Growth-related traits are of great importance for aquatic species as they influence the production directly. As a breeding target, growth-related traits are controlled by many genes and multiple pathways. Some previous studies have identified some candidate genes related to growth, revealing the genetic basis of growth in aquatic species. Growth hormone (GH) gene was one of the earliest genes in growth, development, reproduction, immune function, food conversion and appetite of fish (Quik et al., 2010). GH gene is an ideal candidate for fish growth trait selection, because many growth-related GH genes' single nucleotide polymorphisms (SNPs) have been reported in fish species, such as Atlantic salmon (Gross and Nilsson, 1999), Chinook salmon and yellow croaker (Docker and Heath, 2002; Ni et al., 2012). Additionally, GnRH peptides stimulate GH release in goldfish both in vivo and in vitro, and in common carp in vitro (Lin et al., 1993; Lin et al., 1995). In the process of muscle production and regulation, myogenic regulatory factors activate the transcription of genes involved in skeletal muscle production through multiple pathways, thereby promoting the transformation of quiescent muscle satellite cells and other types of cells into myoblasts and promoting the differentiation of myoblasts into mature cells (Sabourin and Rudnicki, 2000). The paired box protein 3 and 7 (Pax3/7) and several Sox genes also contribute to the control of muscle differentiation (Rescan and Ralliere, 2010).
Variations in DNA sequences are known to be the basis for trait variation. For marker-assisted selection (MAS), both genetic markers and linked genes can be used for selecting fish with specific characteristics. In the initial study, growth traits were mainly localized to the somatotropic axis genes, the myogenic regulatory factor genes, and the transforming growth factor genes (Valente et al., 2013). Some studies suggest that GH, IGF (insulin-like growth factor), and myostatin (MSTN) are candidate genes for marker-assisted selection (De-Santis and Jerry, 2007). Singlenucleotide polymorphisms (SNPs) (Sánchez-Ramos et al., 2012), simple sequence repeats (SSRs) (Almuly et al., 2005), variable number of tandem repeats (VNTRs) and restriction fragment length polymorphisms (RFLPs) have been identified in large numbers in fish growth-related traits (Almuly et al., 2005; Almuly et al., 2008; Sánchez-Ramos et al., 2012). However, identifying growth-related genes one by one is inefficient. With the advent of high-throughput sequencing technology, it becomes accessible for exploring and genotyping a large number of variants in a genomescale to locate quantitative trait locus (QTLs) and trait-related SNPs. A QTL for growth in linkage group 1 was detected in European sea bass, after which two different QTLs were identified for body weight in the same species (Chatziplis et al., 2007; Massault et al., 2010). In Asian seabass, 43 QTLs were detected for growth traits during the development and two QTLs intervals at two stages were overlapped, while the others were mapped onto different positions (Xia et al., 2013). Twenty-two SNP markers and a mitochondrial haplotype were found to be significantly associated with growth traits in rainbow trout with the full transcriptome analysis of RNA sequences (Salem et al., 2012). Moreover, 435 growth traits-related SNPs were identified through transcriptome SNP analysis between fast- and slow-growing turbot (Robledo et al., 2017). Several miRNAs have been known to regulate muscle growth and development. For example, 168 small RNAs are expressed in the muscles of fast myoblasts, and the expression regulation of several small RNAs is related to the transition from hyperplasia to hypertrophy during development (Johnston et al., 2009).
Tongue sole has become a very important maricultural fish in China. At present, although families with fast growth and high female ratio have been screened out, a breeding generation of Chinese tongue sole needs 2–3 years, and selection breeding faces huge time cost. Using 1007 SSR markers, four weight- and body-width-related quantitative trait loci (QTLs) were detected in Chinese tongue sole (Song et al., 2012). IGF-1 and GH genes of Chines tongue sole were cloned (Ma et al., 2011, 2012). Furthermore, the expression levels of these two genes in females were significantly higher than those in males. There was no significant difference in GHRH expression between males and females before the age of 8 months, but GHRH expression in females was significantly higher than that in males during the age of 9–12 months (Ji et al., 2011). These studies are all at the one-gene level to study the characteristics of tongue sole, which is of great significance to the study of the growth of tongue sole and other fish species. However, functional studies related to growth traits at the genome-wide level has not been carried out.
To gain knowledge on the key genes related to growth in Chinese tongue sole, genome-wide resequencing studies were conducted in large and small groups in this study. Two methods, Fisher's exact test combined with FST statistics were used to identify genetic markers associated with growth. GO and KEGG enrichment analysis of genes were further carried out for the candidate genes. The markers and genes identified in our study can be applied to selective breeding of tongue sole and thus will promote the aquaculture industry and increase economic efficiency.
2 Materials and Methods 2.1 DNA ExtractionExtreme-sized individuals of Chinese tongue soles were selected from mixed cultures. Muscle samples were taken from female individuals (both physiological and genetic sex). Body length, width, and weight were measured. Total DNA was extracted using the Phenol-chloroform method following the protocol of our laboratory and dissolved with TE to preserve genomic DNA. The quality and the concentration of extracted DNA were detected using agarose gel electrophoresis and Nanodrop 2000 spectrophotometers. Then DNA samples were frozen at −20℃.
2.2 WGS and Quality Control (QC)The original image data generated by the sequencing machine were converted into sequence data via base calling (Illumina pipeline CASAVA v1.8.2) and then subjected to quality control (QC) procedure to remove unusable reads:
1) The reads contain the Illumina library construction adapters;
2) The reads contain more than 10% unknown bases (N bases);
3) One end of the read contains more than 50% of lowquality bases (sequencing quality value ≤ 5).
2.3 Variant Detection and AnnotationSequencing reads were aligned to the reference genome using BWA-0.7.17 with default parameters (Li and Durbin, 2009). Duplicate removal was performed using samtools and PICARD (http://picard.sourceforge.net).
The raw SNP/InDel sets are called by samtools-1.9 with the parameters as '-q 1 -C 50 -m 2 -F 0.002 -d 1000' (Li et al., 2009). Then we filtered these sets using the following criteria:
1) The mapping quality > 20;
2) The depth of the variate position > 4.
SnpEff v.4.266 was used for the functional annotation of variants with the Cynoglossus semilaevis reference genome (Cingolani et al., 2012; Chen et al., 2014).
The TBtools software was used to plot the statistical results of the variants (Chen et al., 2018).
2.4 Statistical Difference AnalysisDifferential allele frequency and genotype frequency between large group (LG) and small group (SG) samples were obtained by script following the formula 'allele/genotype frequency = (the count of a type of allele/genotype in one locus)/(total count of allele/genotype in one locus)'. The significance of the allele frequency and genotype frequency between the two groups was tested by Fisher's exact test. Those SNPs showing P-values < 0.001 and further MAF > 0.10 were considered significant. GO term and KEGG pathway enrichment analyses of the annotated genes were performed for the selected genes using the OmicShare tools (www.omicshare. com/tools), a free online platform for data analysis.
2.5 Selective Scanning by F StatisticsAn FST value was calculated for each window to compare the difference between any two pools. Before the calculation, the selected windows and defining sweep windows were identified. VCFtools v0.1.13 was applied to calculated Weir & Cockerham's FST values on a 10 kb window size and with 5 kb step size between windows (Danecek et al., 2011).
2.6 Kompetitive Allele Specific PCR (KASP) GenotypingKASP high-throughput genotyping is a SNPline genotyping assay based on KASP technology. KASP is a competitive allele-specific PCR that performs genotyping of SNPs by performing precise biallelic discrimination in a wide range of genomic DNA samples based on the specific matching of primer-terminal bases.
1) Design 2 SNP PCR primers, each corresponding to an SNP allele. The 3' ends are respectively two alleles, and the 5' ends are respectively FAM/HEX tag sequences.
2) Fluorescent probes were designed to be identical to the tag sequence, with a FAM/HEX fluorophore at each of the 5' end. Besides, a quenching probe with a quenching group at the 3' end was designed.
3) Perform specific PCR amplification and perform SNP genotyping by detecting fluorescence.
2.7 Ethics StatementThe study was conducted following the guidelines and regulations established by the Chinese Government Principles for the Utilization and Care of Animals Used in Testing, Research, and Training. All the experimental protocols were permitted and approved by College of Marine Life Science, Ocean University of China (Qingdao, China).
3 Results 3.1 DNA Resequencing OutputWe conducted a resequencing analysis of two groups of Chinese tongue sole among 15 large individuals and 15 small individuals (Fig. 1). The exclusion of pseudo males was carried out following a previously described method (Liu et al., 2014). A total of 30 libraries were constructed using total DNAs from muscle tissue. Each library was sequenced with an Illumina HiseqTM 4000. As a result, a total of 229.86G raw data were generated, for an average of 7.66G per sample, ranging from 5.43G to 12.70G. After filtering, a total of 228.78G clean data was retained, for an average of 7.63G per sample, ranging from 5.39G to 12.67G. The average effective rate is 99.50% (ranging from 98.64% to 99.77%). The high quality of resequencing (Q20 ≥ 94.13%, Q30 ≥ 87.86%) and the normal distribution of GC content (40.47% – 43.69%) means the next step mapping is effective (Table 1). On average, 96.44% of the clean reads align to the Chinese tongue sole genome, ranging from 94.12% to 97.06%. The average depth is 14.07X, ranging from 9.93X to 22.57X (Table 2). The mapping results are qualified, which can be used in subsequent mutation detection and related analysis.
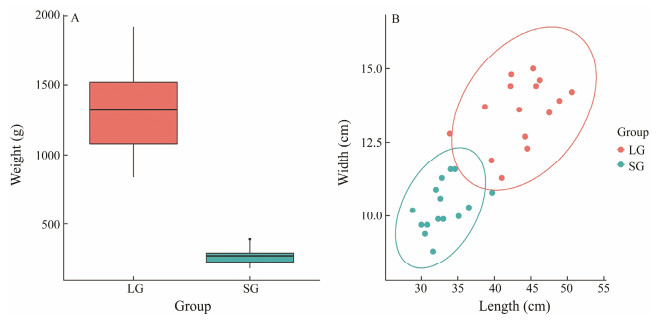
|
Fig. 1 Statistics of two groups of Chinese tongue sole samples for resequencing. A, Body weight of the LG and SG groups; B, Body length and width of the LG and SG groups. |
|
|
Table 1 The statistics of re-sequencing data |
|
|
Table 2 The mapping rate of resequencing data to the genome |
A total of 6545735 SNPs and 1016745 InDels were called based on resequencing data. The sequences containing these SNPs were annotated according to the Chinese tongue sole genome information. Because several effects can be annotated to one locus, a total of 34132325 effected SNPs and 5506066 effected InDels were detected at the level of the entire genome (including repeated annotation). A total of 11846123 effected SNPs (34.71%) and 1796516 effected InDels (32.63%) were found in exonic regions. Among these SNPs, 244456 correspond to non-synonymous variants and 551545 correspond to synonymous variants. There are 34158680 transitions and 21419741 transversions among all SNPs, and the Ts/Tv ratio is 1.59.
3.3 Association AnalysisAllele frequencies and genotype frequencies were estimated for LG (large group) and SG (small group) of Chinese tongue sole, and the significance of each frequency was tested. Significant difference in allele frequencies between LG and SG were detected in 1206 SNPs and 428 Indels, while significant difference in genotype frequencies were found in 830 SNPs and 149 InDels. These 2151 significant variants were located in 1324 genes. Among these variants, 414 significant variants were located in upstream of genes; 170 significant variants were located in exonic regions, which include 58 synonymous variants and 23 non-synonymous variants; 1077 significant variants were located in intron regions; 1027 significant variants were located in non-genetic regions. (A particular type of variant of one gene may be another type of variant of another gene).
3.4 The GO and KEGG Enrichment of Growth-Related MarkersTo better understand the function of these significantly divergent genes between LG and SG groups of Chinese tongue sole, GO term enrichment was conducted. As the results, 669 GO terms with P < 0.05 were significantly enriched in whole divergent genes, and 28 GO terms with FDR < 0.05 were significantly enriched (Fig. 3). Among the 28 most significantly enriched GO terms, there were 17 GO categories within the 'Biological process' and the three most abundant terms were 'cell differentiation', 'single-organism cellular process' and 'cellular developmental process'. There were also a large number of genes being involved in 'single-organism process', 'response to stimulus', 'developmental process', 'single-organism developmental process' and 'single-multicellular organism process'. In the 'Cellular component' division, there were 11 categories and the top three categories were 'cell junction', 'synapse', and 'membrane'. Though there were no most significantly enriched terms in the 'Molecular function' division, the top three categories were 'protein tyrosine kinase activity', 'metalloaminopeptidase activity' and 'transmembrane receptor protein tyrosine kinase activity'.
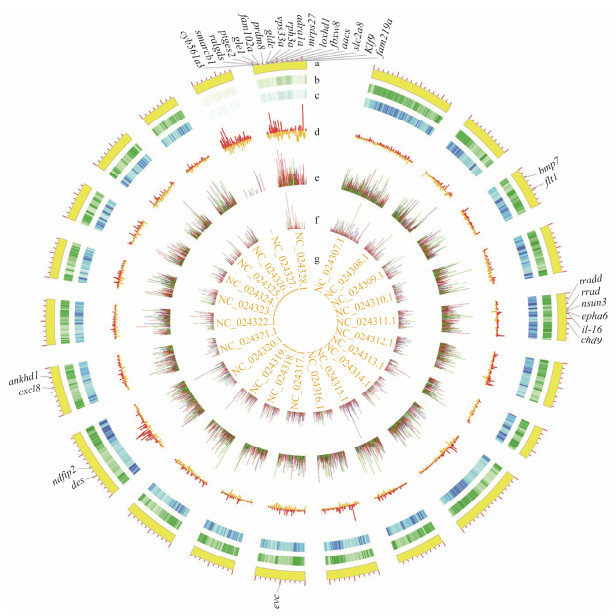
|
Fig. 2 SNP/Indel calling and annotating results. a, Chromosome scale; b, SNP density, the light to dark indicates low to high density; c, Indel density, the light to dark indicates low to high density; d, FST between LG and SG groups; e, Fisher P-value of SNPs; f, Fisher P-value of Indels; g, Chromosome numbers. In e and f, red color stands for allele frequencies; green and blue represent frequencies of different genotypes. |
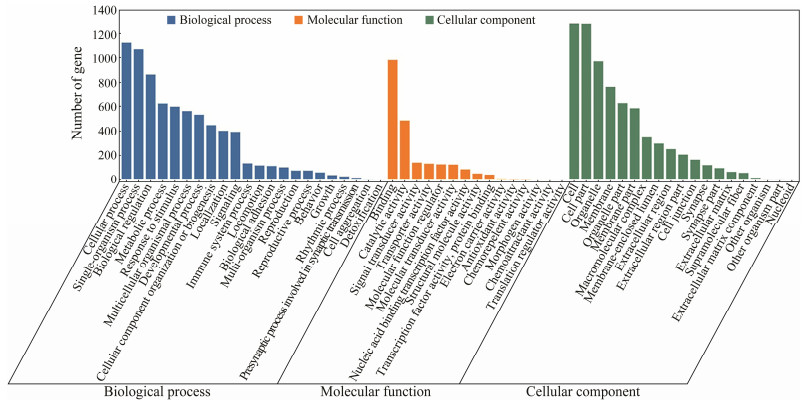
|
Fig. 3 GO enrichment of candidate genes. Enriched GO terms of selected marker genes (P < 0.05). |
Subsequently, KEGG pathway enrichment revealed that 16 pathways were significantly enriched with P < 0.05 (Fig. 4). There were 7 pathways (N-Glycan biosynthesis, other types of O-glycan biosynthesis, glycosphingolipid biosynthesis-lacto and neolacto series, fatty acid biosynthesis, citrate cycle, biotin metabolism, and glycosphingolipid biosynthesis-ganglio series) significantly enriched in metabolism division. In cellular community division, focal adhesion was significantly enriched. In signal transduction division, the Rap1 signaling pathway and Ras signaling pathway were significantly enriched. Among the pathways at the top of the non-significant ranking, salivary secretion, pancreatic secretion, steroid hormone biosynthesis, and GnRH signaling pathway were enriched. There were also a large number of genes being enriched in the PI3K-Akt signaling pathway and the actin cytoskeleton regulation.
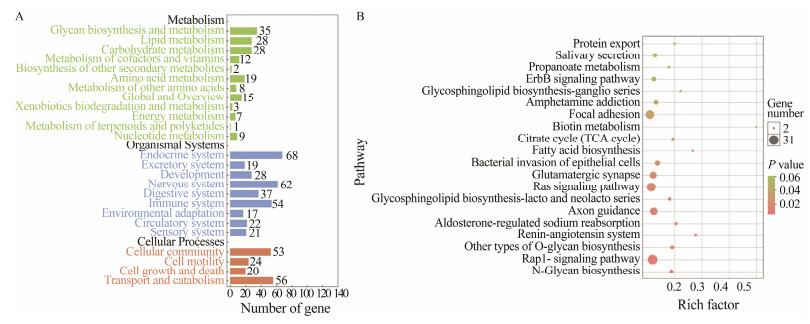
|
Fig. 4 KEGG pathway enrichment of candidate genes. A, Count of enriched KEGG terms of selected marker genes of KEGG A and B class; B, Enriched KEGG pathways of selected marker genes (P < 0.05). The size bar indicates enriched gene numbers of each KEGG term. The color bar indicates P value from low (green) to high (red). |
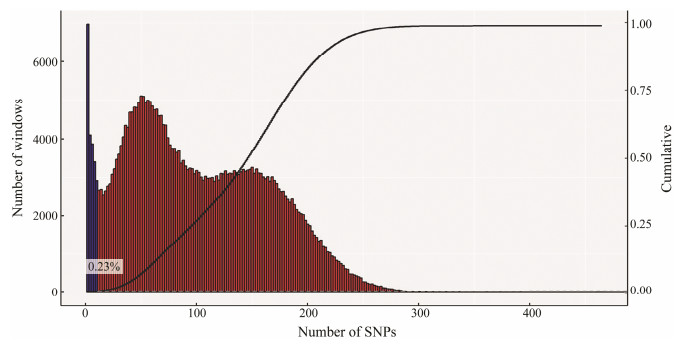
Genome scanning by comparing the single locus estimates of FST between different populations might identify regions of the genome that have been subjected to diversifying selection (Holsinger and Weir, 2009). Among all 91551 windows (10 kb in length, sliding in 5 kb steps across the Chinese tongue sole genome), 87939 windows contained > 10 variants while covering 96.05% of the genome and 99.77% of variants (Fig. 5), which were used to detect signatures of selective sweeps. 4397 windows were selected, and all of them have significantly higher Weir & Cockerham's FST values (5% right tail). Consequently, we identified a total of 42.93 Mb genomic regions (4.80% of the genome) with strong selective sweep signals between LG and SG groups (Fig. 2).
|
Fig. 5 SNP distribution in windows. |
Combining the significant different frequencies, we detected 571 variants. Among these variants, 130 significant variants were located in upstream of genes, 46 significant variants were located in exonic regions, which include 10 synonymous variants and 7 non-synonymous variants, 275 significant variants were located in intron regions, 347 significant variants were located in non-genetic regions. We detected that non-synonymous variants were located at the coding region of Pro-interleukin-16 (il16), Ephrin type-A receptor 6 (epha6), Interleukin-8 (cxcl8), Alpha-1A adrenergic receptor (adra1a), tRNA (cytosine(34)-C(5))-me thyltransferase (nsun3, mitochondrial) and Uncharacterized protein C11orf65 homolog. And we also detected synonymous variants in the coding region of Acetoacetyl-CoA synthetase (aacs), Chromodomain-helicase-DNA-binding protein 9 (chd9), Ellis-van Creveld syndrome protein homolog (evc), F-box/WD repeat-containing protein 8 (fbxw8), Ankyrin repeat and KH domain-containing protein 1 (ankhd), E3 ubiquitin-protein ligase MYCBP2 (mycbp2), tRNA (cytosine(34)-C(5))-methyltransferase (nsun3, mitochondrial), PR domain zinc finger protein 8 (prdm8) and Ral guanine nucleotide dissociation stimulator (rgl4).
In addition to those variants located in coding regions, we also scan the untranslated regions (3', 5' UTR and upstream), and growth-related genes such as Bone morphogenetic protein 7 (bmp7), Vascular endothelial growth factor receptor 1 (flt1), Desmin (des), Muscleblind-like protein 1 (mbnl1), Solute carrier family 2 and facilitated glucose transporter member 8 (slc2a8) were detected.
3.6 Resequencing Results Verified by KASP GenotypingAmong the candidate growth-related genes, we randomly selected 9 extremely significant loci and performed KASP genotyping on the sequenced samples to verify the reliability of the resequencing results. The genotyping results of 8 loci were consistent with the resequencing results, and there was a significant difference between large and small groups (Table 3), which proved that the resequencing results were reliable.
|
|
Table 3 The comparison of P-values between resequencing and KASP genotyping |
In present study, we re-sequenced two groups of female Chinese tongue sole, a large group and a small group, from the same population with different growth rates, and more than two thousand variants were detected. Most of these variants were related to different aspects of growth. A large number of densely distributed divergent variants were clustered at autosome 11 and sex chromosome W. Interestingly, several selected genes are evenly distributed on the sex chromosome W. Quantitative traits located by genome-wide resequencing has been widely used in crop and animal husbandry breeding. And it is very effective and greatly shortens the breeding cycle (Yano et al., 2016).
Growth is coordinated and regulated by different aspect of body systems, including muscle architecture, muscle regeneration and muscle fusion, embryo development, neurological regulation and endocrine regulation, energy metabolism, protein synthesis and degradation. In our work, we detected genes from several aspects related to Chinese tongue sole growth. For instance, there are lots of genes involving in different kinds of metabolism pathways, digestive, excretory, endocrine and immune systems, neurogenesis and sensory formation, regulation of actin cytoskeleton and steroid hormone biosynthesis.
4.1 Muscle ArchitectureUnlike other vertebrates, fish's muscles are separated by discrete layers with different fiber types. Vertebrates have two main types of striated muscle fibers: red and white, which specialize in low-speed swimming and bursts of maximum in fish, respectively (Sanger and Stoiber, 2001). The fat content of fish is generally low, though the speed of muscle growth and the size of muscle fibers are the main indicators to measure the growth of fish. We detected six myosin genes and one intermediate filament gene des was detected in the highly differentiated region by F-statistics. Desmin (des) is a muscle-specific type Ⅲ intermediate filament, which is essential to the structure and function of muscle. It plays an important role in maintaining sarcomere structure, connecting Z disc and forming myofibrils. These genes are not only connected with the cytoskeleton of sarcomere cells, but also with the nucleus and mitochondria, thus providing strength for muscle fibers during activity (Hnia et al., 2015).
4.2 Post-Embryonic GrowthTeleosts often exhibit an indeterminate growth pattern, with body size and muscle mass increasing until mortality or senescence occurs (Johnston et al., 2011). In addition, the body size increases dramatically between embryo and adult in most teleosts. Therefore, there must be certain mechanisms that control the rapid development of the body and the sustained growth of muscles. Since muscle cells are multinucleated, terminal differentiated tissue, postembryonic growth requires a source of proliferative MPC. The adaxial cells of zebrafish differentiate into slow muscle fibers of adult fish, while the lateral presomitic cells remain deep in the muscle and differentiate into fast muscle fibers (Devoto et al., 1996).
Interestingly, we found that a series of selected genes were associated with cellular development, including Chromodomain-helicase-DNA-binding protein 9 (chd9), Ellisvan Creveld syndrome protein homolog (evc) and PR domain zinc finger protein 8 (prdm8). Each of them had a synonymous SNP located in FST selective regions. chd9 participates in the differentiation of progenitor cells during osteogenesis and participates in the relaxed chromatin structure of mouse oocyte, which is a prerequisite for post-fertilization versatility (Shur et al., 2006; Ooga et al., 2018). evc is a component of EvC complex that regulates ciliary Hedgehog (Hh) signaling, and participates in endochondral growth and skeletal development (Ruiz-Perez et al., 2007). prdm8 is a possible histone methyltransferase that preferentially acts on the ninth Lys of Histone H3 (Eom et al., 2009). Steroid-producing marker genes such as cyp17a1 and lhcgr are involved in the control of steroid production through transcriptional inhibition (Ross et al., 2012). It forms a transcriptional inhibitory complex with bhlhe22, which controls genes involved in nerve development and differentiation. In the retina, rod bipolar and type 2 non-conical bipolar cells are required to survive (Jung et al., 2015). Beyond these, bone morphogenetic protein 7 (bmp7) can induce cartilage and bone formation, which is a bone-inducing factor leading to epithelial osteogenesis and plays a role in calcium regulation and bone balance. In addition, bmp7 plays an important role in the development of mammalian skeleton, kidney, eyes and brown adipose tissue (Dudley et al., 1995; Katagiri et al., 1998; Boon et al., 2013). During the development of zebrafish, bmp7b is strongly expressed in the eyes, ears, pronephros and gastrointestinal system (Shawi and Serluca, 2008). Prostaglandin E synthase 2 (ptges2) catalyzes the conversion of PGH2 into prostaglandin E2 (pge2) which is more stable. On one hand, PGE2 production and transport are essential for the formation of cilia and photoreceptor development in zebrafish (Li et al., 2019). On the other hand, PGE2 signal participates in medaka ovulation follicle rupture and results in the cytoskeleton rearrangement of GC actin during ovulation (Takahashi et al., 2018). There is a basement membrane on the outer surface of Schwann cells, which plays an important role in peripheral nerve regeneration. In zebrafish embryos, the lack of gle1, a messenger of gene output, can damage the development of Schwann cells (Seytanoglu et al., 2016). Nucleoporin gle1 is necessary for the nucleus to export poly (a) tail-containing RNA into the cytoplasm. It may be involved in the terminal step of the nuclear pore complex (NPC) in the process of RNA transfer (Bolger et al., 2008), and it is also one of the genes we screened.
In addition to a large number of genes selected for cellular development, we have also screened a series of genes for cell proliferation and differentiation, which may be related to post-embryonic growth. In interleukin-8 (il-8), two non-synonymous SNPs were detected and were consistent with the FST selection regions. il-8 is an autocrine factor that promotes the growth of fibroblasts. In humans, il-8 plays a role in growth and maintenance of ectopic endometrial tissues by chemically attracting and stimulating leucocyte to secrete growth factors and cytokines, and by directly affecting the proliferation of endometrial cells (Arici, 2002). In alpha-1a adrenergic receptor (adra1a), one non-synonymous SNP in FST selection regions was detected. adra1a is a member of the G protein-coupled receptor superfamily. They activate mitotic responses and regulate the growth and proliferation of many cells (Luttrell, 2008). In ral guanine nucleotide dissociation stimulator (ralgds), three upstream SNPs, two 3'UTR variants and one synonymous SNP were detected. ralgds stimulates the separation of ras-related ral-a and Ral-b GTPases from GDP, thus allowing GTP to bind and activate GTPases. ral-a is a multifunctional GTPase involved in many cell processes, including gene expression, cell migration, cell proliferation, carcinogenic transformation and membrane transport. Its multiple functions are realized by interacting with different downstream effectors. At the late stage of cell division, after the formation of bridges between dividing cells is completed, ral-b mediates the recruitment of external cysts to the midbody to drive separation (Cascone et al., 2008). Vascular endothelial growth factor receptor 1 (flt1) is a tyrosine protein kinase. It is the cell surface receptor of vascular endothelial growth factor A, vascular endothelial growth factor B and prostaglandin F, and plays an important role in the development of embryonic vascular system, angiogenesis, cell survival, cell migration, macrophage function, chemotaxis and regulation of cancer cell invasion. It may also play an important role in negative regulation of embryonic angiogenesis by inhibiting endothelial cell proliferation (Kendall and Thomas, 1993). Furthermore, flt1 can promote the expression of GFP in zebrafish arteries and veins as a special enhancer (Bussmann et al., 2010).
4.3 Neurological and Sensory DevelopmentThe development of nerves and senses affects the individual's speed of movement, the ability to find food, the ability to avoid harm, and the regulation of digestion and absorption, so it is inseparable from the individual's growth. A transcription factor for brain development, a vesicle transporter and a secreted glycoprotein involved in regulating chemosensory cilia in olfactory neurons and optic nerve extension were screened by differential expression analysis in turbot (Robledo et al., 2017). A non-synonymous SNP of ephrin type-a receptor 6 (epha6) was detected. It is a tyrosine kinase of EPH receptor. Its membrane-binding ligand ephrins plays a key role in pattern formation and morphogenesis. An important role of EPH receptor and renin is to mediate contact-dependent rejection, which is related to the pathological findings of axons and crest cells and the restriction of cell mixing between the posterior brain segments. Biochemical studies have shown that the degree of EPH receptor polymerization can regulate cell response, and actin cytoskeleton is the main target of EPH receptor-activated intracellular pathway (Wilkinson, 2000). In mice and zebrafish, epha6 was found to be associated with retinal axon guidance, which is one of the eye development receptors, and inhibited by dimer HMX1 (Marcelli et al., 2014). Lipoxygenase homology domain-containing protein 1 (loxhd1) is required for normal function of inner ear hair cells (Grillet et al., 2009). Acetoacetyl-CoA synthetase (aacs) was detected as a synonymous SNP in selected regions. It is a ketone bodyutilizing enzyme for the synthesis of cholesterol and fatty acids and is highly expressed in the brain.
4.4 Hormonal RegulationPituitary growth hormone (GH) is generally considered to be essential for postnatal growth of mammals and is one of the most widely studied genes in the field of fish growth. In fishes, growth hormone induces muscle growth by regulating the expression of several genes of myostatin (MSTN), atrophy, growth hormone and insulin-like growth factor system, and myogenic regulatory factors (MRFs). IGFs stimulates myogenic cell proliferation, differentiation and protein synthesis through MAPK/ERK and PI3K/ Akt/Tor signaling pathways and eliminates protein degradation and atrophy through PI3K/Akt/FoxO signaling pathway (Fuentes et al., 2013). On one hand, molecular variation markers related to growth hormone receptor gene were detected in several animals between fast-growing species and slow-growing species (Zhang et al., 2017). On the other hand, growth hormone transgenic strains may have higher growth rate (Chen et al., 2015). In our research, we detected a few genes associated with some other growthrelated hormones and their receptors, such as solute carrier family 2, facilitated glucose transporter member 8 (slc2a8), which is an insulin-regulated facilitative glucose transporter. Binding cytochalasin B in a glucose-inhibitable manner (Ibberson et al., 2000), protein FAM102A (fam102a) plays a role in estrogen action (Wang et al., 2004). Insulin receptor substrate 1 (IRS1), insulin receptor (insr), parathyroid hormone-related protein (pthlh), Parathyroid hormone/parathyroid hormone-related peptide receptor (pth1r) and acidic fibroblast growth factor intracellular-binding protein (fibp) were all detected in divergent variants analysis, but not in selective scanning analysis.
4.5 Protein DegradationProteins in the body are constantly decomposed and replaced. In many teleosts, skeletal muscle also undergoes a process of accelerating protein decomposition during the seasonal stages of fasting and gonadal maturation. When protein degradation far exceeds protein synthesis, atrophy occurs. Protein degradation is complex, including ubiquitin-proteasome system, calpain, NF-KB pathway and lysosome. Damaged proteins and proteins with short halflives are modified by ubiquitination, resulting in their degradation by a multi-catalytic protease complex called proteasome (Johnston et al., 2011). We detected two genes related to protein degradation. One is tumor necrosis factor receptor type 1-associated DEATH domain protein (tradd), which contains binding molecules interacting with TNFRSF1a/TNFR1 and mediating programmed cell death signals and activation of NF-kappab. The other is the F-BOX/WD repeat containing protein 8 (FBXW8) that mediates the degradation of MAP4K1/hpk1, through which affecting cell proliferation and differentiation (Wang et al., 2014).
4.6 Sexual DimorphismThe genetic sex of Chinese tongue sole is determined by W and Z chromosomes and exhibits female-biased sexual size dimorphism. 64.35% of the module genes were found on the W chromosome in the WGCNA analysis of the body size sexual dimorphism (Chen et al., 2014). In the present study, 58.06% (18/31) genes associated with growth were detected on the W chromosome. Present and previous studies have shown that the W chromosome of Chinese tongue sole not only has genes affecting the male and female body size sexual dimorphism, but also has genes controling the size of female tongue sole. The results were consistent with the results of the body size sexual dimorphism research that a large number of genes that led to male and female body size dimorphism bias were located on the W chromosome (Weir and Cockerham, 1984).
Although we have screened a large number of potential growth-related loci, further validation work needs to be done. This is probably the result of polygenic interaction, and its mechanism needs further research. This study provides a large number of potential research options for the study of genes related to growth traits in Chinese tongue sole, whereas further validation work needs to be done to investigate the gene functions.
5 ConclusionsInstead of random sampling, we selected individuals with extreme weight in population for sampling and used statistical difference analysis to screen out a large number of growth-related mutation sites. Assuming that extreme groups come from different populations, FST method was used to further screen regions with significant differences between two groups in the genome.The results of statistical difference analysis were intersected to further locate genes and loci significantly related to growth traits. It was found that there were a large number of sex-independent but growth-related genes on the W chromosome of the Chinese tongue sole. It was the presence of these genes that led to the larger body size of female (ZW) than male (ZZ) at the same age.
AcknowledgementThis work was supported by the National Natural Sci- ence Foundation of China (No. 31402292).
Almuly, R., Poleg-Danin, Y., Gorshkov, S., Gorshkova, G., Rapoport, B., Soller, M., Kashi, Y. and Funkenstein, B., 2005. Characterization of the 5' flanking region of the growth hormone gene of the marine teleost, gilthead sea bream Sparus aurata: analysis of a polymorphic microsatellite in the proximal promoter. Fisheries Science, 71(3): 479-490. DOI:10.1111/j.1444-2906.2005.00991.x (  0) 0) |
Almuly, R., Skopal, T. and Funkenstein, B., 2008. Regulatory regions in the promoter and first intron of Sparus aurata growth hormone gene: Repression of gene activity by a polymorphic minisatellite. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 3(1): 43-50. DOI:10.1016/j.cbd.2006.12.006 (  0) 0) |
Arici, A., 2002. Local cytokines in endometrial tissue: The role of interleukin-8 in the pathogenesis of endometriosis. Annals of the New York Academy of Sciences, 955(1): 101-109. DOI:10.1111/j.1749-6632.2002.tb02770.x (  0) 0) |
Bolger, T. A., Folkmann, A. W., Tran, E. J. and Wente, S. R., 2008. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell, 134(4): 624-633. DOI:10.1016/j.cell.2008.06.027 (  0) 0) |
Boon, M. R., van den Berg, S. A., Wang, Y., van den Bossche, J., Karkampouna, S., Bauwens, M., De Saint-Hubert, M., van der Horst, G., Vukicevic, S. and and de Winther, M. P., 2013. BMP7 activates brown adipose tissue and reduces diet-induced obesity only at subthermoneutrality. PLoS One, 8(9): e74083. DOI:10.1371/journal.pone.0074083 (  0) 0) |
Bussmann, J., Bos, F. L., Urasaki, A., Kawakami, K., Duckers, H. J. and Schulte, -Merker S., 2010. Arteries provide essential guidance cues for lymphatic endothelial cells in the zebrafish trunk. Development, 137(16): 2653-2657. DOI:10.1242/dev.048207 (  0) 0) |
Cascone, I., Selimoglu, R., Ozdemir, C., Del, Nery E., Yeaman, C., White, M. and Camonis, J., 2008. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. The EMBO Journal, 27(18): 2375-2387. DOI:10.1038/emboj.2008.166 (  0) 0) |
Chatziplis, D., Batargias, C., Tsigenopoulos, C. S., Magoulas, A., Kollias, S., Kotoulas, G., Volckaert, F. A. and Haley, C. S., 2007. Mapping quantitative trait loci in European sea bass (Dicen-trarchus labrax): The BASSMAP pilot study. Aquaculture, 272: S172-S182. DOI:10.1016/j.aquaculture.2007.08.022 (  0) 0) |
Chen, C., Xia, R., Chen, H. and He, Y., 2018. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv: 289660. DOI:10.1101/289660 (  0) 0) |
Chen, J., Luo, Q., Bao, H., Liao, L., Li, Y., Zhu, Z., Wang, Y. and Hu, W., 2015. The integration characteristics of the exogenous growth hormone gene in a transgenic common carp (Cyprinus carpio L.) with fast-growth performance. Science Bulletin, 60(19): 1654-1660. DOI:10.1007/s11434-015-0893-x (  0) 0) |
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., Song, W., An, N., Chalopin, D. and Volff, J. N., 2014. Wholegenome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nature Genetics, 46(3): 253. (  0) 0) |
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., Land, S. J., Lu, X. and Ruden, D. M., 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6(2): 80-92. (  0) 0) |
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., Handsaker, R. E., Lunter, G., Marth, G. T. and Sherry, S. T., 2011. The variant call format and VCFtools. Bio-informatics, 27(15): 2156-2158. DOI:10.1093/bioinformatics/btr330 (  0) 0) |
De-Santis, C. and Jerry, D. R., 2007. Candidate growth genes in finfish——Where should we be looking?. Aquaculture, 272(1-4): 22-38. DOI:10.1016/j.aquaculture.2007.08.036 (  0) 0) |
Devoto, S. H., Melançon, E., Eisen, J. S. and Westerfield, M., 1996. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development, 122(11): 3371-3380. DOI:10.1101/gad.10.22.2935 (  0) 0) |
Docker, M. F. and Heath, D. D., 2002. PCR-based markers detect genetic variation at growth and immune function-related loci in chinook salmon (Oncorhynchus tshawytscha). Molecu-lar Ecology Notes, 2(4): 606-609. DOI:10.1046/j.1471-8286.2002.00315.x (  0) 0) |
Dudley, A. T., Lyons, K. M. and Robertson, E. J., 1995. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes & Develop-ment, 9(22): 2795-2807. DOI:10.1101/gad.9.22.2795 (  0) 0) |
Eom, G. H., Kim, K., Kim, S. M., Kee, H. J., Kim, J. Y., Jin, H. M., Kim, J. R., Kim, J. H., Choe, N. and Kim, K. B., 2009. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochemical and Biophysical Research Commu-nications, 388(1): 131-136. DOI:10.1016/j.bbrc.2009.07.134 (  0) 0) |
Fuentes, E. N., Valdés,, J., A., Molina, A. and Björnsson, B. T., 2013. Regulation of skeletal muscle growth in fish by the growth hormone-insulin-like growth factor system. General and Comparative Endocrinology, 192: 136-148. DOI:10.1016/j.ygcen.2013.06.009 (  0) 0) |
Grillet, N., Schwander, M., Hildebrand, M. S., Sczaniecka, A., Kolatkar, A., Velasco, J., Webster, J. A., Kahrizi, K., Najmabadi, H. and Kimberling, W. J., 2009. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. American Journal of Human Genetics, 85(3): 328-337. DOI:10.1016/j.ajhg.2009.07.017 (  0) 0) |
Gross, R. and Nilsson, J., 1999. Restriction fragment length polymorphism at the growth hormone 1 gene in Atlantic salmon (Salmo salar L.) and its association with weight among the offspring of a hatchery stock. Aquaculture, 173(1-4): 73-80. DOI:10.1016/S0044-8486(98)00470-0 (  0) 0) |
Hnia, K., Ramspacher, C., Vermot, J. and Laporte, J., 2015. Destumin in muscle and associated diseases: Beyond the strucral function. Cell and Tissue Research, 360(3): 591-608. DOI:10.1007/s00441-014-2016-4 (  0) 0) |
Holsinger, K. E. and Weir, B. S., 2009. Genetics in geographically structured populations: Defining, estimating and interpreting F(ST). Nature Reviews Genetics, 10(9): 639. DOI:10.1038/nrg2611 (  0) 0) |
Ibberson, M., Uldry, M. and Thorens, B., 2000. GLUTX1, a novel mammalian glucose transporter expressed in the central nervous system and insulin-sensitive tissues. Journal of Biolo-gical Chemistry, 275(7): 4607-4612. DOI:10.1074/jbc.275.7.4607 (  0) 0) |
Ji, X. S., Liu, H. W., Chen, S. L., Jiang, Y. L. and Tian, Y. S., 2011. Growth differences and dimorphic expression of growth hormone (GH) in female and male Cynoglossus semilaevis after male sexual maturation. Marine Genomics, 4(1): 9-16. DOI:10.1016/j.margen.2010.11.002 (  0) 0) |
Johnston, I. A., Bower, N. I. and Macqueen, D. J., 2011. Growth and the regulation of myotomal muscle mass in teleost fish. Journal of Experimental Biology, 214(10): 1617-1628. DOI:10.1242/jeb.038620 (  0) 0) |
Johnston, I. A., Lee, H. T., Macqueen, D. J., Paranthaman, K., Kawashima, C., Anwar, A., Kinghorn, J. R. and Dalmay, T., 2009. Embryonic temperature affects muscle fibre recruitment in adult zebrafish: Genome-wide changes in gene and microRNA expression associated with the transition from hyperplastic to hypertrophic growth phenotypes. Journal of Experimental Bio-logy, 212(12): 1781-1793. DOI:10.1242/jeb.029918 (  0) 0) |
Jung, C. C., Atan, D., Ng, D., Ploder, L., Ross, S. E., Klein, M., Birch, D. G., Diez, E. and McInnes, R. R., 2015. Transcription factor PRDM8 is required for rod bipolar and type 2 OFF-cone bipolar cell survival and amacrine subtype identity. Pro-ceedings of the National Academy of Sciences, 112(23): E3010-E3019. DOI:10.1073/pnas.1505870112 (  0) 0) |
Katagiri, T., Boorla, S., Frendo, J. L., Hogan, B. L. and Karsenty, G., 1998. Skeletal abnormalities in doubly heterozygous Bmp4 and Bmp7 mice. Developmental Genetics, 22(4): 340-348. DOI:10.1002/(SICI)1520-6408(1998)22:4<340::AID-DVG4>3.0.CO;2-6 (  0) 0) |
Kendall, R. L. and Thomas, K. A., 1993. Inhibition of vascular endothelial cell growth factor activity by an endogenously encoded soluble receptor. Proceedings of the National Academy of Sciences, 90(22): 10705-10709. DOI:10.1073/pnas.90.22.10705 (  0) 0) |
Li, H. and Durbin, R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14): 1754-1760. DOI:10.1093/bioinformatics/btp324 (  0) 0) |
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G. and Durbin, R., 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25(16): 2078-2079. DOI:10.1093/bioinformatics/btp352 (  0) 0) |
Li, W., Jin, D. and Zhong, T. P., 2019. Photoreceptor cell development requires prostaglandin signaling in the zebrafish retina. Biochemical and Biophysical Research Communications, 510(2): 230-235. DOI:10.1016/j.bbrc.2019.01.073 (  0) 0) |
Lin, H., Lu, M., Lin, X., Zhang, W., Sun, Y. and Chen, L., 1995. Effects of gonadotropin-releasing hormone (GnRH) analogs and sex steroids on growth hormone (GH) secretion and growth in common carp (Cyprinus carpio) and grass carp (Ctenopha-ryngodon idellus). Aquaculture, 135(1-3): 173-184. DOI:10.1016/0044-8486(95)01012-2 (  0) 0) |
Lin, X. W., Lin, H. R. and Peter, R. E., 1993. Growth hormone and gonadotropin secretion in the common carp (Cyprinus carpio L.): In vitro interactions of gonadotropin-releasing hormone, somatostatin, and the dopamine agonist apomorphine. Gene-ral and Comparative Endocrinology, 89(1): 62-71. DOI:10.1006/gcen.1993.1009 (  0) 0) |
Liu, Y., Chen, S., Gao, F., Meng, L., Hu, Q., Song, W., Shao, C. and Lv, W., 2014. SCAR-transformation of sex-specific SSR marker and its application in half-smooth tongue sole (Cyno-glossus semiliaevis). Journal of Agricultural Biotechnology, 22(6): 787-792. (  0) 0) |
Luttrell, L. M., 2008. Reviews in molecular biology and biotechnology: Transmembrane signaling by G protein-coupled receptors. Molecular Biotechnology, 39(3): 239-264. DOI:10.1007/s12033-008-9031-1 (  0) 0) |
Ma, Q., Liu, S. F., Zhuang, Z. M., Lin, L., Sun, Z. Z., Liu, C. L., Ma, H., Su, Y. Q. and Tang, Q. S., 2012. Genomic structure, polymorphism and expression analysis of the growth hormone (GH) gene in female and male half-smooth tongue sole (Cy-noglossus semilaevis). Gene, 493(1): 92-104. DOI:10.1016/j.gene.2011.11.015 (  0) 0) |
Ma, Q., Liu, S. F., Zhuang, Z. M., Sun, Z. Z., Liu, C. L., Su, Y. Q. and Tang, Q. S., 2011. Molecular cloning, expression analysis of insulin-like growth factor I (IGF-I) gene and IGF-I serum concentration in female and male Tongue sole (Cynoglos-sus semilaevis). Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 160(4): 208-214. DOI:10.1016/j.cbpb.2011.08.008 (  0) 0) |
Marcelli, F., Boisset, G. and Schorderet, D. F., 2014. A dimerized HMX1 inhibits EPHA6/epha4b in mouse and zebrafish retinas. PLoS One, 9(6): e100096. DOI:10.1371/journal.pone.0100096 (  0) 0) |
Massault, C., Hellemans, B., Louro, B., Batargias, C., Van Houdt, J., Canario, A., Volckaert, F., Bovenhuis, H., Haley, C. and De Koning, D., 2010. QTL for body weight, morphometric traits and stress response in European sea bass Dicentrarchus la-brax. Animal Genetics, 41(4): 337-345. DOI:10.1111/j.1365-2052.2009.02010.x (  0) 0) |
Ni, J., You, F., Xu, J., Xu, D., Wen, A., Wu, Z., Xu, Y. and Zhang, P., 2012. Single nucleotide polymorphisms in intron 1 and intron 2 of Larimichthys crocea growth hormone gene are correlated with growth traits. Chinese Journal of Oceanology and Limnology, 30(2): 279-285. DOI:10.1007/s00343-012-1078-y (  0) 0) |
Ooga, M., Funaya, S., Hashioka, Y., Fujii, W., Naito, K., Suzuki, M. G. and Aoki, F., 2018. Chd9 mediates highly loosened chromatin structure in growing mouse oocytes. Biochemical and Biophysical Research Communications, 500(3): 583-588. DOI:10.1016/j.bbrc.2018.04.105 (  0) 0) |
Quik, E. H., van Dam, P. S. and Kenemans, J. L., 2010. Growth hormone and selective attention: A review. Neuroscience & Biobehavioral Reviews, 34(8): 1137-1143. DOI:10.1016/j.neubiorev.2010.01.001 (  0) 0) |
Rescan, P. Y. and Ralliere, C., 2010. A Sox5 gene is expressed in the myogenic lineage during trout embryonic development. International Journal of Developmental Biology, 54(5): 913-918. DOI:10.1387/ijdb.092893pr (  0) 0) |
Robledo, D., Rubiolo, J. A., Cabaleiro, S., Martínez,, P, . and Bouza, C., 2017. Differential gene expression and SNP association between fast- and slow-growing turbot (Scophthalmus maximus). Scientific Reports, 7(1): 12105. DOI:10.1038/s41598-017-12459-4 (  0) 0) |
Ross, S. E., McCord, A. E., Jung, C., Atan, D., Mok, S. I., Hemberg, M., Kim, T. K., Salogiannis, J., Hu, L. and Cohen, S., 2012. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron, 73(2): 292-303. DOI:10.1016/j.neuron.2011.09.035 (  0) 0) |
Ruiz-Perez, V. L., Blair, H. J., Rodriguez-Andres, M. E., Blanco, M. J., Wilson, A., Liu, Y. N., Miles, C., Peters, H. and Goodship, J. A., 2007. Evc is a positive mediator of Ihh-regulated bone growth that localises at the base of chondrocyte cilia. Development, 134(16): 2903-2912. DOI:10.1242/dev.007542 (  0) 0) |
Sabourin, L. A. and Rudnicki, M. A., 2000. The molecular regulation of myogenesis. Clinical Genetics, 57(1): 16-25. DOI:10.1034/j.1399-0004.2000.570103.x (  0) 0) |
Salem, M., Vallejo, R. L., Leeds, T. D., Palti, Y., Liu, S., Sabbagh, A., Rexroad, III C. E. and Yao, J., 2012. RNA-Seq identifies SNP markers for growth traits in rainbow trout. PLoS One, 7(5): e36264. DOI:10.1371/journal.pone.0036264 (  0) 0) |
Sánchez-Ramos, I., Cross, I., Mácha,, J, ., Martínez-Rodríguez,, G, ., Krylov, V. and Rebordinos, L., 2012. Assessment of tools for marker-assisted selection in a marine commercial species: Significant association between MSTN-1 gene polymorphism and growth traits. The Scientific World Journal, 2012: 369802. DOI:10.1100/2012/369802 (  0) 0) |
Sanger, A. and Stoiber, W., 2001. Muscle fiber diversity and plasticity. Fish Physiology, 18: 187-250. DOI:10.1016/S1546-5098(01)18008-8 (  0) 0) |
Seytanoglu, A., Alsomali, N. I., Valori, C. F., McGown, A., Kim, H. R., Ning, K., Ramesh, T., Sharrack, B., Wood, J. D. and Azzouz, M., 2016. Deficiency in the mRNA export mediator Gle1 impairs Schwann cell development in the zebrafish embryo. Neuroscience, 322: 287-297. DOI:10.1016/j.neuroscience.2016.02.039 (  0) 0) |
Shawi, M. and Serluca, F. C., 2008. Identification of a BMP7 homolog in zebrafish expressed in developing organ systems. Gene Expression Patterns, 8(6): 369-375. DOI:10.1016/j.gep.2008.05.004 (  0) 0) |
Shur, I., Socher, R. and Benayahu, D., 2006. In vivo association of CReMM/CHD9 with promoters in osteogenic cells. Jour-nal of Cellular Physiology, 207(2): 374-378. DOI:10.1002/jcp.20586 (  0) 0) |
Song, W., Li, Y., Zhao, Y., Liu, Y., Niu, Y., Pang, R., Miao, G., Liao, X., Shao, C. and Gao, F., 2012. Construction of a highdensity microsatellite genetic linkage map and mapping of sexual and growth-related traits in half-smooth tongue sole (Cynoglossus semilaevis). PLoS One, 7(12): e52097. DOI:10.1371/journal.pone.0052097 (  0) 0) |
Takahashi, T., Hagiwara, A. and Ogiwara, K., 2018. Prostaglandins in teleost ovulation: A review of the roles with a view to comparison with prostaglandins in mammalian ovulation. Mole-cular and Cellular Endocrinology, 461: 236-247. DOI:10.1016/j.mce.2017.09.019 (  0) 0) |
Valente, L. M., Moutou, K. A., Conceição, L. E., Engrola, S., Fernandes, J. M. and Johnston, I. A., 2013. What determines growth potential and juvenile quality of farmed fish species?. Reviews in Aquaculture, 5: S168-S193. DOI:10.1111/raq.12020 (  0) 0) |
Wang, D. Y., Fulthorpe, R., Liss, S. N. and Edwards, E. A., 2004. Identification of estrogen-responsive genes by complementary deoxyribonucleic acid microarray and characterization of a novel early estrogen-induced gene: EEIG1. Molecular Endo-crinology, 18(2): 402-411. DOI:10.1210/me.2003-0202 (  0) 0) |
Wang, H., Chen, Y., Lin, P., Li, L., Zhou, G., Liu, G., Logsdon, C., Jin, J., Abbruzzese, J. L. and Tan, T. H., 2014. The CUL7/ F-box and WD repeat domain containing 8 (CUL7/Fbxw8) ubiquitin ligase promotes degradation of hematopoietic progenitor kinase 1. Journal of Biological Chemistry, 289(7): 4009-4017. DOI:10.1074/jbc.M113.520106 (  0) 0) |
Weir, B. S. and Cockerham, C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution, 38(6): 1358-1370. DOI:10.1111/j.1558-5646.1984.tb05657.x (  0) 0) |
Wilkinson, D. G., 2000. Eph receptors and ephrins: Regulators of guidance and assembly. International Review of Cytology, 196: 177-244. DOI:10.1016/S0074-7696(00)96005-4 (  0) 0) |
Xia, J. H., Lin, G., He, X., Liu, P., Liu, F., Sun, F., Tu, R. and Yue, G. H., 2013. Whole genome scanning and association mapping identified a significant association between growth and a SNP in the IFABP-a gene of the Asian seabass. BMC Genomics, 14(1): 295. DOI:10.1186/1471-2164-14-295 (  0) 0) |
Yano, K., Yamamoto, E., Aya, K., Takeuchi, H., Lo, P. C., Hu, L., Yamasaki, M., Yoshida, S., Kitano, H. and Hirano, K., 2016. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nature Genetics, 48(8): 927. DOI:10.1038/ng.3596 (  0) 0) |
Zhang, B., Shang, P., Tao, Z., Qiangba, Y., Wang, Z. and Zhang, H., 2017. Effect of a single nucleotide polymorphism in the growth hormone secretagogue receptor (GHSR) gene on growth rate in pigs. Gene, 634: 68-73. DOI:10.1016/j.gene.2017.09.007 (  0) 0) |
 2021, Vol. 20
2021, Vol. 20


