MicroRNAs (miRNAs) are single-stranded, 19–25 nucleotide non-coding RNAs that bind to target mRNAs and mediate translation repression and/or mRNA degradation (Wahid et al., 2010). Animal genomes contain hundreds of microRNA genes, and each microRNA, in turn, can directly regulate hundreds of target mRNAs (Ambros, 2004; Alvarez-Garcia and Miska, 2005; Bartel, 2009). They play vital regulatory roles in various biological processes, including immune responses (Guan et al., 2019), tumorigenesis (Ma et al., 2014), reproduction (Zhang et al., 2018) and stress resistance (Su et al., 2019), cell signaling and tissue morphogenesis (Kloosterman and Plasterk, 2006).
MiR-430, a well-characterized miRNA expressed at maternal-zygotic transition (MZT) stage, has been shown to target a large number of maternally expressed mRNAs and promote their deadenylation and degradation (Giraldez et al., 2006; Svoboda and Flemr, 2010). During zebrafish embryonic development, miR-430 regulates primordial germ cell (PGC) development through controlling genes sdf1a, cxcr7, tdrd7 and nanos1 expressions (Mishima et al., 2006; Takeda et al., 2009; Staton et al., 2011; Bhattacharya et al., 2017). Furthermore, it can stimulate optimal Nodal signaling activity by inhibiting the expression of the agonist sqt and the antagonist left (Choi et al., 2007), which can keep the balance between Nodals and Leftys and decide the degree of mesoderm formation (Schier, 2003; Schier and Talbot, 2005; Shen, 2007). Recently, some researchers have found the conservation between zebrafish and mouse mechanisms of left-right patterning. Unfortunately, the current studies about miR-430 family are limited to zebrafish and other model organisms. Its roles in early embryonic development remain elusive in other teleosts.
P. olivaceus is one of the most important marine economic fishes. It undergoes metamorphosis, which is prominently characterized by morphological transformation from a bilaterally symmetrical to an asymmetrical body shape accompanied by extensive morphological and physiological remodeling of tissues/organs from larval to juvenile forms (Inui et al., 1995; Schreiber, 2006). Many studies have shown that some miRNAs play important roles in regulating gene expressions during metamorphosis of juvenile flounder, such as miR-1, miR-22a and let-7 microRNAs (Fu et al., 2011; Fu et al., 2013). However, the functions of miR-430 family in metamorphosis of P. olivaceusis have not been studied.
Based on the known knowledge of the functions of miR-430, we hypothesized that miR-430 can regulate the early embryonic development and metamorphosis of P. olivaceusis by targeting and regulating the genes of Nodal signal pathway or other vital pathways involved in early embryonic development in P. olivaceusis.
2 Materials and Methods 2.1 MaterialsThe experimental materials are collected from Shandong Yantai Tianyuan Aquaculture Center. In the spawning season of P. olivaceus, healthy and well developed males and females were selected. By controlling light and temperature, the researchers can make the fish naturally produce fertilized eggs. The fertilized eggs were placed into diluted sea water with a salinity of 17. The sea water was continuously aerotaed and the temperature was kept at 20–22℃. Embryos at different developmental stages (1-, 4-, 32-cell, morula, blastula, gastrula, neurula stage, heartbeating stage, klebsiella vesiclebefore (kv) stage, before hatching stage; n = 50) were collected. All samples were immediately frozen using liquid nitrogen and stored at -80℃ for RNA extraction. MiR-430, LNA-430 and control miRNA were injected into the embryos at the 1-cell stage, respectively. These embryos developed to gastrula stage about 10 h after injection and to tailbud stage about 25 h after injection. Embryos from these two developmental stages were sampled for subsequent RNA extraction and morphological observation.
2.2 Isolation of Total RNA and cDNA SynthesisTotal RNA was extracted using Trizol Reagent (Invitrogen, CA, USA) following the manufacturer's protocol and then frozen at -80℃. The cDNA was reversely transcribed from total RNA using the Reverse Transcriptase M-MLV Kit (TaKaRa, Dalian, China). The extraction of miRNA and the synthesis of cDNA were performed using mirVana miRNA I solution Kit Procedure (Ambion, USA) following the manufacturer's protocol. The quality and quantity of total RNA were evaluated via 1.5% (w/v) agarose gel electrophoresis and spectrophotometry using Nano Photometer Pearl (Implen, Munich, Germany).
2.3 Molecular Cloning of Polefty-3'UTR Fragment and Sequence AnalysisOn the basis of the transcriptome data for Japanese flounder, we found lefty gene, namely Polefty by TBL-ASTx. A target site of miR-430 in Polefty-3'UTR region was predicted by RNAhybrid software. Multiple alignments of the miR-430 nucleotide sequences with other known fishes were performed using Clustal X (Larkin et al. 2007). The Polefty-3'UTR region was amplified by PCR with degenerated primer (Polefty-3'UTR-Fw/Rv, Table 1) designed according to IDT (https://sg.idtdna.com/Primerquest/Home/Index). PCR conditions were as follows: 1 cycle at 95℃ for 3 min; 34 cycles at 95℃ for 30 s, 52℃ for 30 s, and 72℃ for 45 s; and a final extension of 7 min at 72℃. All amplified PCR products were identified by agarose gel electrophoresis and purified with the Gel DNA Recovery Kit (Zymoclean, CA, USA), and ligated into the pMD-19T vector (TaKaRa) for sequencing.
|
|
Table 1 Primers used in this study |
The expression patterns of lefty, sqt, ntl, gsc and miR430 were analyzed by qRT-PCR. Specific primers were described in Table 1. Transcription mRNA abundance of several genes was normalized using 18S and U6 as the reference genes. qRT-PCR was performed on Light-Cycler Roche 480 (Roche Applied Science, Mannheim, Germany) in a 20 µL reaction volume containing 10 µL 2×SYBR Premix Ex Taq II (TaKaRa, Dalian, China), 0.4 µL of each primer (10 µmol L-1), 2 µL cDNA (5 ng µL-1) and 7.2 µL nuclease-free water. The PCR included 1 cycle at 95℃ for 5 min for pre-incubation, followed by 45 cycles at 95℃ for 15 s and 60℃ for 45 s. Melting curves were generated following each cycle to confirm the specificity of the amplicons. Three biological replicates of each sample were analyzed. The relative expression levels of target genes were calculated by the 2-ΔΔCt comparative Ct method.
2.5 Plasmid Construct Preparation and TransfectionThe Polefty-3'UTR, which contains one putative miR-430 binding site, was amplified by PCR and cloned into the pMcherry-C1 vector (Promega) through T4 ligase after digested by XhoI and BamHI enzymes. The binding site for miR-430 within the Polefty-3' UTR was deleted to generate a mutant control. Embryonic cells of P. olivaceus were transfected with miR-430 and plasmids of mcherry-lefty-3'UTR vector or mcherry-lefty-3'UTR-mutant vector using Lipofectamine3000 (Invitrogen) in 48-well plates. The fluorescence of the cells was observed at regular intervals using inverted fluorescence microscopy (Nikon). The transfection experiments were performed in triplicate.
2.6 MiRNA Synthesis and MicroinjectionThe miRNAs used in this experiment (Table 2) were synthesized by Gemma company, and the lock nucleic acid (LNA) reagents were synthesized by Sonny company. All reagents were dissolved in DEPC-treated water and diluted to a certain gradient. The fertilized embryos were collected and injected with 10 pg of miRNA at the 1-cell stage. After microinjection, the embryos were keep at 20– 22℃ and the developmental states of the embryos were observed every 30 min. Some embryos at midgastrula stage and tailbud stage were placed in an RNA-free EP tube, rinsed with sterilized phosphate-buffered saline (PBS), fixed in RNAwait reagent overnight at 4℃ and then stored at -80℃ for RNA extraction. Some embryos were collected in sterilized PBS buffer to be directly used for morphological observation. Each experiment was conducted with triplicate. The embryos injected with 10 pg of control miRNA or control LNA were analyzed as control group.
|
|
Table 2 miRNA and LNA sequences |
The larva began to deflect the right eye about 22 days post hatching (dph). From then on, the larva were treated with 30 mg mL-1 Thiourea (TU) during metamorphosis until 35 dph. The samples were collected and fixed in RNAwait regent overnight at 4℃ and then stored at -80℃ for qRTPCR. The larva without any treatment were collected at the same developmental stage as control.
2.8 Statistical AnalysisData were expressed as means ± standard error of the mean (SEM), and were analyzed by one-way analysis of variance. All statistical analyses were performed using the SPSS 19.0 software (IBM, New York, USA). P < 0.05 indicated statistical significance.
3 Results 3.1 Lefty is a Target of MiR-430 3.1.1 Amplification of P. olivaceus 3'UTR (Polefty-3'UTR) FragmentSince miR-430 has been shown to inhibit lefty in the Nodal signaling pathway in zebrafish, and it has an impact on the early development of the embryos (Choi et al., 2007), we want to explore whether miR-430 also regulates lefty in P. olivaceus. Two types of miR-430, miR-430a and miR-430c, were found in the microRNA database of P. olivaceus. A 380 bp fragment of Polefty-3'UTR was amplified, which contained the target sites of miR-430a and miR-430c (Figs. 1A and 1B). The target sites were very conservative among different fish species (Fig. 2). We speculated that miR-430 has a certain regulation effect on lefty through these two sites.

|
Fig. 1 Complementary pairing of the sequence between miR-430 and its target sites in the Polefty: A, the complementary pairing between miR-430a and the predicted target site; B, the complementary pairing between miR-430c and the predicted target site. The bases of the complementary pairing between miR-430 and the predicted target sites were highlighted with the red color. |

|
Fig. 2 Multiple alignment of miR-430a and miR-430c stem-loop sequence in different fish species. Identical sequences are in dark grey background. The conserved regions where miR-430 may bind to the Polefty-3'UTR are indicated with red boxes. The sequences were obtained with the following GeneBank accession and miRBase accession numbers: Danio rerio (Dr, NR_034234.2 and NR_045220.1); Oryzias latipes (Ol, MI0013008 and MI0013009); Petromyzon marinus (Pm, MI0017130 and MI0017132); Gadus morhua (Gm, MI0036111 and MI0036013) and Cyprinus carpio (Cc, MI0023398). |
In order to verify the region of target sites in Polefty by miR-430, we designed two expression vectors: mcherry-lefty-3'UTR vector with full Polefty-3'UTR region and mcherry-lefty-3'UTR mutant vector with deleted target site of miR430 (including miR430a and miR430c) on the Polefty-3'UTR fragment. Two kinds of vectors were cotransfected into embryonic cells of P. olivaceus together with exogenous miR-430, respectively. The effect of miR-430 on Polefty were detected by observing the fluorescence strength of the cells. In the cells transfected with the mcherry-lefty-3'UTR vector or the mcherry-lefty-3'UTR mutant vector with control miRNA, red fluorescence signals could be observed. This indicates that the constructed vector can be successfully transcribed and translated in the cells without miR-430 interference, which was employed as a positive control (Figs. 3A1 and 3C1). Red fluorescence were observed in the cells transfected with the mcherry-lefty-3'UTR vector with control miRNA (Fig.3A1), while it was quenched in the cells co-transfected with the mcherry-lefty-3'UTR vector and miR-430 (Fig. 3B1), which indicated that the expression of mcherry-lefty-3'UTR was silenced by the miR-430. Nevertheless, when the cells were transfected with both mcherry-lefty-3'UTR mutant vector and control miRNA, or both mcherry-lefty-3'UTR mutant vector and miR-430, the red fluorescence could be observed in the cells, indicating that transcribed mRNA containing the mcherry-lefty-3'UTR mutant fragment was not recognized by miR-430 and was not inhibited (Figs. 3C1 and 3D1). From the results, we concluded that lefty was indeed the target gene of miR-430 in P. olivaceus.

|
Fig. 3 Co-transfection miRNA (miR-430 and control miRNA) and two kinds of mcherry-lefty-3'UTR vectors (wild type and mutant type) into P. olivaceus embryonic cell. The transfection reagents are as follows: A1, mcherry-lefty-3'UTR vector + control miRNA; B1, mcherry-lefty-3'UTR vector + miR-430; C1, mcherry-lefty-3'UTR mutant vector + control miRNA; D1, mcherry-lefty-3'UTR mutant vector + miR-430. A2–D2 are the views of the above picture under white light. |
The temporal expression patterns of miR-430a and miR-430c during early embryonic development stages were analyzed by qRT-PCR (Fig. 4). The expression trends of miR-430a and miR-430c are very similar. MiR-430a and miR-430c mRNA both persisted throughout the embryogenesis until hatching, and higher expression was detected from 1-cell stage to blastula stage, indicating that miR-430a and miR-430c were maternally inherited. With the embryonic development, the amount of miR-430a and miR-430c transcripts decreased gradually after gastrula, reaching the minimum level at the heart-beating stage, then started to rise before the hatching stage.

|
Fig. 4 Embryonic level of miR-430a and miR-430c mRNA quantified by qRT-PCR. Data are shown as mean ± SEM (n = 3). Values with different superscripts indicate statistical significance (P < 0.05). |
Lefty, a member of the TGF-β super-family, is a transforming growth factor. At the same time, as a Nodal signaling pathway downstream gene, it can play important role in the balance of the Nodal signaling pathway by repressing Nodal expression. Many studies have shown that it plays an important regulatory role during the development of embryonic mesoderm, the formation of gastrula and the asymmetry of vertebrate embryos. Since lefty was the target gene of miR-430, we assumed that miR-430 was involved in the regulation of Nodal signaling pathway and resulted in the successful mesoderm formation in P. olivaceus. To detect this hypothesis, qRT-PCR was used to test the transcription levels of genes related to mesoderm formation, including lefty, sqt, ntl and gsc, after the overexpression of miR-430 in the cells (Fig. 5). The expression level of lefty and sqt were significantly inhibited after injection of miRNA-430 at midgastrula and tailbud stages (P < 0.05) (Figs. 5A and 5B). MiR-430 can inhibit the early embryonic development of P. olivaceus by suppressing lefty, and the inhibition of sqt shows the abnormal Nodal signaling pathway. It can be observed that miR-430 plays an important role in the early mesoderm formation of P. olivaceus.

|
Fig. 5 The relative expression of genes at the stage of 50% epiboly and tailbud after the overexpression of miR-430. A, the relative expression of Polefty at midgastrula and tailbud stage; B, the relative expression of Posqt at midgastrula and tailbud stage; C, the relative expression of Pontl at midgastrula and tailbud stage; D, the relative expression of Pogsc at midgastrula and tailbud stage; All datas are shown as mean ± SD (n = 3), the symbol of '*' represents the significantly different (0.01 < P < 0.05), and the symbol of '**' represents the difference is especially striking (P < 0.01). |
Mesoderm marker gene ntl was also selected to explore the effect of miR-430 on mesodermal development in P. olivaceus. The trancription level of ntl was evaluated at the midgastrula stage and tailbud stage (Fig. 5C). The results showed that the inhibitory effect of miR-430 on ntl was not significant at the midgastrula stage, while it was completely inhibited at the tailbud stage (P < 0.05), which suggested that the effect of miR-430 on ntl may play an essential role in the late stage of embryonic development in P. olivaceus.
However, for the expression level of gsc, no significant difference was observed between the cells injected with miR-430 and the control cells at the midgastrula and tailbud stages (Fig. 5D), indicating that a series of gene expression changes caused by miR-430 did not affect the transcription level of gsc. As a tissue-specific marker gene, gsc is associated with mesodermal neurodevelopment. So, the results showed miR-430 would not affect and regulate the neurodevelopment of mesoderm.
For verifying the effects of miR-430 on development of P. olivaceus embryos, the embryonic phenotype was observed after the upregulation and downregulation of miR-430 (Fig. 6). With the overexpression of miR-430, the head of the embryo was enlarged, the spine was slightly curved (Fig. 6C1), and the tail had a bifurcation (Fig. 6B1). Additionally, after inhibiting miR-430, the head of the embryo was malformed, the spine was swollen (Fig. 6B2), and the tail was deformed (Fig. 6C2). This indicated that miR-430 played an essential role in regulating the development of the head and tail, the organization of central axis, and the left-right asymmetry of the embryos. These results confirmed that stable miR-430 was necessary for the early embryo development.

|
Fig. 6 Morphological observation of P. olivaceus embryos at tailbud stage after the overexpression and the inhibition expression of miR-430. A1, A2 for the control group; B1, B2 for the injection of miR-430 group; C1, C2 for the injection of LNA group. Sample numbers for each group are about 30 zygotes. The head, tail and spine of the embryo was marked with dotted line. |
Previous studies have focused on the stages of metamorphosis according to the changes of eyes in P. olivaceus (Okada et al., 2001). P. olivaceus larvae began to deflect the right eye at 22 dph when it started the metamorphosis development. The right eye moved near the top at 27 dph. Then it deflected to the left side of the fish absolutely at 35 dph. Therefore, we chose these time points to explore the effects of miR-430 on metamorphosis of P. olivaceus. We use (TU) to interfere the normal metamorphosis process of P. olivaceus because some reports have confirmed it can inhibit the metamorphosis of P. olivaceus (Inui et al., 1995).
By qRT-PCR, the levels of miR-430a and miR-430c were identified at different developmental stages between the TU-treated and the control groups (Fig. 7). The miR-430a expression was significantly increased after treatment with TU in the middle of metamorphosis (P < 0.05). However, miR-430c did not show a significant increase. It implied miR-430a might play an important role in larvae metamorphosis development, while miR-430c does not have a corresponding function.
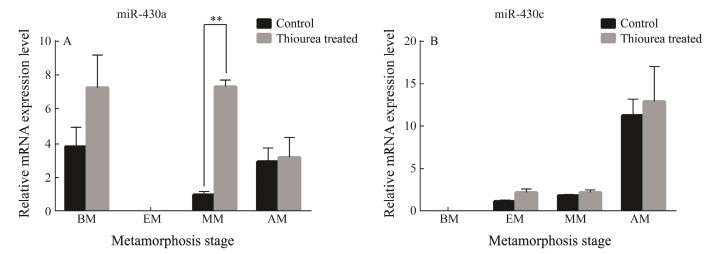
|
Fig. 7 The analysis of expression profiles of miR-430a and miR-430c after larval were treated by TU. All data are shown as mean ± SD (n = 3), the symbol of '*' represents the significantly different (P < 0.05). BM, before metamorphosis; EM, early metamorphosis; MM, middle metamorphosis; AM, after metamorphosis. |
MiR-430 family is highly expressed during early zebrafish development, targets hundreds of mRNAs, and is required for embryonic morphogenesis and clearance of maternal mRNAs (Choi et al., 2007). Recent studies have shown that the zebrafish genome contains more than 70 copies of miR-430 genes, which might facilitate the early boost of the miR-430 transcript during embryogenesis (Thatcher et al., 2008). Among different classes of zebrafish, miR-430, miR-430a, miR-430b, and miR-430c are particularly abundant in early embryos (Giraldez et al., 2006). Since miR-430 family is involved in the early embryonic development, we speculate that it may play certain role in embryonic development in other teleosts. In this study, the injection of miR-430 into the embryos of P. olivaceus at 1-cell stage caused swollen spine, malformed head and deformed tail of embryos (Fig. 6). These results indicate that excessive miR-430 disrupt normal embryogenesis in P. olivaceus.
MiR-430 has been reported to be a well-characterized miRNA expressed at maternal-zygotic transition (MZT) stage in zebrafish (Giraldez et al., 2006). Interestingly, we found miR-430a and miR-430c were also maternal miRNAs in P. olivaceus. The expressions of miR-430a and miR-430c were both up-regulated from zygotic stage to blastrula stage, and gradually became lower since gastrula stage. Considering the sequence alignment results (Fig. 2), we concluded that miR-430 may be functionally conserved between zebrafish and Japanese flounder due to its similar stem-loop sequence.
Previous study has proved that Nodal signal pathway is crucial for the mesoderm formation and the establishment of asymmetrical forms (Liu et al., 2013). Lefty and sqt are both regulators of Nodal signal pathway. Lefty antagonists mediated feedback regulation of Nodal signal to shape left-side morphology (Li et al., 2017). Sqt is a maternal RNA encoding Nodal factor. It is only expressed in two of four cells at the 4-cell stage, and predicts embryonic dorsal (Gore et al., 2005). In this study, we provided the direct evidence that lefty was one of miR-430 target genes in P. olivaceus. Overexpression of miR-430 initiates robust inhibitory effect on lefty and ntl, thereby causing the severe malformation in P. olivaceus. Thus, we speculated that miR-430 plays a vital role in the establishment of proper temporal and spatial expression profile of lefty and ntl, which can directly disturb the normal Nodal signal pathway and then influence mesoderm formation during early embryonic development.
Sqt was believed to have a long-range effect: it may act as a morphogen and directly activate target gene expression at a distance, or activate a relay signal which then acts on distant cells (Chen and Schier, 2001). As a downstream gene of sqt, ntl was more easily induced than gsc by sqt (Gritsman et al., 2000). We hypothesized that ntl was inhibited after midgastrula because of signal transmission from sqt. Choi et al. (2007) found that miR-430 can dampen Nodal signaling by repressing sqt and enhance Nodal signaling by dampening lft2. However, the induction of gsc was not strongly affected in sqt/lft2-TPmiR-430 embryos and MZdicer mutants (Choi, 2009). These results were consistent with our present research results, as the overexpression of miR-430 strongly inhibited the expressions of lefty and sqt rather than that of gsc in midgastrula.
Fu et al. (2013) identified many differently expressed miRNAs during metamorphosis of Japanese flounder. For example, miR-17 and its target gene cdc42 may play a role in flounder metamorphosis development through the metamorphic changes that contribute to cell apoptosis, cell proliferation and cranial cartilage (Zhang et al., 2016). Some researchers propose a mechanism for the mediation of lateralization by the nodal-lefty-pitx2 (NLP) pathway in flounders, in which pitx2, the final left-right determinant of the NLP pathway, is re-expressed in the left habenula at pre-metamorphosis (Suzuki et al., 2009). Previous studies have found that lefty was involved in embryonic organ lateralization and was re-expressed in the larva during flatfish metamorphosis (Schreiber, 2013). So we speculated that miR-430 plays a vital role in metamorphosis as an indirect regulator because it can inhibit lefty as miR-17 can inhibit cdc42 (Zhang et al., 2016).
We analyzed the expression level of miR-430 during metamorphosis. The expression level of miR-430a was significantly increased in the mid-metamorphosis and remained at a relatively high level after metamorphosis. This high expression level suggests that miR-430 plays an important role in the mid and late stages of metamorphosis of P. olivaceus.
5 ConclusionsIn conclusion, lefty has been verified as one of the target genes of miR-430 in P. olivaceus embryos. The excess expression of miR-430 can affect the genes related to Nodal signaling pathway, resulting in the early embryonic developmental malformations. However, excessive miR-430 expression has no significant effect on gsc, the marker gene of neurodevelopment. After disrupting the normal deflection of P. olivaceus by TU treatment, we found the expression of miR-430 family genes change in the metamorphosis stage. Our results showed miR-430a, other than miR-430c, plays an important role in the metamorphosis. This research provides an experimental basis for further studying the relationship between miR-430 and the Nodal signaling pathway.
AcknowledgementsThis study was supported by the National Natural Science Foundation of China (No. 31372511) and the Fundamental Research Funds for the Central Universities (No. 201822026).
Alvarez-Garcia, I. and Miska, E. A., 2005. MicroRNA functions in animal development and human disease. Development, 132(21): 4653-4662. DOI:10.1242/dev.02073 (  0) 0) |
Ambros, V., 2004. The functions of animal microRNAs. Nature, 431(7006): 350. DOI:10.1038/nature02871 (  0) 0) |
Bartel, D. P., 2009. MicroRNAs: Target recognition and regulatory functions. Cell, 136(2): 215-233. DOI:10.1016/j.cell.2009.01.002 (  0) 0) |
Bhattacharya, M., Sharma, A. R., Sharma, G., Patra, B. C., Nam, J. S., Chakraborty, C. and Lee, S. S., 2017. The crucial role and regulations of miRNAs in zebrafish development. Protoplasma, 254(1): 17-31. DOI:10.1007/s00709-015-0931-1 (  0) 0) |
Chen, Y. and Schier, A. F., 2001. The zebrafish Nodal signal squint functions as a morphogen. Nature, 411(6837): 607-610. DOI:10.1038/35079121 (  0) 0) |
Choi, W. Y., 2009. Regulation of antagonistic signaling components by miR-430 microRNAs. PhD thesis. New York University.
(  0) 0) |
Choi, W. Y., Giraldez, A. J. and Schier, A. F., 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science, 318(5848): 271-274. DOI:10.1126/science.1147535 (  0) 0) |
Fu, Y., Shi, Z., Wang, G., Zhang, J., Li, W. and Jia, L., 2013. Expression of let-7 microRNAs that are involved in Japanese flounder (Paralichthys olivaceus) metamorphosis. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 165(2): 106-113. DOI:10.1016/j.cbpb.2013.03.012 (  0) 0) |
Fu, Y., Shi, Z., Wu, M., Zhang, J., Jia, L. and Chen, X., 2011. Identification and differential expression of microRNAs during metamorphosis of the Japanese flounder (Paralichthys olivaceus). PLoS One, 6(7): e22957. DOI:10.1371/journal.pone.0022957 (  0) 0) |
Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., Enright, A. J. and Schier, A. F., 2006. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science, 312(5770): 75-79. DOI:10.1126/science.1122689 (  0) 0) |
Gore, A. V., Maegawa, S., Cheong, A., Gilligan, P. C., Weinberg, E. S. and Sampath, K., 2005. The zebrafish dorsal axis is apparent at the four-cell stage. Nature, 438(7070): 1030-1035. DOI:10.1038/nature04184 (  0) 0) |
Gritsman, K., Talbot, W. S. and Schier, A. F., 2000. Nodal signaling patterns the organizer. Development, 127(5): 921-932. DOI:10.1007/s004290050019 (  0) 0) |
Guan, X. L., Zhang, B. C. and Sun, L., 2019. pol-miR-194a of Japanese flounder (Paralichthys olivaceus) suppresses type I interferon response and facilitates Edwardsiella tarda infection. Fish & Shellfish Immunology, 87: 220-225. DOI:10.1016/j.fsi.2019.01.017 (  0) 0) |
Inui, Y., Yamano, K. and Miwa, S., 1995. The role of thyroid hormone in tissue development in metamorphosing flounder. Aquaculture, 135(1-3): 87-98. DOI:10.1016/0044-8486(95)01017-3 (  0) 0) |
Kloosterman, W. P. and Plasterk, R. H., 2006. The diverse functions of microRNAs in animal development and disease. Developmental Cell, 11(4): 441-450. DOI:10.1016/j.devcel.2006.09.009 (  0) 0) |
Larkin, M. A., Blackshields, G., Brown, N., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J., Gibson, T. J. and Higgins, D. G., 2007. Clustal W and Clustal X version 2.0. Bioinformatics, 23(21): 2947-2948. DOI:10.1093/bioinformatics/btm404 (  0) 0) |
Li, G., Liu, X., Xing, C., Zhang, H., Shimeld, S. M. and Wang, Y., 2017. Cerberus-Nodal-Lefty-Pitx signaling cascade controls left-right asymmetry in amphioxus. Proceedings of the National Academy of Sciences, 114: 3684-3689. DOI:10.1073/pnas.1620519114 (  0) 0) |
Liu, X., Ma, Y., Zhang, C., Wei, S., Cao, Y. and Wang, Q., 2013. Nodal promotes mir206 expression to control convergence and extension movements during zebrafish gastrulation. Journal of Genetics and Genomics, 40(10): 515-521. DOI:10.1016/j.jgg.2013.07.001 (  0) 0) |
Ma, L., Wang, W., Yang, X., Jiang, J., Song, H., Jiang, H., Zhang, Q. and Qi, J., 2014. Characterization of the Dmrt1 gene in the black rockfish Sebastes schlegeli revealed a remarkable sex-dimorphic expression. Fish Physiology and Biochemistry, 40(4): 1263-1274. (  0) 0) |
Mishima, Y., Giraldez, A. J., Takeda, Y., Fujiwara, T., Sakamoto, H., Schier, A. F. and Inoue, K., 2006. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Current Biology, 16(21): 2135-2142. DOI:10.1016/j.cub.2006.08.086 (  0) 0) |
Okada, N., Takagi, Y., Seikai, T., Tanaka, M. and Tagawa, M., 2001. Asymmetrical development of bones and soft tissues during eye migration of metamorphosing Japanese flounder, Paralichthys olivaceus. Cell and Tissue Research, 304(1): 59-66. DOI:10.1007/s004410100353 (  0) 0) |
Schier, A. F., 2003. Nodal signaling in vertebrate development. Annual Review of Cell and Developmental Biology, 19(1): 589-621. DOI:10.1146/annurev.cellbio.19.041603.094522 (  0) 0) |
Schier, A. F. and Talbot, W. S., 2005. Molecular genetics of axis formation in zebrafish. Annual Review of Genetics, 39: 561-613. DOI:10.1146/annurev.genet.37.110801.143752 (  0) 0) |
Schreiber, A. M., 2006. Asymmetric craniofacial remodeling and lateralized behavior in larval flatfish. Journal of Experimental Biology, 209(4): 610-621. DOI:10.1242/jeb.02056 (  0) 0) |
Schreiber, A. M., 2013. Flatfish: An asymmetric perspective on metamorphosis. Current Topics in Developmental Biology, 103: 167-194. DOI:10.1016/B978-0-12-385979-2.00006-X (  0) 0) |
Shen, M. M., 2007. Nodal signaling: Developmental roles and regulation. Development, 134(6): 1023-1034. DOI:10.1242/dev.000166 (  0) 0) |
Staton, A. A., Knaut, H. and Giraldez, A. J., 2011. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nature Genetics, 43(3): 204-211. DOI:10.1038/ng.758 (  0) 0) |
Su, Y., Xiao, X., Ling, H., Huang, N., Liu, F., Su, W., Zhang, Y., Xu, L., Muhammad, K. and Que, Y., 2019. A dynamic degradome landscape on miRNAs and their predicted targets in sugarcane caused by Sporisorium scitamineum stress. BMC Genomics, 20(1): 57. DOI:10.1186/s12864-018-5400-8 (  0) 0) |
Suzuki, T., Washio, Y., Aritaki, M., Fujinami, Y., Shimizu, D., Uji, S. and Hashimoto, H., 2009. Metamorphic pitx2 expression in the left habenula correlated with lateralization of eye-sidedness in flounder. Development, Growth & Differentiation, 51(9): 797-808. DOI:10.1111/j.1440-169X.2009.01139.x (  0) 0) |
Svoboda, P. and Flemr, M., 2010. The role of miRNAs and endogenous siRNAs in maternal‐to‐zygotic reprogramming and the establishment of pluripotency. EMBO Reports, 11(8): 590-597. DOI:10.1038/embor.2010.102 (  0) 0) |
Takeda, Y., Mishima, Y., Fujiwara, T., Sakamoto, H. and Inoue, K., 2009. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly (A) tail length in zebrafish. PLoS One, 4(10): e7513. DOI:10.1371/journal.pone.0007513 (  0) 0) |
Thatcher, E. J., Bond, J., Paydar, I. and Patton, J. G., 2008. Genomic organization of zebrafish microRNAs. BMC Genomics, 9(1): 253. DOI:10.1186/1471-2164-9-253 (  0) 0) |
Wahid, F., Shehzad, A., Khan, T. and Kim, Y. Y., 2010. Micro-RNAs: Synthesis, mechanism, function, and recent clinical trials. Biochimica et Biophysica Acta (BBA) – Molecular Cell Research, 1803(11): 1231-1243. DOI:10.1016/j.bbamcr.2010.06.013 (  0) 0) |
Zhang, H., Fu, Y., Shi, Z., Su, Y. and Zhang, J., 2016. miR-17 is involved in Japanese flounder (Paralichthys olivaceus) development by targeting the Cdc42 mRNA. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 191: 163-170. DOI:10.1016/j.cbpb.2015.10.005 (  0) 0) |
Zhang, X., Li, L., Jiang, H., Ma, J. E., Li, J. and Chen, J., 2018. Identification and differential expression of microRNAs in testis and ovary of Amur sturgeon (Acipenser schrenckii). Gene, 658: 36-46. DOI:10.1016/j.gene.2018.03.014 (  0) 0) |
 2020, Vol. 19
2020, Vol. 19


