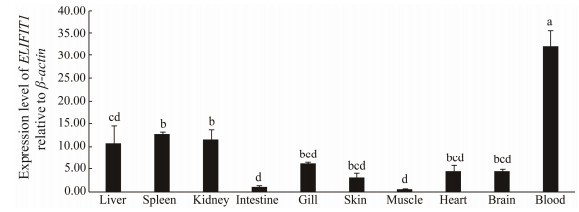
RT-qPCR analysis was performed to investigate the expression pattern of ELIFIT1 mRNA in healthy fish (Fig.6). The ELIFIT1 mRNA was mainly expressed in blood, and the higher mRNA levels were detected in spleen, kidney, and liver, followed by gill, heart, brain, skin, intestine, and muscle.
|
Fig. 6 The expression levels of ELIFIT1 mRNA in different tissues from healthy fish. Gene β-actin was used as an internal control of the RT-qPCR. The values are presented as means ± SE (n = 5) and letters represent significant difference in gene expression among tissues according to Duncan test in the statistical analysis. |
To investigate whether ELIFIT1 participates in immune response, especially during virus infection, RT-qPCR was used to analyze the expression pattern of ELIFIT1 mRNA during SKIV infection. As shown in Fig.7A, compared to 0 h group and PBS group, the transcript levels of ELIFIT1 mRNA were significantly upregulated by 5-fold at 72 h and downregulated by 4-fold at 96 h after infection in the spleen. The change in expression of ELIFIT1 mRNA was even more drastic in the kidney and liver than spleen, and transcript levels of ELIFIT1 mRNA was upregulated approximately by 30-fold at 72 h and downregulated by 5-fold at 96 h after infection (Figs. 7B, C).

|
Fig. 7 ELIFIT1 expression during SKIV infection. Expression of ELIFIT1 mRNA in the spleen (A), kidney (B), and liver (C) of control group (PBS) and SKIV-infected groups. The ELIFIT1 mRNA levels were determined by qRT-PCR at various time points. Values are presented as means ± SE (n = 5). *P < 0.05, **P < 0.01 as compared with 0 h time point. |
In this study, the biological property of ELIFIT1 was analyzed. Sequence analysis showed that ELIFIT1 encodes a 437-amino-acid polypeptide, which share highest identity (82.3%) with E. Coioides IFIT1 protein. As an important component of the IFIT1 family, IFIT1 proteins are conserved in various species, indicating their conservative function. TPR motif was a classical protein structure which composed of 34 amino acids, and scaffolds formed among tandem TPR motifs of IFIT1 mediate protein-protein interactions (Blatch and Lassle, 1999). The number of TPR motifs was 10 in HuIFIT1 (NP_001539) and MuIFIT1 (NP_032357), while it is inconclusive in fish. In our study, the protein sequence of ELIFIT1 shows the highest similarity with EcIFIT1. Zhang et al. (2019 found only three TPR motifs are formed by residues 53-86, 96-129, and 139-174 in EcIFIT1. We retrieved the TPR motifs of EcIFIT1 using TPRpred and found 6 TPR motifs including TPR1 (53-86), TPR2 (96-129), TPR3 (141-174), TPR6 (251-284), TPR7 (294-327), and TPR9 (368- 401). Inside the sequence, residues 196 to 231 was predicted to be SEL1 motif with a P-value of 7.7e−04, and the probability for been SEL1 is 0.09%, so that we concluded that it was a wrong annotate (Fig.3). Hwang et al. (2017 predicted that there are 10 TPR motifs in PoIFIT1 through TPRpred and sequences alignment; however, we found that 'TPR10s' in fish is less than 34 amino acid, and cannot form a complete TPR motif. The similar IFIT1 without TPR10 was observed in large yellow croaker (Larimichthys crocea), Crucian carp (Carassius auratus L.), and pufferfish (Fugu rubrides) (Zhang and Gui, 2004; Wan and Chen, 2008). Using both TPRpred and multiple sequence alignment, we predicted that the number of TPR motifs is 9 in the ELIFIT1, as annotated in Fig.2. Phylogenic analysis showed that marine fish IFIT1 proteins form a distinct clad with the mammalian IFIT1 in Fig.5, suggesting that the mammalian IFIT1 might undergo amplification to 10 TPR motifs in some emergencies with evolution.
Previous studies have shown that the expression level of IFIT1 are higher in immunity-related tissues such as spleen, kidney, and liver in healthy fish (Wan and Chen, 2008; Long and Sun, 2014; Zhang et al., 2019). In our study, it is worth mentioning that the expression level of ELIFIT1 mRNA in the blood was much higher than in visceral tissue, which suggests that ELIFIT1 may function as a safe guarder in the blood for protection against viral infection.
Mammals IFIT family members regulate immune response and restrict viral infections through blocking viral RNA translation. Moreover, IFIT1 regulates both viral and cellular functions. IFIT1 expression was obviously upregulated in other teleosts infected by DNA viruses such as SKIV and megalocytivirus (Long and Sun, 2014; Zhang et al., 2019), or RNA virus such as VHSV, red-spotted grouper nervous necrosis virus, and grass carp hemorr-hage virus (GCHV) (Zhang and Gui, 2004; Hwang et al., 2017; Zhang et al., 2019). The results indicated that ELIFIT1 was significantly increased in the spleen, kidney, and liver after SKIV infection, which is consistant with previous studies. Previous studies have confirmed that IFIT1 is an antiviral protein that binds to free 5'-triphosphate virus RNA or 5' Capped 2'-O unmethylated RNA (Pichlmair et al., 2011; Kimura et al., 2013). It is necessary to investigate the anti-SKIV function of ELIFIT1 in future research.
The presence of interferon motifs functioning as stimulatory response element in the promoters of fish ISG15 and IFIT5 have been confirmed (Liu et al, 2002; He et al., 2017). In our future plan, we will study the transcriptional regulation mechanism of potential response elements in promoter of ELIFIT1. Xie et al. (2016 confirmed that polymorph rs303218 in human IFIT1 gene can predict the IFNα treatment efficiency for Chinese hepatitis B virus infection. Our findings provide a new insight in the teleost fish, while molecular polymorphism still needs further validation.
5 ConclusionsIn this study, the IFIT1 gene from E. lanceolatus was identified for the first time. The predicted ELIFIT1 protein has 9 TRP domains. The multiple sequence alignment and phylogenetic analysis revealed that ElIFIT1 has high similarity with other IFIT1 proteins in teleosts. The expression analysis indicated the important immunity function of ELIFIT1 in the blood and other immune-related tissues. After viral infection, the expression pattern of ELIFIT1 indicated its potential antiviral function, which will contribute to the functional research on IFIT1 proteins in the future.
AcknowledgementsThis work was supported by the Shandong Breeding Project (No. 2016LZGC009), the Projects from Laboratory for Marine Fisheries Science and Food Production Processes, Pilot National Laboratory for Marine Science and Technology (Qingdao) (Nos. 2018-MFS-T08, 2017ASTCP-OS15), the Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD20), and the Central Public-Interest Scientific Institution Basal Research Fund, YSFRI, CAFS (No. 20603022018026).
Blatch, G. L. and Lassle, M., 1999. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays, 21(11): 932-939. DOI:10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N (  0) 0) |
Chebath, J., Merlin,, G,, Metz, R., Benech, P. and Revel, M., 1983. Interferon-induced 56, 000 Mr protein and its mRNA in human cells: Molecular cloning and partial sequence of the cDNA. Nucleic Acids Research, 11(5): 1213-1226. DOI:10.1093/nar/11.5.1213 (  0) 0) |
Chen, X. Y., Shao, T. Y. and Long, X. H., 2018. Evaluation of the effects of different stocking densities on the sediment microbial community of juvenile hybrid grouper (female symbol Epinephelus fuscoguttatus × male symbol Epinephelus lanceo-latus) in recirculating aquaculture systems. PLoS One, 13(12): e208544. DOI:10.1371/journal.pone.0208544 (  0) 0) |
Chen, Z. F., Tian, Y. S., Wang, P. F., Tang, J., Liu, J. C, Ma, W. H., Li, W. S., Wang, X. M. and Zhai, J. M., 2018. Embryonic and larval development of a hybrid between kelp grouper Epinephelus moara ♀ × giant grouper E. lanceolatus ♂ using cry-opreserved sperm. Aquaculture Research, 49(4): 1407-1413. DOI:10.1111/are.13591 (  0) 0) |
Daffis, S., Szretter, K. J., Schriewer, J., Li, J., Youn, S., Errett, J., Lin, T. Y., Schneller, S., Zust, R., Dong, H., Thiel, V., Sen, G. C., Fensterl, V., Klimstra, W. B., Pierson, T. C., Buller, R. M., Gale, Jr. M., Shi, P. Y. and Diamond, M. S., 2010. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature, 468(7322): 452-456. DOI:10.1038/nature09489 (  0) 0) |
D'Andrea, L. D. and Regan, L., 2003. TPR proteins: The versatile helix. Trends in Biochemical Sciences, 28(12): 655-662. DOI:10.1016/j.tibs.2003.10.007 (  0) 0) |
Diamond, M. S., 2014. IFIT1: A dual sensor and effector molecule that detects non-2'-O methylated viral RNA and inhibits its translation. Cytokine & Growth Factor Reviews, 25(5): 543-550. DOI:10.1016/j.cytogfr.2014.05.002 (  0) 0) |
Fensterl, V. and Sen, G. C., 2011. The ISG56/IFIT1 gene family. Journal of Interferon & Cytokine Research, 31(1): 71-78. DOI:10.1089/jir.2010.0101 (  0) 0) |
He, X. M., Du, X., Zhuo, J. S., Jing, X. Y., Yang, X. Q. and Liu, D. K., 2017. Promoter identification and effect on activation of NF-κB of porcine ISG58. Acta Agriculturae Scandinavica, Section A-Animal Science, 67: 1-6. DOI:10.1080/09064702.2017.1330359 (  0) 0) |
Hubel, P., Urban, C., Bergant, V., Schneider, W. M., Knauer, B., Stukalov, A., Scaturro, P., Mann, A., Brunotte, L., Hoffmann, H. H., Schoggins, J. W., Schwemmle, M., Mann, M., Rice, C. M. and Pichlmair, A., 2019. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nature Immunology, 20(4): 493-502. DOI:10.1038/s41590-019-0323-3 (  0) 0) |
Hui, D. J., Bhasker, C. R., Merrick, W. C. and Sen, G. C., 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2. GTP. Met-tRNAi. Journal Biological Chemistry, 278: 39477-39482. DOI:10.1074/jbc.M305038200 (  0) 0) |
Hui, D. J., Terenzi, F., Merrick, W. C. and Sen, G. C., 2005. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. Journal Biological Chemistry, 280: 3433-3440. DOI:10.1074/jbc.M406700200 (  0) 0) |
Hwang, J. Y., Ahn, S. J., Kwon, M. G., Seo, J. S., Hwang, S. D. and Son, M. H., 2017. Interferon-induced protein 56 (IFI56) is induced by VHSV infection but not by bacterial infection in olive flounder (Paralichthys olivaceus). Fish & Shellfish Immunology, 66: 382-389. DOI:10.1016/j.fsi.2017.05.027 (  0) 0) |
Karpenahalli, M. R., Lupas, A. N. and Soding, J., 2007. TPRpred: A tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics, 8: 2. DOI:10.1186/1471-2105-8-2 (  0) 0) |
Kimura, T., Katoh, H., Kayama, H., Saiga, H., Okuyama, M., Oka-, moto, T, ., Umemoto, E., Matsuura, Y., Yamamoto, M., Ta-, keda and K, ., 2013. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5' capped 2'-O unmethylated RNA. Journal of Virology, 87(18): 9997-10003. DOI:10.1128/JVI.00883-13 (  0) 0) |
Kiriyakit, A., Gallardo, W. G. and Bart, A. N., 2011. Successful hybridization of groupers (Epinephelus coioides × Epinephelus lanceolatus) using cryopreserved sperm. Aquaculture, 320(1): 106-112. DOI:10.1016/j.aquaculture.2011.05.012 (  0) 0) |
Lafage, M., Clauss, I., Couez, D., Simonetti, J., Wathelet, M. G. and Huez, G., 1992. The interferon- and virus-inducible IFI-56K and IFI-54K genes are located on human chromosome 10 at bands q23-q24. Genomics, 13(2): 458-460. DOI:10.1016/0888-7543(92)90272-T (  0) 0) |
Liu, M., Reimschuessel, R. and Hassel, B. A., 2002. Molecular cloning of the fish interferon stimulated gene, 15 kDa (ISG15) orthologue: A ubiquitin-like gene induced by nephrotoxic damage. Gene, 298(2): 129-139. DOI:10.1016/S0378-1119(02)00932-0 (  0) 0) |
Liu, Y., Zhang, Y. B., Liu, T. K. and Gui, J. F., 2013. Lineage-specific expansion of IFIT gene family: An insight into coevolution with IFN gene family. PLoS One, 8(6): e66859. DOI:10.1371/journal.pone.0066859 (  0) 0) |
Long, H. and Sun, L., 2014. CsIFIT1, an interferon-induced protein with tetratricopeptide repeat, inhibits viral infection in tongue sole (Cynoglossus semilaevis). Fish & Shellfish Immunology, 41(2): 231-237. DOI:10.1016/j.fsi.2014.09.006 (  0) 0) |
Pichlmair, A., Lassnig, C., Eberle, C. A., Gorna, M. W., Baumann, C. L., Burkard, T. R., Bürckstümmer,, T, ., Stefanovic, A., Krieger, S., Bennett, K. L., Rülicke,, T, ., Weber, F., Colinge, J., Müller,, M, . and Superti-Furga, G., 2011. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nature Immunology, 12: 624-630. DOI:10.1038/ni.2048 (  0) 0) |
Robert, X. and Gouet, P., 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42(Web Server issue): W320-W324. DOI:10.1093/nar/gku316 (  0) 0) |
Robertsen, B., 2006. The interferon system of teleost fish. Fish & Shellfish Immunology, 20(2): 172-191. DOI:10.1016/j.fsi.2005.01.010 (  0) 0) |
Samuel, C. E., 2001. Antiviral actions of interferons. Clinical Microbiology Reviews, 14(4): 778-809. DOI:10.1128/CMR.14.4.778-809.2001 (  0) 0) |
Varela, M., Diaz-Rosales, P., Pereiro, P., Forn-Cuni, G., Costa, M. M., Dios, S., Romero, A., Figueras, A. and Novoa, B., 2014. Interferon-induced genes of the expanded IFIT family show conserved antiviral activities in non-mammalian species. PLoS One, 9(6): e100015. DOI:10.1371/journal.pone.0100015 (  0) 0) |
Wan, X. and Chen, X. H., 2008. Molecular characterization and expression analysis of interferon-inducible protein 56 gene in large yellow croaker Pseudosciaena crocea. Journal of Experimental Marine Biology & Ecology, 364(2): 91-98. DOI:10.1016/j.jembe.2008.07.027 (  0) 0) |
Wathelet, M., Moutschen, S., Defilippi, P., Cravador, A., Collet, M., Huez, G. and Content, J., 1986. Molecular cloning, full-length sequence and preliminary characterization of a 56-kDa protein induced by human interferons. European Journal of Biochemistry, 155(1): 11-17. DOI:10.1111/j.1432-1033.1986.tb09452.x (  0) 0) |
Williams, K. C., 2009. A review of feeding practices and nutritional requirements of postlarval groupers. Aquaculture, 292(3): 141-152. DOI:10.1016/j.aquaculture.2009.04.026 (  0) 0) |
Xie, D. Y., Wang, S. M., Yang, J. M., Wang, L. H., Chen, H. Y., Huai, C., Shang, J., Mao, Q., Lei, C. L., Luo, G. H., Qian, J. and Lu, D. R., 2016. IFIT1 polymorphisms predict interferon-alpha treatment efficiency for hepatitis B virus infection. World Journal of Gastroenterology, 22(44): 9813-9821. DOI:10.3748/wjg.v22.i44.9813 (  0) 0) |
Zahoor, M. A., Xue, G., Sato, H., Murakami, T., Takeshima, S. N. and Aida, Y., 2014. HIV-1 Vpr induces interferon-stimulated genes in human monocyte-derived macrophages. PLoS One, 9(8): e106418. DOI:10.1371/journal.pone.0106418 (  0) 0) |
Zahoor, M. A., Xue, G., Sato, H., Murakami, T., Takeshima, S. N. and Aida, Y., 2015. Genome-wide transcriptional profiling reveals that HIV-1 Vpr differentially regulates interferon-stimulated genes in human monocyte-derived dendritic cells. Virus Research, 208: 156-163. DOI:10.1016/j.virusres.2015.06.017 (  0) 0) |
Zhang, Y. B. and Gui, J. F., 2004. Identification and expression analysis of two IFN-inducible genes in crucian carp (Carassius auratus L.). Gene, 325: 43-51. DOI:10.1016/j.gene.2003.09.039 (  0) 0) |
Zhang, Y., Wang, Y. X., Liu, Z. T., Zheng, J. Y., Huang, Y. H., Huang, X. H. and Qin, Q. W., 2019. Grouper IFIT1 inhibits iridovirus and nodavirus infection by positively regulating interferon response. Fish & Shellfish Immunology, 94: 81-89. DOI:10.1016/j.fsi.2019.08.075 (  0) 0) |
Zhou, Q., Gao, H. Y., Zhang, Y., Fan, G. Y., Xu, H., Zhai, J. M., Xu, W. T., Chen, Z. F., Zhang, H., Liu, S. S., Niu, Y. P., Li, W. S., Li, W. M., Lin, H. R. and Chen, S. L., 2019. A chromosome-level genome assembly of the giant grouper (Epinephelus lanceolatus) provides insights into its innate immunity and rapid growth. Molecular Ecology Resources, 19(5): 1322-1332. DOI:10.1111/1755-0998.13048 (  0) 0) |
Zou, J. and Secombes, C. J., 2011. Teleost fish interferons and their role in immunity. Developmental and Comparative Immunology, 35(12): 1376-1387. DOI:10.1016/j.dci.2011.07.001 (  0) 0) |
 2021, Vol. 20
2021, Vol. 20


