2) Key Laboratory of Applied Marine Biotechnology, Ministry of Education, Ningbo University, Ningbo 315211, China
The genus Scylla includes four species of mud crabs: the purple mud crab (S. tranquebarica), the giant mud crab (S. serrata), the orange mud crab (S. olivacea), and the green mud crab (S. paramamosain) (Keenan et al., 1998; Fazhan et al., 2020). These species are distributed across the Indo-Western Pacific, including regions such as Australia, China, Japan, and several Southeast Asian countries (Alberts-Hubatsch et al., 2016; Zhao et al., 2016). In China, they are particularly prevalent along the southeast coastlines. Mud crabs thrive in brackish water ecosystems, moving from high salinity to estuarine environment during their juvenile phases (Rabbani and Zeng, 2005).
Mud crabs are economically significant in aquaculture due to their rapid growth and desirable meat quality, leading to high market value and increased demand (Hamasaki et al., 2011; Ogawa et al., 2012; Ikhwanuddin et al., 2015). Despite advancements in breeding techniques, most crabs are still obtained from the wild (Mirera et al., 2015; Ikhwanuddin et al., 2018). The global seafood market is expected to surpass $730.28 billion by 2030, with significant contributions from segments like fish, crustaceans, and mollusks, driven by increasing health-consciousness and consumption trends (SkyQuest Technology, 2024). Crustaceans contribute significantly to this growth, accounting for 94% of the anticipated increase in global fisheries (Boenish et al., 2021). Major producers of mud crabs include Indonesia, the Philippines, Thailand, and China (FAO, 2018).
High-resolution melting (HRM) analysis is a modern technique used for SNP detection and genotype analysis, characterized by its simplicity, speed, and accuracy (Rengmark et al., 2006; Vandersteen et al., 2007). HRM technology has been extensively utilized in SNP molecular marker studies due to its efficiency and reliability (Maurischat et al., 2015).
The nutritional value of mud crabs is influenced by their muscle, gonads, and biochemical composition, including amino acids and fatty acids (Naczk et al., 2004; Jiang et al., 2014; Sreelakshmi et al., 2016). Studies have shown that mud crabs contain significant amount of polyunsaturated fatty acids (PUFAs), which are beneficial for human health (Skonberg and Perkins, 2002; Çelik et al., 2004). Additionally, essential amino acids found in crab muscles contribute to their nutritional profile and flavour (Jiang et al., 2014; Meng et al., 2017).
The integration of molecular identification with the analysis of amino acid and fatty acid content in crab meat is essential for accurate species-specific nutrition profiling. This approach ensures that nutritional benefits are correctly attributed and leveraged for improving aquaculture practices, ultimately contributing to the sustainable management and utilization of Scylla species (Jiang et al., 2014; Fazhan et al., 2020).
Given the increasing consumer demand for high-quality seafood, the current study aims to develop a rapid identification method using molecular marker technology to distinguish mud crab species in China. Additionally, this research compares the nutritional composition of different wild populations of mud crabs, examining the variations across species, and analyzing free amino acids and fatty acids to provide a comprehensive evaluation of the nutriational components of Scylla species. The findings will support germplasm identification, nutritional assessment, and sustainable resource development of mud crabs.
2 Materials and Methods 2.1 Experimental Materials and Instruments 2.1.1 Samples collectionThe four Scylla species used in this experiment, namely S. tranquebarica, S. serrata, S. olivacea, and S. paramamosain, were bought from three sites, Sanmen, Jiantiao harbour and Lulin market located in Zhejiang Province, China. The number of samples for each species was 30 and a total of 120 samples (average weight of 193.75 g) were transported to the laboratory alive. The body was weighed alive.
2.1.2 Ethical approvalThe four Scylla species were sampled and treated following the guidelines set forth by the Animal Research Institute Committee of Ningbo University, China, ensuring that ethical standards were maintained in animal experimentation. Approval from the esteemed Committee of Animal Research Institute, Ningbo University, China, was obtained to validate the legitimacy of the study.
2.1.3 DNA extraction and nutrition analysisGenomic DNA was extracted using the General Novel Animal Tissue DNA Extraction Kit following the manufacturer's instructions. After DNA extraction, the value of OD260/OD280 of DNA samples were measured with a NanoDrop2000 ultramicro photometer to determine the concentration of DNA. Then the concentration was adjusted to 100 ng μL−1 and the samples were stored at −80℃. Crabs were dissected following ice bath anaesthesia, and the muscle tissues were taken from the legs and internal body mass. Ten individuals from each species were randomly mixed into one sample for nutrition analysis and 2.0 g was taken from each individual. Then the samples were freeze-dried for about 48 h, mashed and mixed to be measured. For each species, the analysis was conducted with three replicates, and a totally of 30 individuals were selected for each species.
2.2 Experimental Methods 2.2.1 Morphological identificationVernier callipers were used to determine the morphological characters of all four species following the procedure of Abbas et al. (2016). Colour and carapace size are two main factors used to describe the exterior morphology of crabs. The median axis was followed to the posterior edge of the carapace to determine the length of the carapace. Grooves or teeth on the carapace's anterolateral border were identified.
2.2.2 Key instrumentsAmong all the experimental instruments, the column of the gas chromatography-mass spectrometer is DB-23 (30 m × 320 μm × 0.25 μm), and the column of the liquid chromatography-mass spectrometer is BEH C18 (1.7 μm, 100 mm × 2.1 mm).
2.2.3 Molecular identification and sequencingThe morphological features of species are complex and changeable. To get more accuracy, we used the Single Nucleotide Polymorphism (SNPs)-based reliable and cost-effective High-Resolution Melting (HRM) technology (Chen et al., 2020). The HRM curve analysis was conducted using LightCycler® 480 High-Resolution Melting Master Kit and LightCycler® 480 Multiwell Plate-96 PCR reaction plate (Roche, Germany). Genomic DNAs were extracted from muscle tissues using DNA extraction kit GK4002 (Generay Bioengineering Co., Shanghai), and the concentration of DNA was determined with NanoDrop2000. DNA samples were stored at −80℃ before further analyses.
Primers were designed using Primer Premier (5.0) software based on the conserved sequences among the four different mud crab species and all the pre-requisite adjustments were made.
The HRM genotyping was determined using 30 samples from each of the four species of the Scylla genus. PCR amplification and HRM analysis were performed with the primers after primary screening on LightCycler® 480 realtime fluorescent quantitative PCR instrument. After the HRM analysis, the standardized melting curve, standardized reference shift and standard melting curve peak diagrams were exported.
For sequencing validation analysis, about eight samples from each of the four Scylla species were randomly selected and sent to Shanghai Huada Gene Technology Co., Ltd. for sequencing. The sequences obtained were adjusted and assembled using the DNA star v.7.1 software package and were BLAST-analyzed for species validation.
2.2.4 Amino acid determinationThe procedures utilized for analyzing the amino acid profiles were based on the methodologies outlined in a prior research study conducted by Fang et al. (2021). To 100 mg of crab powder (whole crab), 1 mL of acetonitrile-water was added, followed by a 30-min ultrasonic extraction. After precipitating the protein with 9x acetonitrile, centrifuge the supernatant once more. With acetonitrile-water, the supernatant was diluted 100 times before being evaluated on the apparatus. A high-performance liquid chromatography-tandem mass spectrometry system (HPLC-MS/MS) constituted of an LC-20AD system (Shimadzu Corporation) was connected to a 5500 QTRAP LC-MS/MS spectrometer, and was used to examine free amino acids (AB). Mobile phase A is water, whereas mobile phase B is acetonitrile. Mobile phases A and B each contains 0.01% heptafluorobutyric acid and 0.1% formic acid. Separation was conducted with BEH-C18 columns (pore size 1.7 m, 100 mm, 2.1 mm, after the addition of 1 μL aliquots of sample extract). The column temperature was maintained at 50℃ during analysis.
2.2.5 Fatty acid determinationThe assessment of fatty acids was based on a procedure defined by Xie et al. (2015). A certain grade of crab flesh taken from legs and internal body mass was weighed, 2 mL of n-hexane was added and the sample was shaken at 50℃ for 30 min. Then 3 mL of a 0.4 mol L−1 KOH methanol solution was added, and the sample was shaken at 50℃ for another 30 min. Finally, 1 mL of water and 2 mL of n-hexane were added, and the sample was shaken for 20 min at room temperature. By using a split ejector and gas chromatography-mass spectrometry (GC-MS), conventional fatty acids were identified (1:5 ratio). The capillary column (DB23) diameters were 30 m, 320 μm, and 0.25 μm, respectively. To each capillary column, 1 μL aliquot of the sample extract was added. The injection port temperature was kept at 250℃, and the detector temperature was kept at 230℃. Helium (He) was used as the carrier gas. The starting column box temperature was held at 50℃ for one minute. Then the temperature was raised to 175℃ at a rate of 25℃ min and was gradually raised to 230℃ at 4℃ min−1. The temperature was held at 230℃ for 25 min.
2.3 Statistical AnalysisThe data were analyzed statistically using SPSS19.0 and were presented with mean ± SD. The one-way ANOVA and difference significance test was performed to examine the differences between groups using the Duncan multiple comparison technique. The difference was considered significant when P < 0.05. Principal component analysis (PCA) of amino acids and fatty acids for each species was carried out using SIMCA-P software (v.11.0). The SPSS Euclidean shortest distance method and R software were used to compare the degree of similarity between different species' polyunsaturated fatty acids and flavour amino acids (v.3.0.0).
3 Results 3.1 Morphological DescriptionFour species of the Scylla genus, S. paramamosain, S. olivacea, S. serrata, and S. tranquebarica, were identified morphologically in the current study by matching them with the phenotypic description provided by Keenan et al. (1998). The dorsal view of each species is shown in Fig.1. It can be observed that the frontal lobe spines of S. serrata and S. tranquebarica are blunt whereas the S. paramamosain has a triangular frontal lobe spine and the S. olivacea has a rounded lobe. Carpus and Propodus spines are enhanced in all of the species. Concerning colour identification, S. serrata is characterized by yellow to green colouration, S. paramamosain with dark green to brown, S. olivacea with green to orange and S. tranquebarica with green to blue colouration pattern possessing specific polygonal patterns.
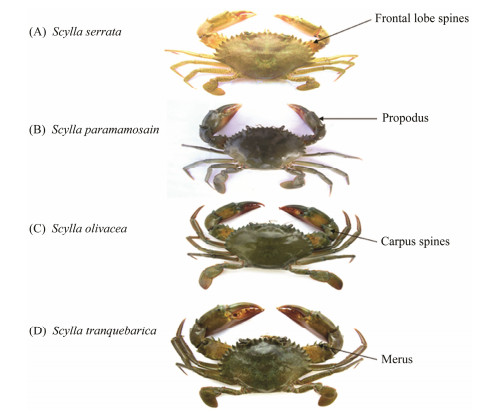
|
Fig. 1 Dorsal view of four mentioned Scylla species. |
Download the mitochondrial whole genome of four species at http://www.ncbi.nlm.nih.gov/ (Accession nos.: FJ 827761.1, FJ827758.1, FJ827760.1, FJ827759.1) and save in FASTA format for sequence alignment using ClustalX software (Larkin et al., 2007). Based on conserved sequences between species, primers were designed using Primer Premier 5.0 software. The primer design needs to consider the following conditions: there is a difference in GC content between different sequences of the target fragment; the length of the target product ranges from 100 – 250 bp; the annealing temperature range is 50 – 60℃. The primer synthesis was completed by Shanghai BGI Technology Co., Ltd.
3.2.2 Primer screeningThe DNA of two samples of each species of blue crab was randomly selected, the synthesized primers were amplified through PCR respectively, and each PCR product was preliminarily screened by 1% agarose gel electrophoresis to confirm whether the target band of the primers was single, and the optimal Tm value of each pair of primers was finally determined. A total of 51 primer pairs were designed. After detection analysis, it was found that one pair of primers (CSN9) could identify S. serrata, S. paramamosain, S. olivacea and S. tranquebarica. The genotyping of four kinds of mud crabs and CSN9 primer information is shown in Table 1.
|
|
Table 1 Information of primers used for SNP genotyping by HRM |
After HRM detection, it was found that the melting curves of HRM analysis were classified into these four species of the Scylla genus. In HRM analysis, the difference in the shape of the melting curve between samples indicates that the DNA samples differ in their sequence composition (Liew et al., 2004). Tm Calling analysis showed that the Tm of S. serrata and S. paramamosain were 79.55℃ and 80.12℃, respectively, while the Tm values of S. tranquebarica and S. olivacea appeared at 78.12℃ and 82.04℃, and at 78.42℃ and 84.40℃, respectively (Fig.2). It can be seen that the melting peaks of the amplification products of the four species at the CSN9 locus are clearly distinguished, indicating that there are differences in the amplicon sequences of the primers, which can identify the genotypes of the four species and effectively combine the four amplification products. Melting curves and melting profiles are distinguished.
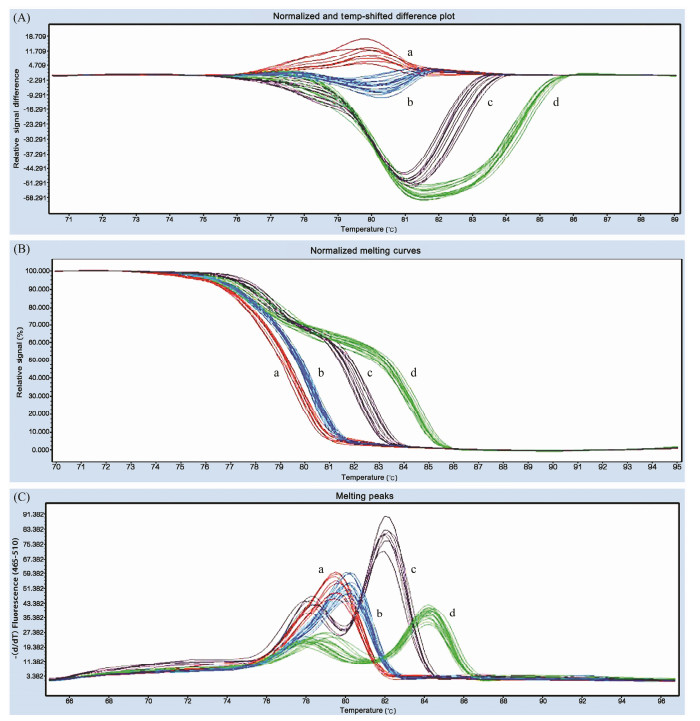
|
Fig. 2 Discriminations of single-nucleotide polymorphisms (SNPs) among four Scylla species using high-resolution melting analysis. (a), S. serrate; (b), S. paramamosain; (c), S. tranquebarica; (d), S. olivacea. |
Thirty samples were sequenced following HRM typing
to verify the accuracy of the HRM typing results. The DnaSP 6 software of Rozas et al. (2017) was used to identify the haplotypes of 24 samples that have been sequenced, and a total of 9 haplotypes were identified including 2 types of Scylla serrata, 3 types of S. olivacea, 2 types of S. paramamosain and 2 types of S. tranquebarica. The results confirmed a total of 28 SNP sites from the 203 bp amplicons of the target region, of which 27 sites were found to have a base conversion and one site had a base inversion, further validating the HRM genotyping results. The nine haplotype sequences were aligned and analyzed using the MegAlign program in the DNAStar (v.7.1) software package, showing 100% consistency with HRM results, as shown in Fig.3.

|
Fig. 3 Alignment of haplotype fragments from the mitochondrial DNA sequence of the four species using primer CSN9. Nucleotides are identical to the sequence on the top and are shown with dots. |
The muscle tissue of four different species of mud crabs contains a wide variety of amino acids, including a total of 20 common free amino acids. Cysteine (Cys) was not found in S. paramamosain and S. olivacea. The most prevalent amino acids in the four species of Scylla are glycine (Gly), arginine (Arg), glutamine (Gln), alanine (Ala), proline (Pro), and glutamic acid (Glu). Among these amino acids, the concentration of arginine is the highest in S. paramamosain (40.64 mg g−1), whereas the concentration of glycine is the highest in the other three species. The total free amino acid contents (TFAA) were from 135.7 to 149.65 mg g−1. The ratio of total flavour amino acid (TDAA) concentration to total free amino acids concentration (TDAA/TFAA) was 53.21% ± 4.92%. It was the highest in S. serrata (57.71% ± 1.58%) followed by S. tranquebarica (57.31% ± 1.46%) (Table 2).
|
|
Table 2 Amino acid compositions and contents (mg g−1 dry weight) of the four species of Scylla |
Total free amino acid content, non-essential amino acid content, and flavour amino acid content in muscle tissue did not differ significantly between the four species (P > 0.05). The average amount of total essential amino acid (TEAA) was (7.64 ± 1.2) mg g−1, while the S. Serrata and S. tranquebarica had the highest amounts (P < 0.05; Table 2). In terms of total non-essential amino acids (TNEAA), the four species had an average content of (142.94 ± 41.04) mg g−1 and S. olivacea with (135.30 ± 18.22) mg g−1, possessing the highest content however S. tranquebarica possessed slightly the lowest contents. The S. Serrata had the highest average flavour amino acid content of (75.88 ± 7.24) mg g−1, whereas S. paramamosain had the lowest average flavour amino acid content (Fig.4).
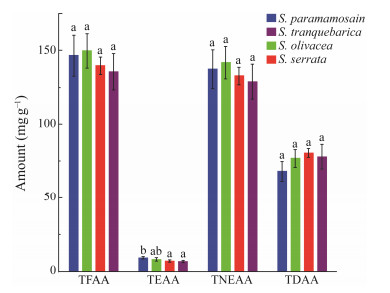
|
Fig. 4 Comparison of amino acids among the four species of Scylla. |
Table 3 shows that 26 kinds of fatty acids were discovered from the muscle tissue of four species of Scylla, ranging in concentrations from 5.98 to 10.80 mg g−1, with S. paramamosain having the greatest concentration (P < 0.05). Of these fatty acids, polyunsaturated fatty acids (PUFA) were the most abundant, followed by saturated fatty acids (SFA), with an average content of 1.72 to 3.40 mg g−1, accounting for 23.45% to 31.77% of the total fatty acids. Monounsaturated fatty acids (MUFAs) were the least abundant, with an average content of 1.05 to 1.88 mg g−1, accounting for 14.29% to 19.17% of the total fatty acids.
|
|
Table 3 Fatty acid compositions and contents (mg g−1 dry weight) of the four species of Scylla |
Eicosapentaenoic acid (EPA; C20:5n-3), one of the 26 identified fatty acids, accounted for 16.17% to 23.26%. Another key fatty acid identified was C18:1N9c (oleic acid).
The S. paramamosain had the greatest concentration of EPA (1.17 mg g−1), making it the species with the highest fatty acid content overall. S. paramamosain also had the greatest concentration of saturated fatty acid C18:0 (1.79 mg g−1), which was substantially higher than that of the other three species (P < 0.05). The concentrations of C16:0 (palmitoleic acid), C22:6n3 (DHA) and C20:4n6 (ARA) were also higher in the same manner. The S. paramamosain, in particular, had ω3-PUFA and ω6-PUFA (ω3/ω6) with a ratio of 1.10 to 3.75, which was significantly higher than that of the other three species (P < 0.05).
There were some variations among species, but the overall amount of different fatty acids in the muscular tissues of the mentioned four species of mud crabs was reasonable. Data analysis showed that the DHA and EPA contents, as well as the total fatty acid content (TFA), total saturated fatty acids (TSFA), total monounsaturated fatty acids (TMUFA), and total polyunsaturated fatty acids (TPUFA) were higher in the S. paramamosain than in the other three species (Fig.5). Additionally, the amounts of TPUFA and TUFA were significantly higher in S. paramamosain and S. tranquebarica (P < 0.05), whereas TMUFA concentration was significantly higher in S. serrata and S. olivacea (P < 0.05) in comparison to the other two Scylla species.
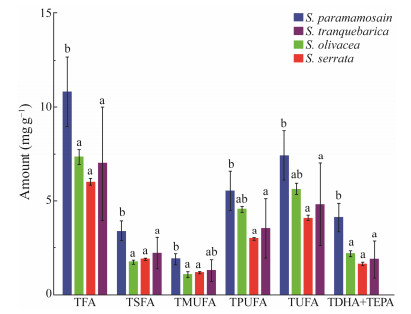
|
Fig. 5 Graphical representation of the fatty acid composition of four species of Scylla. |
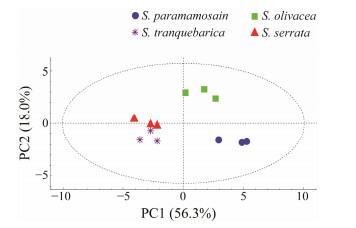
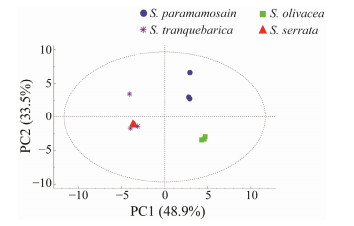
The amino acid and fatty acid compositions/contents of the four Scylla species were evaluated with principal component analysis (PCA; Figs.6 and 7). The three independent replication group samples from all of the populations were within the confidence interval. The distribution area of the four species was obvious and the amino acid and fatty acid PCA score maps were consistent. S. olivacea and S. paramamosian were distributed on the upper and lower sides of PC2, and the compositions of flavour amino acids and polyunsaturated fatty acids were different. The distribution of S. serrata and S. tranquebarica in the left section of PC1, which was comparatively near and farther away from the first two species, indicating that the makeup and level of amino acids and fatty acids were less distinct.
|
Fig. 6 PCA scores plot of the four species of Scylla based on the amino acid contents. |
|
Fig. 7 PCA scores plot of the four species of Scylla based on fatty acids contents. |
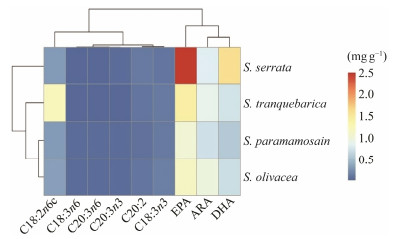
SPSSv.19.0 software was used for cluster analysis. With Euclidean distance as the evaluation index, the intergroup joining method was used for clustering the composition data of flavour amino acids and polyunsaturated fatty acids, and the cluster analysis dendrograms were created (Figs.8 and 9). The flavour amino acid clustering heat map of the four mud crabs (Fig.8) and the polyunsaturated fatty acid cluster heat map (Fig.9) showed that glycine (Gly) was the most abundant flavour amino acid of the four mud crabs, followed by a proline (Pro) and alanine (Ala). EPA (C20: 5n3) was the most abundant PUFA in the four Scylla species, however, DHA (C22:6n3) and ARA (C20:4n6) were significantly abundant in S. paramamosian. Moreover, linoleic acid (C18:2n-6) had the maximum PUFA level in S. olivacea, while the EPA, was considerably higher amount in S. paramamosain than in the other three species (P < 0.05).

|
Fig. 8 Heatmap of the four species of Scylla based on the flavour amino acids contents. |
|
Fig. 9 Heatmap of the four species of Scylla based on the polyunsaturated fatty acids contents. |
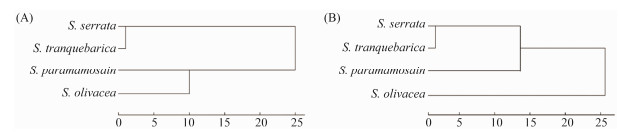
The phylogenetic dendrograms were generated from the four mentioned mud crab species based on amino acid and fatty acid contents (Figs.10A and B). Fig.10A shows the copolymerization of four samples into two categories 'S. serrata and S. tranquebarica' and 'S. olivacea, and S. paramamosain', which were clustered into one class displaying that the amino acid compositions/contents of S. serrata and S. tranquebarica are similar, while those of S. olivacea and S. paramamosain are similar.
|
Fig. 10 Cluster tree plot of the four species of Scylla based on the delicious amino acids (A) and polyunsaturated fatty acids (B) contents. |
On the other hand, Fig.10B shows the copolymerization of polyunsaturated fatty acid composition/content of four mud crab species into three branches. The S. serrata and S. tranquebarica gathered into one, then gathered with S. olivacea and finally clustered with S. paramamosain. The results showed that the contents of polyunsaturated fatty acids and flavour amino acids of S. serrata and S. tranquebarica were similar, while the compositions of polyunsaturated fatty acids were significantly different between S. paramamosain and the other three Scylla species.
4 Discussion 4.1 Morphological Analysis of Scylla SpeciesThe Scylla species is a tasty and nutrient-rich aquatic food that is a vital resource for global fisheries and aquaculture. Early research on morphological traits gave a straightforward and obvious way for the classification of Scylla species. Keenan et al. (1998) previously combined several morphological studies to determine species identification, particularly the Scylla species. The current investigation confirmed that the morphological traits of all four Scylla species were identical to those described by previous studies. The identification characteristics included colouration, frontal lobe spines, carpus, and propodus spines. The current study can help identify new species, particularly if only a limited number of physical traits are accessible for evaluation.
4.2 Molecular Identification of Scylla SpeciesIn this study, the single nucleotide polymorphism (SNP) sites of four different types of mud crab mitochondrial genes were analyzed. The high-resolution melting curve (HRM) technology was employed to create a pair of SNP markers that can reliably and effectively distinguish the four species. The 203 bp segment of the tagged product included 28 mutation sites. According to the sequencing data, the mitochondrial DNA gene changes across species were significantly greater than those within species. The results of the current study were very consistent with the findings of Lin et al. (2007), demonstrating that mt DNA can be employed to identify different Scylla species in a precise and reliable manner.
4.3 Amino Acids and Fatty Acids Analysis of Four Species of ScyllaStudies have shown that S. paramamosain (Meng et al., 2017), S. olivacea (Karim et al., 2016), S. serrata and S. tranquebarica (Sreelakshmi et al., 2016) are all high-protein, low-fat marine economic species. The current study has shown free amino acids in the muscles of four Scylla species which are rich in lysine (Lys), tryptophan (Tyr), phenylalanine (Phe), histidine (His), threonine (Thr), valine (Val), leucine (Ile), isoleucine (Leu). The total content of essential amino acids is the highest in S. paramamosain, which is significantly higher than those of S. serrata and S. tranquebarica (P < 0.05). Among these amino acids, arginine (Arg) and glycine (Gly) are the most abundant amino acids in S. paramamosain, however, glycine is the highest content amino acid in S. olivacea, S. serrata and S. tranquebarica. It is important to clarify that the discussion primarily focuses on free amino acids due to their significant role in nutrition and flavour. While structural amino acids also contribute to the overall amino acid profile, the emphasis in this study is on free amino acids because they are more directly related to the taste and nutritional quality of crab meat. Several studies have confirmed that arginine has a variety of physiological functions such as promoting wound healing, immune regulation and intestinal development, and is a raw material for the synthesis of creatine and NO which can improve muscle strength and pumping during training (Tenenbaum et al., 1998; Pahlavani et al., 2014). Gly has a clear protective effect against liver damage caused by multiple causes (Lee and Kim, 2019). Compared with large animals such as livestock and poultry, marine crabs have a higher content of polyunsaturated fatty acids (PUFAs) and higher nutritional value of fatty acids (Hu et al., 2017).
From the composition and content results of flavour amino acids and polyunsaturated fatty acids in the current study, it can be comprehended that these four kinds of mud crab are all aquatic products with delicious meat and unique flavour. From the perspective of amino acid composition, the content of six flavour amino acids (Glu), Asp, Gly, Ala, Ser and Pro was high (TDAA/TFAA average value was 53.21%), which was much higher than those in other aquatic species such as Sepioteuthis lessoniana (Satjarak et al., 2021), abalone (Venter et al., 2018), and even higher than aquatic economic crabs such as swimming crab, Chinese mitten crab (Jiang et al., 2014), clams (Liew et al., 2004) and marine oysters (Wang et al., 2020). Gly, Pro and Ala are the main contributors of sweetness (Guo et al., 2014) and Glu and Asp are characteristic amino acids with umami taste (Ma et al., 2006), and their content affects the taste of crab muscles. The high content of polyunsaturated fatty acids and flavour amino acids makes the meat of different species of Scylla delicious with unique aroma and tenderness.
4.4 Comparative Analysis of Nutritional Differences Among Four Species of ScyllaStatistical analysis of free amino acid data showed that there was no significant difference in muscle amino acid content, non-essential amino acid content, and flavour amino acid content amongst the mentioned Scylla species (P > 0.05). The composition and content of flavour amino acids (Glu, Asp, Gly, Ala, Ser and Pro) in muscle affect the freshness of meat taste (Rabie et al., 2014). The cluster tree analysis (Fig.10A) and the cluster heat map (Figs.8 and 9) analysis of flavour amino acids showed that the composition content of flavour amino acids of S. serrata and S. tranquebarica were relatively similar in comparison to the other two species.
Statistical analysis of fatty acid data showed that the content of fatty acid in S. paramamosain was higher than that of S. serrata, S. olivacea, and S. tranquebarica. In the current study, the content of C18:2n6 fatty acid in S. paramamosain was significantly higher than that of the other three species, which to some extent reflects the differences of food sources from different environments. Romano et al. (2014) studied the response of Scylla serrata juveniles to the gradual decrease in salinity and found that when the salinity decreased eicosapentaenoic acid (EPA), arachidonic acid (ARA), ω3-PUFA and Long-chain polyunsaturated fatty acids (PUFA) were significantly increased (P < 0.05), and the decrease of these fatty acids are considered to maintain Na+/K+-ATPase activity by increasing membrane fluidity to adapt to salinity changes. Zafar et al. (2004) conducted a one-year sampling study on S. serrata in the waters of Chakaria Sundarban of Bangladesh, Cox's Bazar. He determined that protein and fatty acid contents of female S. serrata were positively correlated with pH, dissolved oxygen (DO), salinity and transparency of the seawater, while those of male S. serrata were negatively correlated with DO and salinity of seawater. Species of the genus Scylla have strong saline adaptability, wide-ranging temperatures, and are widely distributed in different sea waters. The environment of their habitats and the source of bait organisms may be one of the reasons for the difference in nutrient composition. The specific relationship between genetic background, feeding mode and nutritional composition needs to be further investigated in detail.
5 ConclusionsThe recognition of the four Scylla species (S. olivacea, S. tranquebarica, S. paramamosain, and S. serrata) was confirmed in the current study with the help of morphological and molecular differentiation techniques. A pair of mitochondria-specific primers (CSN9) was successfully identified through high-resolution melting analysis (HRM), which can simultaneously identify four species of Scylla. Sequencing analysis was used for the confirmation of the CSN9 product which was 203 bp. A total of 9 haplotypes were identified in 24 experimental samples and 28 SNP sites were detected, of which 27 sites were found to have a single base transversion and only one site had the base inversion mutation. A base transversion is consistent with HRM genotyping results. No significant difference was found in the amino acid composition of the four species. S. paramamosain in comparison to the other three species has shown obvious dominance regarding fatty acid contents, saturated fatty acid, ω3/ω6 value and the sum of DHA and EPA. However, the contents of unsaturated fatty acids and saturated fatty acids were significantly higher in S. serrata and S. paramamosain, whereas that of monounsaturated fatty acids was significantly higher in Scylla serrata and S. olivacea. The current study will provide a theoretical basis for the development and rational utilization of economic species resources of the genus Scylla.
AcknowledgementsThis study was funded by the Key Scientific and Technological Grant of Zhejiang (No. 2021C02069-6), the Earmarked Fund (No. CARS-48) and K.C. Wong Magana Fund in Ningbo University.
Author Contributions
Imtiaz Nida: methodology, formal analysis; investigation, validation, visualization, writing-original draft preparation. Runze Jin: investigation; methodology, formal analysis. Yongliang Li: data curation. Qingyang Wu: supervision, conceptualization, data curation, formal analysis, project administration. Changkao Mu: writing-review and editing. Chunlin Wang: visualization. Ronghua Li: conceptualization, writing-original draft preparation, funding acquisition.
Data Availability
The data are available from the corresponding author based on reasonable requests.
Declarations
Ethics Approval and Consent to Participate
This study was conducted in accordance with the recommendations in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The experimental protocols were approved by the Animal Care and Use Committee of Ningbo University (NBU20220079).
Consent for Publication
Informed consent for publication was obtained from all participants.
Conflict of interest
The authors declare they have no competing interests in the study.
Aaqillah-Amr, M. A., Hidir, A., Noordiyana, M. N., and Ikhwanuddin, M., 2018. Morphological, biochemical and histological analysis of mud crab ovary and hepatopancreas at different stages of development. Animal Reproduction Science, 195: 274-283. DOI:10.1016/j.anireprosci.2018.06.005 (  0) 0) |
Abbas, E. M. H., Abdelsalam, K. M., Mohammed-Geba, K., Ahmed, H. O., and Kato, M., 2016. Genetic and morphological identification of some crabs from the Gulf of Suez, Northern Red Sea, Egypt. The Egyptian Journal of Aquatic Research, 42(3): 319-329. DOI:10.1016/j.ejar.2016.08.003 (  0) 0) |
Alberts-Hubatsch, H., Lee, S. Y., Meynecke, J., Diele, K., Nordhaus, I., and Wolff, M., 2016. Life-history, movement, and habitat use of Scylla serrata (Decapoda, Portunidae): Current knowledge and future challenges. Hydrobiologia, 763(1): 5-21. DOI:10.1007/s10750-015-2393-z (  0) 0) |
Amin-Safwan, A., Muhd-Farouk, H., Nadirah, M., and Ikhwanuddin, M., 2016. Effect of water salinity on the external morphology of ovarian maturation stages of orange mud crab, Scylla olivacea (Herbst, 1796) in captivity. Pakistan Journal of Biological Sciences, 19(5): 219-226. DOI:10.3923/pjbs.2016.219.226 (  0) 0) |
Boenish, R., Kritzer, J. P., Kleisner, K., Steneck, R. S., Werner, K. M., Zhu, W., et al., 2021. The global rise of crustacean fisheries. Frontiers in Ecology and the Environment, 20(2): 102-110. DOI:10.1002/fee.2431 (  0) 0) |
Celik, M., Türeli, C., Çelik, M., Yanar, Y., Erdem, Ü., and Küçükgülmez, A., 2004. Fatty acid composition of the blue crab (Callinectes sapidus Rathbun, 1896) in the north eastern Mediterranean. Food Chemistry, 88(2): 271-273. DOI:10.1016/j.foodchem.2004.01.038 (  0) 0) |
Chen, X. Q., Li, R. H., Wang, C. L., Mu, C. K., Song, W. W., Liu, L., et al., 2020. An effective method for identification of three mussel species and their hybrids based on SNPs. Conservation Genetics Resources, 12(1): 5-8. DOI:10.1007/s12686-018-1051-y (  0) 0) |
Fang, F., Yuan, Y., Jin, M., Shi, B., Zhu, T. T., Luo, J. X., et al., 2021. Hepatopancreas transcriptome analysis reveals the molecular responses to different dietary n-3 PUFA lipid sources in the swimming crab Portunus trituberculatus. Aquaculture, 543: 737016. DOI:10.1016/j.432aquaculture.2021.737016 (  0) 0) |
Fang, W., Liu, L., Chang, W., Liu, Z., Yang, J., Liu, X., et al., 2022. Comparative analysis of burrowing crab growth and nutrient composition under saline-alkali water and mariculture. Journal of Fisheries of China, 46(11): 2143-2157. (  0) 0) |
FAO, 2018. FAO Yearbook: Fishery and Aquaculture Statistics, 2018. FAO, Rome, 110pp.
(  0) 0) |
Fatihah, S. N., Julin, H. T., and Chen, C. A., 2017. Survival, growth, and molting frequency of mud crab Scylla tranquebarica juveniles at different shelter conditions. Aquaculture, Aquarium, Conservation & Legislation, 10 (6): 1581-1589, http://www.bioflux.com.ro/aacl.
(  0) 0) |
Fazhan, H., Ikhwanuddin, M., Quinitio, E. T., Baylon, J. C., Fujaya, Y., Rukminasari, N., et al., 2020. Morphological descriptions and morphometric discriminant function analysis reveal an additional four groups of Scylla spp. PeerJ, 8: e8066. DOI:10.7717/peerj.8066 (  0) 0) |
Ghazali, A., Azra, M. N., Noordin, N. M., Abol-Munafi, A. B., and Ikhwanuddin, M., 2017. Ovarian morphological development and fatty acids profile of mud crab (Scylla olivacea) fed with various diets. Aquaculture, 468: 45-52. DOI:10.1016/j.aquaculture.2016.09.038 (  0) 0) |
Guo, Y., Gu, S., Wang, X., Zhao, L., and Zheng, J., 2014. Comparison of fatty acid and amino acid profiles of steamed Chinese mitten crab. Fisheries Science, 80(3): 621-633. DOI:10.1007/s12562-014-0738-6 (  0) 0) |
Hamasaki, K., Matsui, N., and Nogami, M., 2011. Size at sexual maturity and body size composition of mud crabs Scylla spp. caught in Don Sak, Bandon Bay, Gulf of Thailand. Fisheries Science, 77(1): 49-57. DOI:10.1007/s12562-010-0307-6 (  0) 0) |
Ikhwanuddin, M., Fazhan, H., Quinitio, E. T., Baylon, J. C., Fujaya, Y., Azmie, G., et al., 2018. Larval rearing of mud crab (Scylla): What lies ahead. Aquaculture, 493: 37-50. DOI:10.1016/j.aquaculture.2018.04.047 (  0) 0) |
Ikhwanuddin, M., Mustaqim, M., Fazhan, H., Norfaizza, W. I. W., and Megat, F. H., 2015. Mating behaviour of the orange mud crab, Scylla olivacea: The effect of sex ratio and stocking density on mating success. Aquaculture Reports, 2: 50-57. DOI:10.1016/j.aqrep.2015.08.004 (  0) 0) |
Jiang, K., Zhang, F., Pi, Y., Jiang, L., Yu, Z., Zhang, D., et al., 2014. Amino acid, fatty acid, and metal compositions in edible parts of three cultured economic crabs: Scylla paramamosain, Portunus trituberculatus, and Eriocheir sinensis. Journal of Aquatic Food Product Technology, 23(1): 73-86. DOI:10.1080/10498850.2012.695761 (  0) 0) |
Karim, M. Y., Azis, H. Y., and Muslimin, T. A., 2016. Nutrient content of body and growth as physiological responses of mud crab Scylla olivacea reared male monosex in mangrove. International Journal of PharmTech Research, 9(6): 336-338. (  0) 0) |
Keenan, C., Davie, P. J., and Mann, D. L., 1998. A revision of the genus Scylla de Haan, 1833 (Crustacea: Decapoda: Brachyura: Portunidae). The Raffles Bulletin of Zoology, 46: 217-245, http://era.daf.qld.gov.au/id/eprint/8476/.
(  0) 0) |
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al., 2007. Clustal W and Clustal X version 2.0. Bioinformatics, 23(21): 2947-2948. DOI:10.1093/bioinformatics/btm404 (  0) 0) |
Lee, D. Y., and Kim, E., 2019. Therapeutic effects of amino acids in liver diseases: Current studies and future perspectives. Journal of Cancer Prevention, 24(2): 72-78. DOI:10.15430/jcp.2019.24.2.72 (  0) 0) |
Liew, M., Pryor, R. G. L., Palais, R., Meadows, C., Erali, M., Lyon, E., et al., 2004. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clinical Chemistry, 50(7): 1156-1164. DOI:10.1373/clinchem.2004.032136 (  0) 0) |
Lin, Q., Li, S. J., Li, Z., and Wang, G., 2007. Species composition in genus Scylla from the coast of southeast China. Journal of Fisheries of China, 31(2): 211-219. (  0) 0) |
Lu, J., Shu, M. A., Xu, B., Liu, G., Ma, Y., Guo, X., et al., 2015. Mud crab Scylla paramamosain glutamate dehydrogenase: Molecular cloning, tissue expression and response to hyposmotic stress. Fisheries Science, 81(1): 175-186. DOI:10.1007/s12562-014-0828-5 (  0) 0) |
Ma, A. J., Liu, X. F., Zhai, Y. X., Liu, X. Z., and Zhuang, Z. M., 2006. Biochemical composition in muscle of wild and cultivated tongue sole (Cynoglossus semilaevis Gunther). Marine Fisheries Research, 27(2): 49-54. (  0) 0) |
Maurischat, S., Szabo, I., Baumann, B., and Malorny, B., 2015. Rapid real-time PCR methods to distinguish Salmonella Enteritidis wildtype field isolates from vaccine strains Salmovac SE/Gallivac SE and AviPro SALMONELLA VAC E. Journal of Microbiological Methods, 112: 92-98. DOI:10.1016/j.mimet.2015.03.015 (  0) 0) |
Meng, F., Huanan, G., Tang, X., Wang, A., Yao, X., Liu, C., et al., 2017. Biochemical composition of pond-cultured vs. wild gravid female mud crab Scylla paramamosain in Hainan, China: Evaluating the nutritional value of cultured mud crab. Journal of Shellfish Research, 36(2): 445-452. DOI:10.2983/035.036.0216 (  0) 0) |
Mirera, D. O., and Moksnes, P., 2015. Comparative performance of wild juvenile mud crab (Scylla serrata) in different culture systems in East Africa: Effect of shelter, crab size and stocking density. Aquaculture International, 23(1): 155-173. DOI:10.1007/s10499-014-9805-3 (  0) 0) |
Musundire, M. T., Halimani, T. E., and Chimonyo, M., 2017. Physical and chemical properties of meat from scavenging chickens and helmeted guinea fowls in response to age and sex. British Poultry Science, 58(4): 390-396. DOI:10.1080/00071668.2017.1313961 (  0) 0) |
Naczk, M., Williams, J., Brennan, K., Liyana-Pathirana, C. M., and Shahidi, F., 2004. Compositional characteristics of green crab (Carcinus maenas). Food Chemistry, 88(3): 429-434. DOI:10.1016/j.foodchem.2004.01.056 (  0) 0) |
Ogawa, C. Y., Hamasaki, K., Dan, S., Obata, Y., and Kitada, S., 2012. Species composition, reproduction, and body size of mud crabs, Scylla spp., caught in Urado Bay, Japan. Journal of Crustacean Biology, 32(5): 762-768. DOI:10.1163/193724012x649787 (  0) 0) |
Pahlavani, N., Jafari, M., Alizadeh, M., Rezaei, M., Rasad, H., Keikha, M., et al., 2014. L-arginine supplementation and risk factors of cardiovascular diseases in healthy men: A doubleblind randomized clinical trial. F1000Research, 3: 306. DOI:10.12688/f1000research.5877.2 (  0) 0) |
Rabbani, A. G., and Zeng, C., 2005. Effects of tank colour on larval survival and development of mud crab Scylla serrata (Forskal). Aquaculture Research, 36(11): 1112-1119. DOI:10.1111/j.1365-2109.2005.01328.x (  0) 0) |
Rabie, M., Peres, C., and Malcata, F. X., 2014. Evolution of amino acids and biogenic amines throughout storage in sausages made of horse, beef and turkey meats. Meat Science, 96(1): 82-87. DOI:10.1016/j.meatsci.2013.05.042 (  0) 0) |
Rengmark, A. H., Slettan, A., Skaala, Ø., Lie, Ø., and Lingaas, F., 2006. Genetic variability in wild and farmed Atlantic salmon (Salmo salar) strains estimated by SNP and microsatellites. Aquaculture, 253(1-4): 229-237. DOI:10.1016/j.aquaculture.2005.09.022 (  0) 0) |
Romano, N., Wu, X., Zeng, C., Genodepa, J., and Elliman, J., 2014. Growth, osmoregulatory responses and changes to the lipid and fatty acid composition of organs from the mud crab, Scylla serrata, over a broad salinity range. Marine Biology Research, 10(5): 460-471. DOI:10.1080/17451000.2013.819981 (  0) 0) |
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al., 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12): 3299-3302. DOI:10.1093/molbev/msx248 (  0) 0) |
Satjarak, J., Thongprajukaew, K., Kaewtapee, C., Rodjan, P., and Preedaphol, K., 2021. Morphological characteristics and nutriative value of wild and cultured bigfin reef squid (Sepioteuthis lessoniana). Journal of Food Composition and Analysis, 107: 104356. DOI:10.1016/j.jfca.2021.104356 (  0) 0) |
Hu, S., Wang, J., Han, T., Pan, Q., Jiang, Y., and Wang, C., 2017. Effects of dietary DHA/EPA ratios on growth performance, survival and fatty acid composition of juvenile swimming crab (Portunus trituberculatus). Aquaculture Research, 48(3): 1291-1301. DOI:10.1111/are.12971 (  0) 0) |
Skonberg, D. I., and Perkins, B. D., 2002. Nutrient composition of green crab (Carcinus maenus) leg meat and claw meat. Food Chemistry, 77(4): 401-404. DOI:10.1016/s0308-8146(01)00364-8 (  0) 0) |
SkyQuest Technology, 2024. Seafood market: Global trends and insights. Retrieved from https://www.skyquestt.com/sample-request/seafood-market.
(  0) 0) |
Sreelakshmi, K. R., Manjusha, L., Vartak, V. R., and Venkateswarlu, G., 2016. Variation in proximate composition and fatty acid profiles of mud crab meat with regard to sex and body parts. Indian Journal of Fisheries, 63(2): 147-150. DOI:10.21077/ijf.2016.63.2.34511-23 (  0) 0) |
Tenenbaum, A., Fisman, E. Z., and Motro, M., 1998. L-Arginine: Rediscovery in progress. Cardiology, 90(3): 153-159. DOI:10.1159/000006837 (  0) 0) |
Vandersteen, J. G., Bayrak-toydemir, P., Palais, R. A., and Wittwer, C. T., 2007. Identifying common genetic variants by highresolution melting. Clinical Chemistry, 53: 1191-1198. DOI:10.1373/clinchem.2007.085407 (  0) 0) |
Venter, L., Loots, D. T., Vosloo, A., Van Rensburg, P. J., and Lindeque, J. Z., 2018. Abalone growth and associated aspects: Now from a metabolic perspective. Reviews in Aquaculture, 10(2): 451-473. DOI:10.1111/raq.12181 (  0) 0) |
Wang, H., Wei, H., Tang, L., and Lu, J., 2018. A proteomics of gills approach to understanding salinity adaptation of Scylla paramamosain. Gene, 677: 119-131. DOI:10.1016/j.gene.2018.07.059 (  0) 0) |
Wang, X., Yu, H., Xing, R., Liu, S., Chen, X., and Li, P., 2020. Optimization of oyster (Crassostrea talienwhanensis) protein hydrolysates using response surface methodology. Molecules, 25(12): 2844. DOI:10.3390/molecules25122844 (  0) 0) |
Xie, D., Wang, S., You, C., Chen, F., Tocher, D. R., and Li, Y., 2015. Characteristics of LC-PUFA 518 biosynthesis in marine herbivorous teleost Siganus canaliculatus under different ambient 519 salinities. Aquaculture Nutrition, 21(5): 541-551. DOI:10.1111/anu.12178 (  0) 0) |
Zafar, M. A., Siddiqui, M. S., and Hoque, M. M., 2004. Biochemical composition in Scylla serrata (Forskal) of Chakaria Sundarban area, Bangladesh. Pakistan Journal of Biological Sciences, 7(12): 2182-2186. DOI:10.3923/pjbs.2004.2182.2186 (  0) 0) |
Zhao, J., Wen, X., Li, S., Zhu, D., and Li, Y., 2016. Effects of different dietary lipid sources on tissue fatty acid composition, serum biochemical parameters and fatty acid synthase of juvenile mud crab Scylla paramamosain (Estampador 1949). Aquaculture Research, 47(3): 887-899. DOI:10.1111/are.12547 (  0) 0) |
 2025, Vol. 24
2025, Vol. 24


