Hyaluronidase (HAase) is a class of β-N-acetyl-galactosaminase that can degrade hyaluronic acid, and widely distributed in prokaryotic and eukaryotic species in nature. It plays an important role in microbial invasion of the host as well as in the physiological and pathological processes of animals. Hyaluronic acid is an important component of extracellular matrix, and it is able to reduce the molecular weight of hyaluronic acid by breaking the glycosidic bond between uronic acid and N-acetyl glucosamine in the hyaluronic acid chain (Wang et al., 2017). Current studies on hyaluronidase mainly focus on humans and mice. Human hyaluronidase family consists of 6 members, including hyal1, hyal2, hyal3, hyal4, hyalp1 and spam1 (sperm adhesion molecule 1). The hyaluronidase family of mice has one more member than that of humans, namely hyal5. HYAL1 is a secreted hyaluronidase, which is the first hyaluronidase isolated and purified from mammalian somatic tissue. It is mainly distributed in serum and urine of mammals, and it is also highly expressed in major parenchymal organs such as heart, liver, spleen and kidney. It was found that the activity of HYAL1 in urine is more than 100 times higher than that in plasma (Ginetzinsky, 1958; Salegui and Pigman, 1967; Csóka et al., 1997; Frost et al., 1997). HYAL2 is expressed in all tissues except for brain. HYAL3 is highly expressed in testis and bone marrow tissues, and its expression level is low in other tissues. Unlike HYAL1 and HYAL2, which nearly are not expressed in brain tissue, HYAL3 has a considerable expression level in adult brain tissue (Csóka et al., 1999). HYAL4, like HYAL2, is also a kind of GPI anchor protein, which mainly exists in human skeletal muscle, testis, placenta and other tissues (Csóka et al., 1999; Bastow et al., 2008; Farrugia et al., 2019). HYALP1 is mainly expressed in testis, but HYALP1 in human is a pseudogene, and there is a termination codon in its mRNA, which makes the translation terminate early (Csóka et al., 1999). Like HYAL2 and HYAL4, SPAM1 is a GPI anchor protein. Research has shown that SPAM1 is mainly anchored to the sperm plasma membrane and acrosome inner membrane through GPI (Hunnicutt et al., 1996). HYAL5 is a unique hyaluronidase in rodents, and is highly expressed in the testis of mouse. It is anchored on the plasma membrane and acrosomal membrane of sperm through GPI. Its function is similar to that of SPAM1, which is to assist sperm to cross the cumulus and mediate sperm-egg binding (Kim et al., 2006; Kimura et al., 2009).
Among these family members, SPAM1 is the most special one, which can be used as a symbol of sperm maturation (Jansen et al., 2001). In mammals, SPAM1 is anchored to the plasma membrane surface and acrosomal membrane of mature sperm by GPI and participates in the fertilization process through the following three pathways (Li et al., 1997; Martin-DeLeon, 2011; Ortiz-Escribano et al., 2016): 1) Enzymatic hydrolysis of hyaluronic acid-rich cumulus matrix outside the egg cell; 2) Binding to Zona Pellucida (ZP) and mediating sperm-egg recognition; 3) Binding to hyaluronic acid and increasing the concentration of Ca2+ in sperm, thus mediating signal transduction between cells.
Sebastes schlegelii, also known as black rockfish (Scorpaeniformes, Scorpaenidae and Sebastes), has a unique method of viviparous reproduction (Kunz et al., 1983). Male black rockfish copulates with female fish after sperm maturation in November and December, but egg maturation occurs in March and April of the next year. Sperm are stored in the ovary for more than 5 months before fertilization (Mori et al., 2003). At present, studies regarding the reproduction of black rockfish mainly focus on the development of gonads in male and female fish, the maturation and localization of sperm before and after mating, and the cryopreservation of sperm (Li et al., 2021; Zhao et al., 2021; Lyu et al., 2022). Little is known about the role of hyaluronidase family genes in the reproductive process of black rockfish. This study identified the hyaluronidase family genes in black rockfish, analyzed the selection pressure of those genes, and further explored the expression profiles of hyaluronidase genes in different tissues. This study laid a foundation for further study on the evolution and function of hyaluronidase in viviparous teleosts.
2 Materials and Methods 2.1 Identification of Hyaluronidase Family Genes in Black RockfishThe hyaluronidase genes of 12 representative species, including Homo sapiens, Mus musculus, Scophthalmus manximus, Gasterosteus aculeatus, Lepisosteus oculatus, Danio rerio, Xenopus tropicalis, Poecilia reticulata, Xiphophorus maculatus, Branchiostoma floridae, Ciona intestinalis and Petromyzon marinus, were collected from NCBI and Ensemble databases, and these hyaluronidase sequences were used as query to blast transcriptome assembly of black rockfish in our lab. BLAST algorithm search was performed and sequences within the e-value of 1e-5 were collected, The open reading frames (ORF) of retrieved sequences were identified by ORF Finder (https://www.ncbi.nlm.nih.gov/gorf/gorf.html) and verified by BLASTP against NCBI nonredundant protein database (Wang et al., 2019). Phylogenetic trees were constructed using hyaluronidase protein sequence from black rockfish and twelve above representative species. The Maximum Likelihood method was utilized to perform phylogenetic analysis by MEGA X. The optimal model for tree construction was General Time Reversible (GTR) and Gamma distributed with Invariant sites (G + I), and the final bootstrap value was set to 1000. Multiple sequence alignments of hyaluronidase sequences were conducted by MUSCLE (Codons).
2.2 Molecular Characters of the Hyaluronidase in Black RockfishThe physiochemical properties of identified hyaluronidase genes, including the molecular weight (Mw) and theoretical isoelectric point (pI), were calculated by ExPasy software. Subsequently, the chromosomal location information of each gene in the hyaluronidase family was obtained based on existing genome annotation files in the laboratory. The exon-intron structure of hyaluronidase genes was determined by comparison of genome and transcriptome sequences following the GT-AG rule. The schematic diagram of gene structure was displayed using the GSDS 2.0 program. Protein motif analysis was carried out with the Motif Elicitation (MEME) program.
2.3 Molecular Evolutionary Analysis of Hyaluronidase in Black RockfishTo determine whether adaptive evolution has occurred on the hyaluronidase family genes of black rockfish, the PAML package with a maximum-likelihood (ML) approach was used to calculate non-synonymous to synonymous rate ratios (ω = dN/dS). The ratios of ω > 1, = 1 and < 1, indicate positive selection, neutrality, and negative selection, respectively. The hyaluronidase coding sequences of 37 teleosts, including Anabas testudineus, Anarrhichthys ocellatus, Anguilla anguilla, Archocentrus centrarchus, Astatotilapia calliptera, Austrofundulus limnaeus, Carassius auratus, Cynoglossus semilaevis, Cyprinus carpio, Danio rerio, Gadus morhua, Gasterosteus aculeatus, Haplochromis burtoni, Hippocampus comes, Lates calcarifer, Lepisosteus oculatus, Mastacembelus armatus, Maylandia zebra, Melanotaenia boesemani, Myripristis murdjan, Nematolebias whitei, Nothobranchius furzeri, Notothenia coriiceps, Oncorhynchus mykiss, Oryzias latipes, Oryzias melastigma, Parambassis ranga, Pimephales promelas, Poecilia reticulata, Pungitius pungitius, Pygocentrus nattereri, Scleropages formosus, Scophthalmus maximus, Sinocyclocheilus rhinocerous, Tachysurus fulvidraco, Takifugu rubripes, and Xiphophorus maculatus, were collected from NCBI and Ensemble databases and used to construct phylogenetic trees by MEGA X with counterpart in black rockfish.
2.4 Synteny Analysis of Spam1 Among VertebratesSynteny analysis was based on the comparison of adjacent genes between the hyaluronidase genes family of black rockfish and corresponding genes of other species (Mus musculus, Homo sapiens, Lepisosteus oculatus, Danio rerio, Gasterosteus aculeatus, Scophthalmus maximus, Xiphophorus maculatus, Xenopus tropicalis and Poecilia reticulata). We used the available data in our laboratory to determine the genes, transcription directions and specific chromosome locations of each member of the hyaluronidase genes family in black rockfish, then searched the linear structure of the genes of the hyaluronidase family of our selected species through the Genomicus database.
2.5 Expression Profiles of Hyaluronidases Genes in Different Tissues of Black RockfishA large number of transcriptome database of tissue was built in black rockfish. In this study, a total of 10 tissue libraries (Heart, Liver, Spleen, Kidney, Brain, Gill, Muscle, Intestine, Testis, and Ovary) were utilized to characterize the expression profiles of black rockfish hyaluronidases genes. The transcriptome data used to analyze the expression of hyaluronidase genes are available at CNSA (CNGB Nucleotide Sequence Archive) under the accession ID CNP 0000222.
2.6 Predicted Three-Dimensional Structure of SPAM1 and Binding Sites Between SPAM1 and ZP ProteinThe amino acid sequences of four SPAM1 of black rockfish were compared and their similarity was analyzed by MUSCLE in MEGA X. The three-dimensional structure of SPAM1 was predicted by software AlphaFlod2 (Version 2.1.0), and the predicted results were visualized by Pymol software. And three-dimensional molecular structure of hyaluronic acid was retrieved from Zinc database (https://zinc.docking.org). The interaction model between SPAM1 and ZP protein was predicted by the Z-DOCK (http://zdock.umassmed.edu). Molecular docking experiment was conducted by Autodock4, a molecular docking software.
2.7 In situ HybridizationIn situ hybridization of testis was performed as previously described (He et al., 2019). The RNA probe was synthesized using the Roche DIG RNA Labeling Kit (SP6/ T7). The image was observed with a Nikon AZ100 microscope.
2.8 Histology and ImmunohistochemistryThe protocol was carried out according to the previous literature (He et al., 2019). In brief, the testis samples in Bouin's fixative were dehydrated in a graded alcohol series and transitioned to 100% ethanol, xylene and paraffin embedding finally. Primary antibody (anti-SPAM1 in 5% Normal Goat Serum: 1:150) was added and incubated with HRP conjugated goat anti-mouse IgG (CWBIO, Taizhou, China), detected with a Metal Enhanced DAB Substrate Kit (Solarbio, Beijing, China). Finally, staining was performed with Modified Lillie-Mayer Hematoxylin solution (Solarbio, Beijing, China). The staining results were observed and photographed with a Nikon Eclipse Ti-U microscope (Nikon, Tokyo, Japan).
3 Results 3.1 Identification of Hyaluronidase Genes in Black RockfishThe hyaluronidase amino acid sequences from some representative species, including H. sapiens, M. musculus, S. maximus, G. aculeatus, L. oculatus, D. rerio, X. tropicalis, P. reticulata, X. maculatus, B. floridae, C. intestinalis and P. marinus, were used to blast the genome and transcriptome database of black rockfish. Phylogenetic relations of hyaluronidase genes among vertebrates showed that the hyaluronidase genes in each subfamily of the black rockfish (marked with red) were clustered with those from other twelve species (Fig. 1). In total, 10 hyaluronidase genes in black rockfish were identified. In addition, in the hyaluronidase family of black rockfish, the subfamilies of hyal2 and spam1 were both expanded. According to the evolutionary tree clustering, hyal2 genes were divided into two clusters, named hyal2a (Ss_10012680) and hyal2b (Ss_10 006096), and spam1 genes were divided into four clusters, named spam1a (Ss_10006905), spam1b (Ss_10006906), spam1c (Ss_10006907) and spam1d (Ss_10006908), respectively.
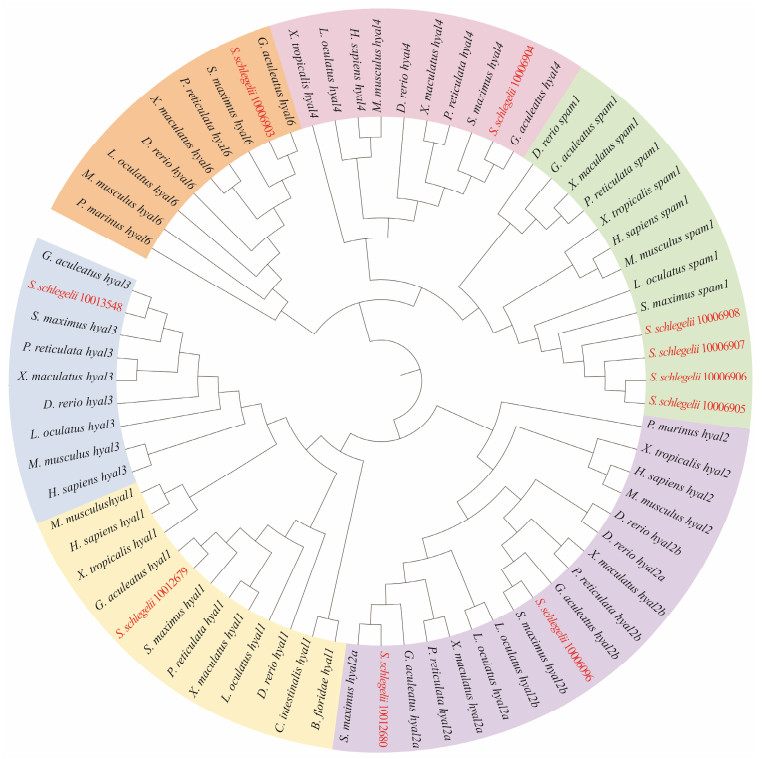
|
图 1 Phylogenetic relations of hyaluronidase genes among vertebrates. The unrooted maximum-likelihood phylogenetic tree was constructed with MEGA X. |
The hyaluronidase family genes were distributed on three chromosomes, among which hyal1, hyal2a and hyal2b were distributed on the chromosome of No.1, hyal3 on No.10, and hyal4, hyal6, spam1a, spam1b, spam1c and spam1d on No.9. The length of coding sequences of hyaluronidase genes ranged from 1323 bp to 2745 bp, with presumed protein sequences length ranging from 440 to 914 amino acids. The molecular weight of hyaluronidase proteins ranged from 49.674 to 103.854 kDa with the isoelectric point ranging from 7.95 to 9.59. The predicted results of conserved domains showed that all the 10 hyaluronidases of black rockfish contained Glyco_hydro_56 hyaluronidase domain. According to the predicted results of PredGPI, it was found that there were GPI sites in most hyaluronidase genes except hyal3, hyal4 and hyal6. It indicated that these three genes might have a different function from other hyaluronidase, especially lose the conventional hyaluronidase function. The detailed information of hyaluronidase genes was summarized in Table 1.
|
|
表 1 Information of hyaluronidases in black rockfish |
The intron-exon structure and motif structure of the hyaluronidase genes of black rockfish were shown in Fig. 2. Hyal3 showed the oldest gene in this phylogenetic tree, and spam1d showed the older gene than other spam1, such as spam1a, spam1b and spam1c (Fig. 2A), with similar results in Fig. 1. These hyaluronidase genes of black rockfish were relatively similar in structure, with seven motifs except hyal2a, hyal4, hyal1, and hyal3, which lose the motif 7 (Fig. 2B), and three exons except hyal4 and hyal3 (Fig. 2C). All the results showed that the original hyaluronidase gene (hyal3) had a loss of motif 7, while the younger gene, spam1, showed insertion of motif 7.
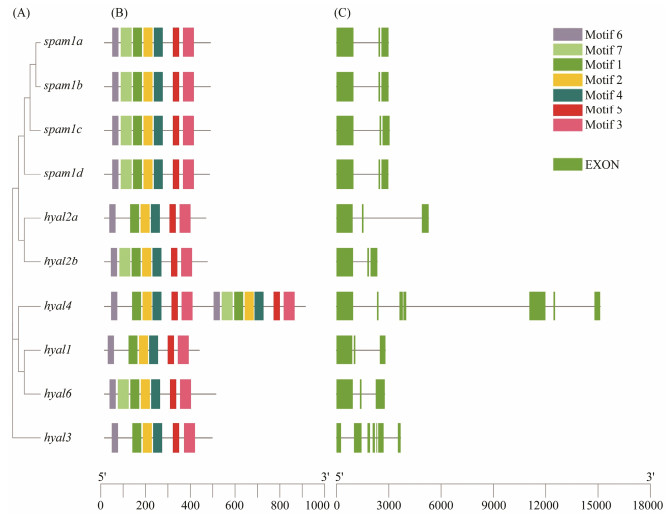
|
图 2 Phylogenetic relationship, gene structure and conserved motif analysis of hyaluronidases genes in black rockfish. (A) Phylogenetic relationship of hyaluronidases genes in black rockfish. The unrooted maximum-likelihood phylogenetic tree was constructed with MEGA X. (B) Distributions of conserved motifs in hyaluronidases genes. Seven putative motifs were indicated in different colored boxes. (C) Exon/intron organization of hyaluronidases genes. Green boxes represent exon and lines represent intron. |
The codeML program in PAML V4.9 was carried out to analyze the selection pressure of hyaluronidase family using the branch model and branch-site model.
3.4.1 Branch model testIn this model, the hyaluronidase genes of black rockfish were taken as the foreground branch, and the other species as the background branch. Except for spam1, the other hyaluronidase genes of black rockfish were not subjected to positive selection pressure in evolution (Table 2). It is worth noting that the rate of evolution of spam1 of black rockfish was significantly faster than that of other teleost (ω1 = 3.16777 > ω0 = 0.17010). The results indicated that the spam1 of black rockfish underwent significant positive selection pressure during evolution.
|
|
表 2 Branch model (BM) tests for detection of hyaluronidase genes among teleosts |
To further explore the positive selective sites in the hyaluronidase genes of black rockfish, the branch-site model was carried out. The results of each hyaluronidase gene were summarized in Table 3. Compared with other teleosts, one or two positive selection sites were found in hyal2 and hyal4, and twenty-one positive selection sites were found in spam1. Subsequently, these predicted positive selective sites were mapped to the functional domain of SPAM1, and two sites, LYS-171 and GLY-164, were located in the neutral hyaluronidase domain of SPAM1. In previous studies, the neutral domain is an enzyme active domain that plays a key role for mammalian sperm to cross the cumulus matrix which is composed of hyaluronic acid. Compared with most teleosts, subjecting positive selection pressure and the existence of positive selection sites may be an adaptive evolution of internal fertilization.
|
|
表 3 Branch-site model (BSM) tests for detection of positively selected sites of hyaluronidase genes among teleosts |
Synteny analysis of hyaluronidase genes of ten species showed that two spam1 in L. oculatus and four spam1 in black rockfish were tandem, while other species had only one spam1 (Fig. 3). Among these hyaluronidase genes, hyal5 is unique in rodents, located downstream of spam1 in mouse chromosome. In addition, experimental studies have proved that hyal5 has similar functions to spam1 in the process of fertilization (Park et al., 2019). In mice with spam1-/-, hyal5 can also function like spam1 to ensure fertilization. Moreover, four extended spam1 were sequentially arranged in tandem, which indicated that spam1 might play an important role in the reproductive process of black rockfish.
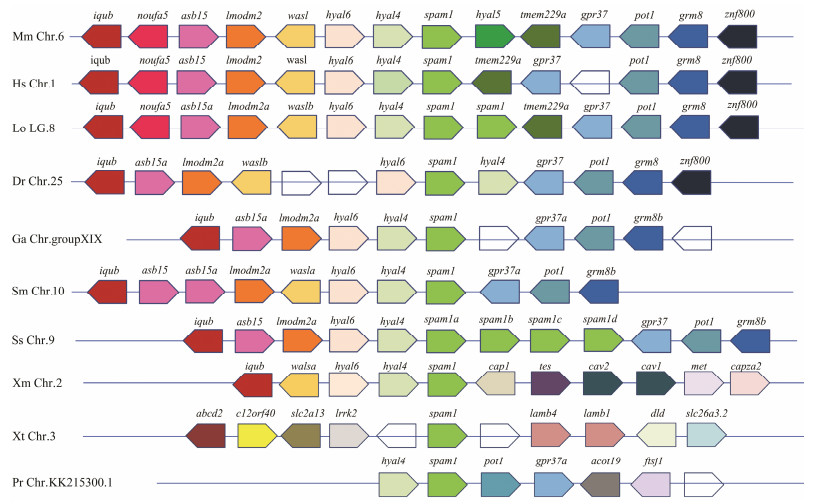
|
图 3 Synteny analysis of spam1 among vertebrates. Mm, Mus musculus; Hs, Homo sapiens; Lo, Lepisosteus oculatus; Dr, Danio rerio; Ga, Gasterosteus aculeatus; Sm, Scophthalmus maximus; Ss, Sebastes schlegelii; Xm, Xiphophorus maculatus; Xt, Xenopus tropicalis; Pr, Poecilia reticulata. |
Hyaluronidase genes were analyzed using transcriptomic data of each tissue in the black rockfish, as shown in Fig. 4. Four spam1 genes were preferentially expressed in the testis, which were similar to the expression pattern in human and mice. Hyal6 also was expressed in testis to a certain extent. Other members of hyaluronidase showed different expression patterns. Hyal1 was mainly expressed in the ovary of female, and gill of male. Hyal2a was in ovary of female, while hyal2b was in female gill. And hyal3 and hyal4 were highly expressed in the spleen of both male and female. These results indicated different hyaluronidase genes might play diverse roles in different tissues.
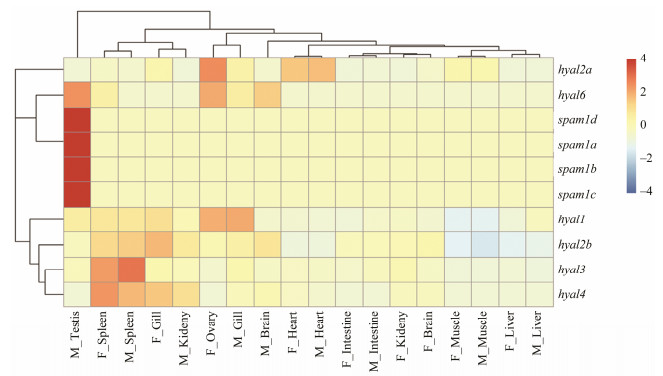
|
图 4 Expression profiles of hyaluronidase genes in different tissues of black rockfish. The color scale represents the TPM value. The red and yellow colors represent the relatively higher and lower TPM values, respectively. |
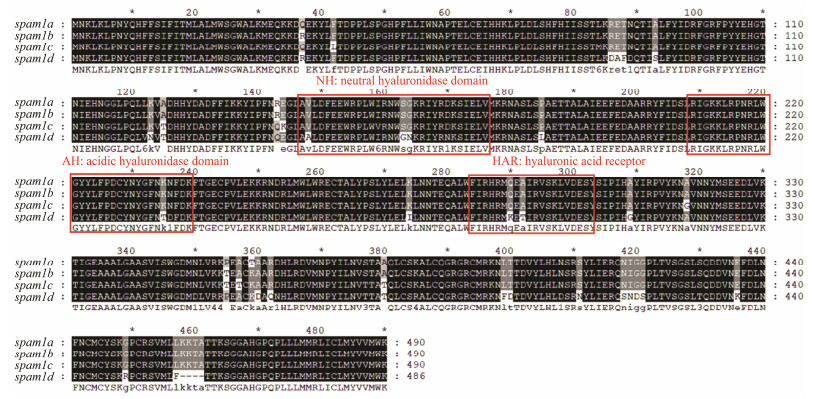
When the amino acid sequences of the inferred four SPAM1s of black rockfish were compared, they showed high similarities. The sequence of SPAM1D was quite different from that of the other three proteins. It had six amino acid residues that are different from the SPAM1A, SPAM1B and SPAM1C in the three functional domains of neutral hyaluronidase domain (NH), acid hyaluronidase domain (AH) and hyaluronic acid receptor (HAR). This might infer that SPAM1D has a new function different from SPAM1A, SPAM1B and SPAM1C (Fig. 5).
|
图 5 Alignment of SPAM1 amino acid in black rockfish. |
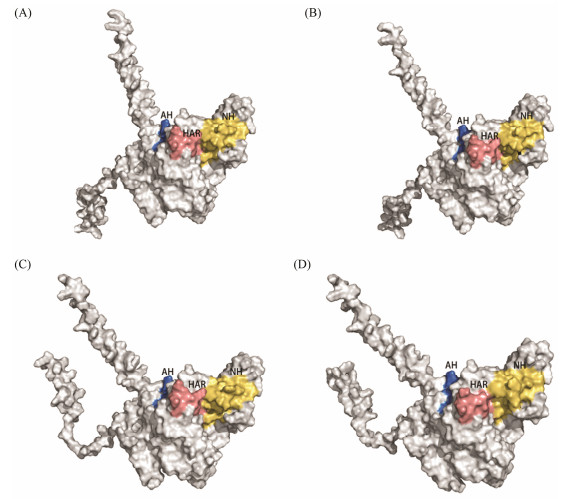
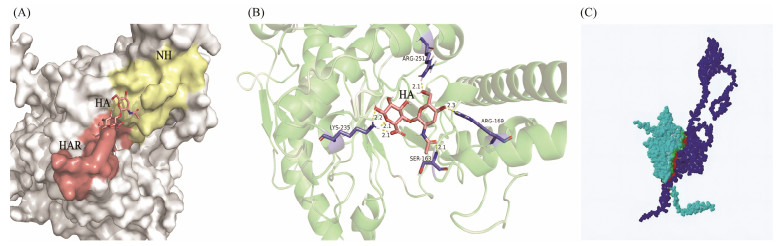
The three-dimensional structures of the four SPAM1 proteins showed that there was a groove on the surface of the SPAM1, which was the site of hyaluronic acid docking so as to hydrolyze hyaluronic acid. NH and AH domains were all completely exposed to external surface, and parts of AH domain were folded inside the protein (Fig. 6). Four amino acid residues (SER-163, ARG-169, LYS-235 and ARG-251) were identified to interact with hyaluronic acid. Among these four amino acid residues, SER-163 and ARG-169 were located in the NH domain, and LYS-235 was located in HAR, which were involved in the interaction between SPAM1 and hyaluronic acid (Fig. 7). The ZPB2a protein in black rockfish is the most similar to that of mammals (Li et al., 2022). After constructing the binding model of SPAM1 and ZPB2a proteins, they can be complementary in spatial conformation and the binding sites between SPAM1 and ZPB2a were located in the groove of SPAM1 surface (Fig. 7C).
|
图 6 Three-dimensional structures of SPAM1 predicted by AlphaFlod2. (A) – (D) represent the subfamily of SPAM1, i.e., SPAM1A, SPAM1B, SPAM1C, and SPAM1D. Neutral hyaluronidase domain (NH) is marked by yellow, acidic hyaluronidase domain (AH) is marked by blue, and hyaluronic acid Receptor (HAR) is marked by pink. |
|
图 7 Docking of SPAM1 with hyaluronic acid in black rockfish. (A) shows as surface; (B) shows as Cartoon. NH, neutral hyaluronidase domain; HA, hyaluronic acid; HAR, hyaluronic acid receptor. (C) Binding model of SPAM1 and ZPB2a. Light blue represents SPAM1, dark blue represents ZPB2a. |
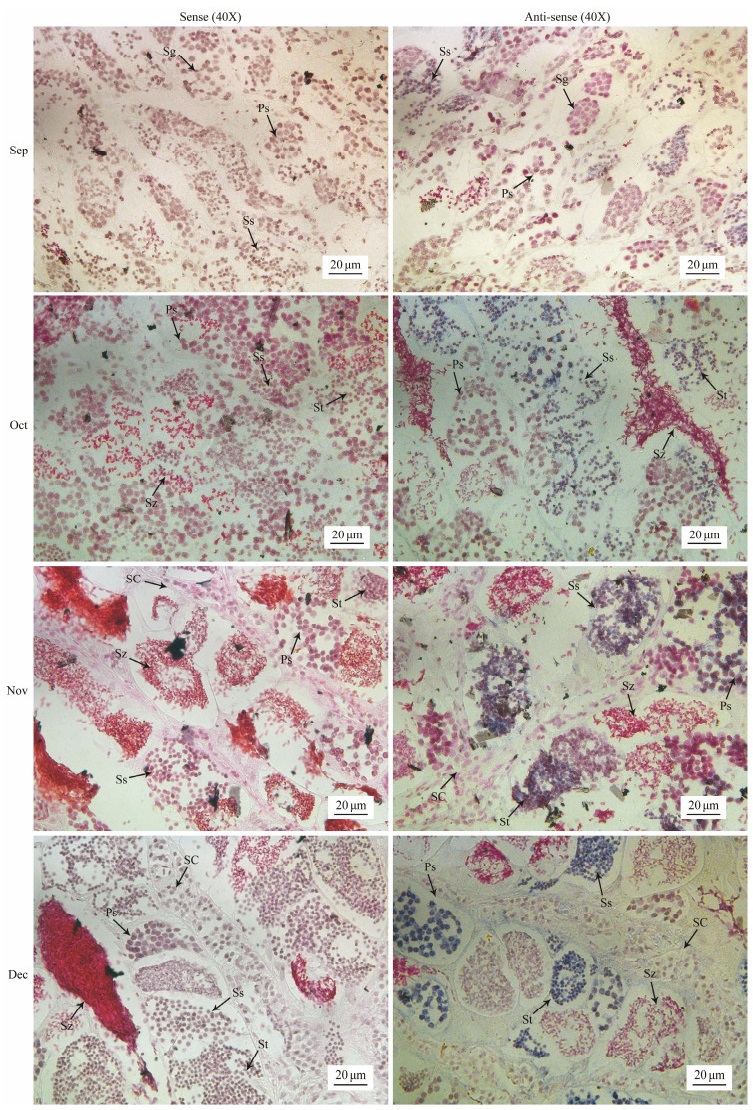
In order to further investigate the detailed spatial expression location of spam1 in the testis of black rockfish, in situ hybridization experiment was performed in testis from September to December, and the results showed that spam1 expression was mainly in primary spermatocytes, secondary spermatocytes and spermatid, but neither in spermatozoa of metaphase nor in mature spermatozoa (Fig. 8).
|
图 8 Spatial expression of spam1 in testis of black rockfish. Sg, spermatogonium; Ps, primary spermatocyte; Ss, secondary spermatocyte; St, spermatid; Sz, spermatozoa; Sc, sertoli cell. |
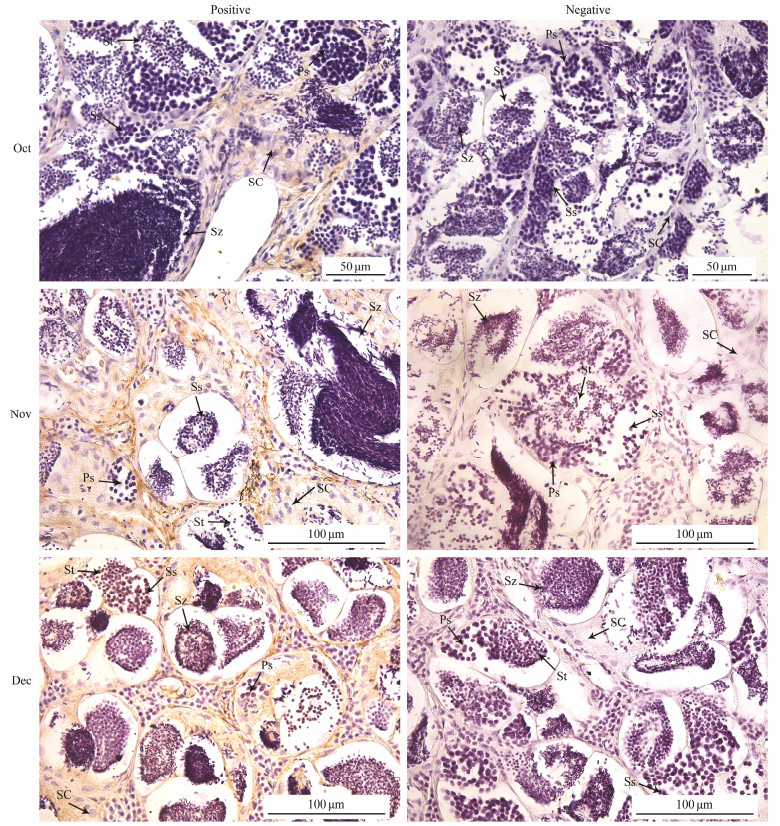
In order to detect the expression of SPAM1 at different developmental stages in testis on the protein level, SPAM1 protein was able to be identified by immunohistochemistry. The result showed that it was mainly located in the interstitial cell and gradually increased during the spermatogenesis (November to December) (Fig. 9).
|
图 9 Localization of SPAM1 in testis of black rockfish at different developmental stages. Sg, spermatogonium; Ps, primary spermatocyte; Ss, secondary spermatocyte; St, spermatid; Sz, spermatozoa; Sc, sertoli cell. |
We identified 10 hyaluronidase genes in the genome of black rockfish, and further analyzed the gene structure, conserved domain, motif and GPI in this study. The results showed that these hyaluronidase genes have conserved Glyco_hydro_56 domain. Among them, hyal1, hyal2a, hyal2b and spam1 were predicted to have GPI sites, which was consistent with the current reported studies (Csóka et al., 1999), indicating that these proteins could be anchored on the plasma membrane of the cell through GPI to play similar biological functions. In addition, we found that spam1 were expanded in black rockfish. Some studies show that, in human and mice, the promoter of spam1 originated from an ancient Endogenous retrovirus (ERV) (Dunn and Mager, 2005). However, ERV transposon could not be found in the promoter of spam1 in black rockfish, suggesting that the expansion of spam1 may be caused by gene replication other than mediated by ERV transposon.
To further investigate whether these hyaluronidase genes were affected by positive selection pressure during evolution, 38 hyaluronidase genes from teleosts were chosen for selection pressure analysis. The results showed that spam1 in black rockfish was subjected to positive selection and had positive selection sites compared with other hyaluronidase genes. Further analyses of these positive selection sites revealed that LYS-171 and GLY-164 in these sites were located in the neutral hyaluronidase domain of SPAM1. SPAM1 has three functional domains, including two hyaluronidase catalytic domains and one domain that can bind with hyaluronic acid (Cherr et al., 2001). The neutral hyaluronidase domain participates in the process of crossing through cumulus cells for sperm (Bleil and Wassarman, 1983; Lin et al., 1994). Moreover, the key peptide that affects the activity of hyaluronic acid at neutral pH may be an acidbase catalytic structure of SPAM1 in rhesus monkeys (Cherr et al., 2001). The binding regions of hyaluronic acid, which promotes the increase of Ca2+ concentration in sperm by binding to hyaluronic acid at the outside of oocytes, and mediates cell-to-cell signaling. At present, the research on the reproductive effect of spam1 is mainly focused on mammals, such as humans and mice. Studies on spam1 have pointed out that it is mainly expressed in germ cells in testes and epithelial cells in epididymis (Deng et al., 2000; Evans et al., 2003). In this study, spam1 of the ten hyaluronidase family genes showed expansion, and SPAM1 protein was expressed in testis. The specific expression patterns of these four spam1 in testicular tissue were similar to those in mice, humans and other mammals (Evans et al., 2003; Morales et al., 2004).
It has been shown that the receptor protein on the sperm surface binds to the zona pellucida after the spermatozoa cross the radial crown to reach the zona pellucida (Tumova et al., 2021), which includes the hyaluronidase SPAM1 (Primakoff et al., 1985; Meyers, 2001). The binding of sperm to zona pellucida at the outside of the oocyte is divided into two main stages, including primary and secondary bindings (Bleil and Wassarman, 1983; Bleil et al., 1988), and the zona pellucida proteins involved in their combination are different in these two stages. SPAM1 plays a role in primary binding and mediates the acrosome reaction after binding to zona pellucida protein 3 (ZP3). The various acrosomal enzymes released can digest the zona pellucida and allow sperm to enter the perivitelline space (Yudin et al., 1999). It was found that sperm bind to ZP3 without acrosome reaction, which is the primary binding of sperm to the zona pellucida. Secondary binding occurs when sperm undergo acrosome reaction, which binds to zona pellucida protein 2 (ZP2). Similar results were found in the guinea pig, where both nonacrosome reacting and acrosome reacting sperm could bind to the zona pellucida in vitro (Huang et al., 1981; Myles et al., 1987). At present, there is no report on whether there is acrosome reaction in spermatozoa of black rockfish. However, four expanded SPAM1 proteins were found, especially the SPAM1D, which was apart from others. Based on the binding model of SPAM1 and ZPB2a proteins, the spatial fitting of two proteins indicated SPAM1 acts through the ZPB2a, which is consistent with the function of SPAM1 that it can interact with ZP to mediate sperm-egg recognition. Whether it is related to the mechanism of viviparous reproduction remains to be further explored in the future.
AcknowledgementsThis study was supported by the National Natural Science Foundation of China (Nos. 32273133, 31970492). We are grateful to Sheng Hang Aquatic Co. (Weihai, Shandong, China) for providing the samples of black rockfish.
Bastow, E., Byers, S., Golub, S., Clarkin, C., Pitsillides, A., and Fosang, A., 2008. Hyaluronan synthesis and degradation in cartilage and bone. Cellular and Molecular Life Sciences, 65: 395-413. DOI:10.1007/s00018-007-7360-z (  0) 0) |
Bleil, J. D., and Wassarman, P. M., 1983. Sperm-egg interactions in the mouse: Sequence of events and induction of the acrosome reaction by a zona pellucida glycoprotein. Developmental Biology, 95(2): 317-324. DOI:10.1016/0012-1606(83)90032-5 (  0) 0) |
Bleil, J. D., Greve, J. M., and Wassarman, P. M., 1988. Identification of a secondary sperm receptor in the mouse egg zona pellucida: Role in maintenance of binding of acrosome-reacted sperm to eggs. Developmental Biology, 128(2): 376-385. DOI:10.1016/0012-1606(88)90299-0 (  0) 0) |
Cherr, G. N., Yudin, A. I., and Overstreet, J. W., 2001. The dual functions of GPI-anchored PH-20: Hyaluronidase and intracellular signaling. Matrix Biology, 20(8): 515-525. DOI:10.1016/S0945-053X(01)00171-8 (  0) 0) |
Csóka, A. B., Scherer, S. W., and Stern, R., 1999. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics, 60(3): 356-361. DOI:10.1006/geno.1999.5876 (  0) 0) |
Csóka, T. B., Frost, G. I., Wong, T., and Stern, R., 1997. Purification and microsequencing of hyaluronidase isozymes from human urine. FEBS letters, 417(3): 307-310. DOI:10.1016/j.febslet.2004.04.014 (  0) 0) |
Deng, X., Yuyan, H. E., and Martin-Deleon, P. A., 2000. Mouse Spam1 (PH-20): Evidence for its expression in the epididymis and for a new category of spermatogenic-expressed genes. Journal of Andrology, 21(6): 822-832. DOI:10.1002/j.1939-4640.2000.tb03412.x (  0) 0) |
Dunn, C. A., and Mager, D. L., 2005. Transcription of the human and rodent SPAM1/PH-20 genes initiates within an ancient endogenous retrovirus. BMC Genomics, 6: 1-14. DOI:10.1186/1471-2164-6-47 (  0) 0) |
Evans, E. A., Zhang, H., and Martin-DeLeon, P. A., 2003. SPAM1 (PH-20) protein and mRNA expression in the epididymides of humans and macaques: Utilizing laser microdissection/RTPCR. Reproductive Biology and Endocrinology, 1(1): 54. DOI:10.1186/1477-7827-1-54 (  0) 0) |
Farrugia, B. L., Mizumoto, S., Lord, M. S., O'Grady, R. L., Kuchel, R. P., Yamada, S., et al., 2019. Hyaluronidase-4 is produced by mast cells and can cleave serglycin chondroitin sulfate chains into lower molecular weight forms. Journal of Biological Chemistry, 294(30): 11458-11472. DOI:10.1074/jbc.RA119.008647 (  0) 0) |
Frost, G. I., Csóka, T., Wong, T., and Stern, R., 1997. Purification, cloning, and expression of human plasma hyaluronidase. Biochemical and Biophysical Research Communications, 236(1): 10-15. DOI:10.1006/bbrc.1997.6773 (  0) 0) |
Ginetzinsky, A., 1958. Role of hyaluronidase in the re-absorption of water in renal tubules: The mechanism of action of the antidiuretic hormone. Nature, 182: 1218-1219. DOI:10.1038/1821218a0 (  0) 0) |
He, Y., Chang, Y., Bao, L. S., Yu, M. J., Li, R., Niu, J. J., et al., 2019. A chromosome-level genome of black rockfish, Sebastes schlegelii, provides insights into the evolution of live birth. Molecular Ecology Resources, 19(5): 1309-1321. DOI:10.1101/527036 (  0) 0) |
Huang, T., Fleming, A., and Yanagimachi, R., 1981. Only acrosome-reacted spermatozoa can bind to and penetrate zona pellucida: A study using the guinea pig. Journal of Experimental Zoology, 217(2): 287-290. DOI:10.1002/jez.1402170215 (  0) 0) |
Hunnicutt, G. R., Primakoff, P., and Myles, D. G., 1996. Sperm surface protein PH-20 is bifunctional: One activity is a hyaluronidase and a second, distinct activity is required in secondary sperm-zona binding. Biology of Reproduction, 55(1): 80-86. DOI:10.1095/biolreprod55.1.80 (  0) 0) |
Jansen, S., Ekhlasi-Hundrieser, M., and Töpfer-Petersen, E., 2001. Sperm adhesion molecules: Structure and function. Cells Tissues Organs, 168(1-2): 82-92. DOI:10.1159/000016809 (  0) 0) |
Kim, E., Baba, D., Kimura, M., Yamashita, M., Kashiwabara, S., and Baba, T., 2006. Identification of a hyaluronidase, Hyal5, involved in penetration of mouse sperm through cumulus mass. Proceedings of the National Academy of Sciences, 102(50): 18028-18033. DOI:10.1073/pnas.0506825102 (  0) 0) |
Kimura, M., Kim, E., Kang, W., Yamashita, M., Saigo, M., Yamazaki, T., et al., 2009. Functional roles of mouse sperm hyaluronidases, HYAL5 and SPAM1, in fertilization. Biology of Reproduction, 81(5): 939-947. DOI:10.1095/biolreprod.109.078816 (  0) 0) |
Kunz, Y. W., Ennis, S., and Wise, C., 1983. Ontogeny of the photoreceptors in the embryonic retina of the viviparous guppy, Poecilia reticulata P. (Teleostei) An electron-microscopical study. Cell and Tissue Research, 230: 469-486. DOI:10.1007/BF00216193 (  0) 0) |
Li, J., Lyu, L., Wen, H., Li, Y., Wang, X., Zhang, Y., et al., 2021. Comparative transcriptomic analysis of gonadal development and renewal in the ovoviviparous black rockfish (Sebastes schlegelii). BMC Genomics, 22(1): 1-17. DOI:10.1186/s12864-021-08169-x (  0) 0) |
Li, M. W., Cherr, G. N., Yudin, A. I., and Overstreet, J. W., 1997. Biochemical characterization of the PH-20 protein on the plasma membrane and inner acrosomal membrane of cynomolgus macaque spermatozoa. Molecular Reproduction and Development: Incorporating Gamete Research, 48(3): 356-366. DOI:10.1002/(sici)1098-2795(199711)48:3<356::aid-mrd9>3.0.co;2-q (  0) 0) |
Li, R., Qu, J. B., Huang, D., He, Y., Niu, J. J., and Qi, J., 2022. Expression analysis of ZPB2a and its regulatory role in spermbinding in viviparous teleost black rockfish. International Journal of Molecular Sciences, 23(16): 9498. DOI:10.3390/ijms23169498 (  0) 0) |
Lin, Y., Mahan, K., Lathrop, W. F., Myles, D. G., and Primakoff, P., 1994. A hyaluronidase activity of the sperm plasma membrane protein PH-20 enables sperm to penetrate the cumulus cell layer surrounding the egg. Journal of Cell Biology, 125(5): 1157-1163. DOI:10.1002/2014JC010225 (  0) 0) |
Lyu, L., Wang, R., Wen, H., Li, Y., Li, J., Wang, X., et al., 2022. Cyclooxygenases of ovoviviparous black rockfish (Sebastes schlegelii): Cloning, tissue distribution and potential role in mating and parturition. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 257: 110677. DOI:10.1016/j.cbpb.2021.110677 (  0) 0) |
Martin-DeLeon, P. A., 2011. Germ-cell hyaluronidases: Their roles in sperm function. International Journal of Andrology, 34(5pt2): e306-e318. DOI:10.1111/j.1365-2605.2010.01138.x (  0) 0) |
Meyers, S., 2001. Equine sperm-oocyte interaction: The role of sperm surface hyaluronidase. Animal Reproduction Science, 68(3-4): 291-303. DOI:10.1016/S0378-4320(01)00166-X (  0) 0) |
Morales, C. R., Badran, H., El-Alfy, M., Men, H., Zhang, H., and Martin-Deleon, P. A., 2004. Cytoplasmic localization during testicular biogenesis of the murine mRNA for Spam1 (PH-20), a protein involved in acrosomal exocytosis. Molecular Reproduction and Development: Incorporating Gamete Research, 69(4): 475-482. DOI:10.1002/mrd.20177 (  0) 0) |
Mori, H., Nakagawa, M., Soyano, K., and Koya, Y., 2003. Annual reproductive cycle of black rockfish Sebastes schlegeli in captivity. Fisheries Science, 69(5): 910-923. DOI:10.1046/J.1444-2906.2003.00707.X (  0) 0) |
Myles, D. G., Hyatt, H., and Primakoff, P., 1987. Binding of both acrosome-intact and acrosome-reacted guinea pig sperm to the zona pellucida during in vitro fertilization. Developmental Biology, 121(2): 559-567. DOI:10.1016/0012-1606(87)90191-6 (  0) 0) |
Ortiz-Escribano, N., Smits, K., Piepers, S., Van den Abbeel, E., Woelders, H., and Van Soom, A., 2016. Role of cumulus cells during vitrification and fertilization of mature bovine oocytes: Effects on survival, fertilization, and blastocyst development. Theriogenology, 86(2): 635-641. DOI:10.1016/j.theriogenology.2016.02.015 (  0) 0) |
Park, S., Kim, Y. H., Jeong, P. S., Park, C., Lee, J. W., Kim, J. S., et al., 2019. SPAM1/HYAL5 double deficiency in male mice leads to severe male subfertility caused by a cumulus-oocyte complex penetration defect. FASEB Journal, 33(12): 14440-14449. DOI:10.1096/fj.201900889RRR (  0) 0) |
Primakoff, P., Hyatt, H., and Myles, D. G., 1985. A role for the migrating sperm surface antigen PH-20 in guinea pig sperm binding to the egg zona pellucida. Journal of Cell Biology, 101(6): 2239-2244. DOI:10.1083/jcb.101.6.2239 (  0) 0) |
Salegui, M. D., and Pigman, W., 1967. The existence of an acidactive hyaluronidase in serum. Archives of Biochemistry Biophysics, 120(1): 60-67. DOI:10.1016/0003-9861(67)90598-X (  0) 0) |
Tumova, L., Zigo, M., Sutovsky, P., Sedmikova, M., and Postlerova, P., 2021. Ligands and receptors involved in the spermzona pellucida interactions in mammals. Cells, 10(1): 133. DOI:10.3390/cells10010133 (  0) 0) |
Wang, B., Wang, H., Song, H., Jin, C., Peng, M., Gao, C., et al., 2019. Evolutionary significance and regulated expression of Tdrd family genes in gynogenetic Japanese flounder (Paralichthys olivaceus). Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 31: 100593. DOI:10.1016/j.cbd.2019.05.003 (  0) 0) |
Wang, W. S., Wang, J. H., and Li, F. C., 2017. Hyaluronidase and chondroitinase. Protein Reviews, 925: 75-87. DOI:10.1007/5584_2016_54 (  0) 0) |
Yudin, A. I., Vandevoort, C. A., Li, M. W., and Overstreet, J. W., 1999. PH-20 but not acrosin is involved in sperm penetration of the macaque zona pellucida. Molecular Reproduction and Development: Incorporating Gamete Research, 53(3): 350-362. DOI:10.1002/(SICI)1098-2795(199907)53:3<350::AID-MRD11>3.0.CO; (  0) 0) |
Zhao, H. X., Wang, X. Y., Du, T. F., Gao, G., Wu, L. L., Xu, S. H., et al., 2021. Sperm maturation, migration, and localization before and after copulation in black rockfish (Sebastes schlegelii). Theriogenology, 166: 83-89. DOI:10.1016/j.theriogenology.2021.01.001 (  0) 0) |
2. Key Laboratory of Tropical Aquatic Germplasm of Hainan Province, Sanya Oceanographic Institute, Ocean University of China, Sanya 572025, China
 2024, Vol. 23
2024, Vol. 23


