2) Ningbo Academy of Oceanology and Fishery, Ningbo 315048, China
With the global population projected to increase by two billion, reaching 9.7 billion by 2050, the food production sector is faced with a significant challenge, meeting the growing demand for food within the constraints of limited land resources (FAO, 2018). Aquaculture is poised to play a pivotal role in this attempt, leveraging diverse aquatic resources to produce a variety of food organisms through the integration of intensive practices. However, these practices are not without their environmental implications. Aquaculture wastewater contains high levels of nitrogen, phosphorus, and organic matter, posing a threat to water bodies and aquatic life (Martínez-Córdova et al., 2022). The release of wastewater into the environment can lead to various problems, such as negatively affecting aesthetics, causing eutrophication, reducing photosynthetic activity and dissolved oxygen in water bodies, and leading to the bioaccumulation of toxins in aquatic ecosystems. Therefore, proper wastewater treatment is crucial to mitigate these adverse environmental impacts (Oyebamiji et al., 2019). The integrated aquaculture wastewater bioremediation system (IAWBS) presents a promising alternative for nutrient enrichment minimization, marking a crucial step in the regard of successful restoration of aquaculture ecosystems (Nicholaus et al., 2019; Lukwambe et al., 2020). However, limited researches have been conducted on the effects of bio-filters on microbial communities (Lukwambe et al., 2019), and nutrient exclusion (Li et al., 2010; Wang et al., 2018), in the IAWBS. There is still potential for improving the technical process of the IAWBS (Wei et al., 2024).
Photosynthetic bacteria (PSB) have the ability to remove organic nutrients and contaminants due to their diverse metabolic methods and high tolerance (Chen et al., 2019). PSB (e.g., Rhodopseudomonos, Rhodobacteria, and Rhodococcus) have been widely used for pollutant treatment. Rhodopseudomonas plays a crucial role in wastewater treatment by effectively mineralizing organic waste, purifying water from hydrogen sulfide, and participating in denitrification processes (Wu et al., 2021). It has been shown to enhance the growth of purple non-sulfur bacteria biomass in various wastewater sources, leading to the removal of chemical oxygen demand (COD) and reducing the concentrations of various ions and compounds in the water (Tarabas et al., 2019). Additionally, Rhodopseudomonas strains have been found to improve aquaculture water quality, increase disease resistance, and enhance the growth and yield of fish species (Xu et al., 2012). Rhodococcus play a crucial role in pollutant treatment due to their high biodegradation capabilities and diverse metabolic activities (Ivshina et al., 2022; Hosseini et al., 2023; Wang et al., 2020). However, there are certain limitations in the treatment effects of single strain in practical applications. Compound strains have more advantages, especially in the removal of complex contaminants (Aparicio et al., 2018). A reagent made from Rhodobacter blasticus and Rhodobacter capsulatus (1:1) reduced 83.3% COD of swine wastewater, which was 19.3% higher than when using Rhodobacter blasticus and 10.6% higher than when using Rhodobacter capsulatus separately (Wen et al., 2016). The compound bacteria composed of Bacillus cereus, Bacillus amyloliquefaciens and Pseudomonas stutzeri were effective in reducing NH4+-N, nitrate (NO3−-N) and nitrite (NO2−-N), in aquaculture wastewater (Jones et al., 2001). These findings collectively highlight the significant potential of PSB in bioremediation strategies for addressing environmental pollution challenges. In addition, interactions between PSB and bacterioplankton hold promise for shedding light on their coexistence and antagonistic relations, thereby offering valuable indices for assessing the stability of BCs within IAWBS. BCs not only maintain and balance biogeochemical processes but also contribute to nutrient cycling and energy flow within the aquatic food web, serving as bio-indicators of environmental limitations in ecological approaches (Labbate et al., 2016; Dai et al., 2017; Liu et al., 2018). The ability of BC to fulfill these tasks hinges on physiological adaptability, species diversity (i.e., flexibility and resistance), and the effects of interspecies interactions (Xiong et al., 2015; Deng et al., 2016; Jamoneau et al., 2018).
In this study, the compound PSB composed of Rhodopseudomonos, Rhodobacteria, Rhodococcus were added to the IBSAW to investigate its pollutants removal efficiency and the relevant mechanisms. The aims of this study were 1) to investigate the nutrients removal performance of IAWBS with PSB; 2) to characterize the microbial community composition and diversity, focusing on identifying key microorganism which are responsible for nutrient transformation including PSB; 3) to provide insights into optimizing the design and management of IAWBS with PSB for enhanced nutrient removal efficiency and environmental sustainability.
2 Materials and Methods 2.1 Experimental DesignThe study was conducted at the Pilot Test Base of Ningbo University at Meishan, where an IAWBS with four treatment units (sedimentation, biofilm, shellfish and macro-algae) was established. Every unit was housed in a white polyethylene barrel with a capacity of 70 L through which wastewater flowed. The biofilm unit carries polyethylene brushes (diameter 0.2 m, length 0.5 m) with a density of 4 brushes per barrel. One end of the brushes was fixed in the bottom to prevent them floating over the water surface, so they were suspended vertically in the canter in the center of the barrel. In the shellfish unit, the clams Tegillarca granosa were added with the density of 70 ind.m−2 while in the macro-algae unit Gracilaria lemaneiformis was added with the density of 0.2 g L−1. The wastewater used for the experiment was taken from a high-density greenhouse shrimp farm (Haohai Aquaculture farm) which is situated in Yinzhou District, Ningbo City, Zhejiang province, China. To simulate the wastewater treatment process, 20 L of wastewater was transferred to the subsequent treatment unit every two days in the following order: sedimentation, biofilm, shellfish and macro-algae.
Rhodopseudomonos, Rhodobacteria, and Rhodococcus were in the light incubator at 30 after the activation of the culture under 3000 – 4000 LX light conditions. In the treatment group, 1‰ PSB which included Rhodopseudomonos, Rhodobacteria and Rhodococcus were added to the shellfish area every 4 d. As the clams' feeds, the Rhodoyeast with 10 mL per bucket was added in the shellfish unit. The concentrations of the Rhodopseudomonos, Rhodobacteria, and Rhodococcus was 2:2:3 which was arranged from BeNa culture collection center. The study was prolonged for one month.
2.2 Sample Collection and Water Quality Index AnalysisAfter thoroughly mixing the water column from top to bottom, water and microbial samples were collected before and after each water change. Water samples were filtered through a 0.45 μm polycarbonate membrane to measure ammonia nitrogen (NH4+-N), nitrate (NO3−-N), nitrite (NO2−-N), and orthophosphate (PO43−-P). Unfiltered water samples were used to determine total nitrogen (TN), total phosphorus (TP), and chemical oxygen demand (CODMn). Samples were stored at −20℃ until analysis. Microbial samples were filtered through 0.22 μm polycarbonate membranes using a vacuum pump and stored at − 80℃ for subsequent DNA extraction and sequencing.
NH4+-N, NO3−-N, NO2−-N, PO43−-P, TN, and TP were quantified using an automatic discontinuous chemical analyzer (SmartChem). Specifically, NH4+-N, TN, TP, NO3−-N, NO2−-N, PO43−-P and CODMn were measured using hypobromite oxidation, potassium persulfate oxidation, cadmium column reduction, naphthalene ethylenediamine spectrophotometry, phosphomolybdenum blue spectrophotometry, and alkaline potassium permanganate methods, respectively.
2.3 DNA Extraction and SequencingDNA extraction kit (Minkgene water DNA kit) was used for the extraction of bacterial DNA and Nanodrop One spectrophotometer (Thermo Fisher Scientific, MA USA) is used for the purpose of concentrating and purification of bacterial DNA. The universal primer 338F (5'-ACT CCTACGGGAGGCAGCA-3') and 806R (5'-ACTCCT ACGGGAGGCAGCA-3') was used for the amplification of V3 and V4 region of the 16S rRNA. Paired-end sequences of 250 bp were generated on the Illumina HiSeq 2500 platform, courtesy of Guangdong Magigene Biotechnology Co., Ltd., Guangzhou, China. Guangzhou MeiGe Gene Company undertook the high-throughput sequencing on the Illumina MiSeq platform. Bioinformatic processing of sequencing data was conducted in USEARCH (v11.0.667_I8). Sequences were denoised with the UNOI SE3 algorithm (unoise_alpha = 2, midsize = 8, default settings), errors were corrected, chimeras were removed, and zero-radius operational taxonomic units (ZOTUs) were generated. ZOTUs were quantified and taxonomically classified by comparison to the SILVA database. α-diversity metrics including richness, Shannon diversity index, and β-diversity (weighted UniFrac distance and Bray-Curtis dissimilarity) were calculated for the prokaryotic communities.
2.4 Statistical AnalysisNutrients data were exposed to a one-way ANOVA, followed by Tukey's post-hoc test to facilitate multiple comparisons. We conducted an analysis of mean differences among treatment groups at each sampling point using SPSS and then visualized the results using R software (Caporaso et al., 2010). To assess community variations between these groups and the impact of environmental determinants, we employed Permutational Multivariate Analysis of Variance (PERMANOVA) utilizing the 'vegan' package (Oksanen et al., 2016). For an in-depth examination of bacterial composition similarity among the groups, we employed cluster analysis via the 'hclust()' function within the 'vegan' package. To provide an overarching view of the bacterial community structure, we utilized Non-Metric Multidimensional Scaling (NMDS) by employing the 'cmdscale()' function from the 'ape' package and the 'ddply()' function from the 'plyr' package. Additionally, we conducted a Canonical Correspondence Analysis (CCA) with a threshold of (VIF > 20) to assess the correlation between bacterial composition and abiotic as well as biotic factors (Oksanen et al., 2016).
To identify statistically significant biomarkers, we employed a metagenomics biomarker analysis algorithm and conducted linear discriminant analysis (LDA) on the Galaxy Hutlab website. This analysis allowed us to identify biomarkers down to the genus level, characterize them by their statistically significant differences in bacterial composition. Finally, we estimated the parametric measure of effect size and conducted a meta-analysis for each significantly abundant genus using the LDA function (Segata et al., 2011).
3 Results 3.1 Nutrient Dynamics and Removal EfficienciesNutrient concentrations displayed variations across different units of the system, signifying the influence of PSB and consequent shifts over time and system components (Table 1). Significant nutrient removal efficiencies were observed: CODMn stood at 71.42%, while removal rates for NO2−-N, NO3−-N, PO43−-P, and NH4+-N were 87.20%, 88.80%, 91.72%, and 91.37%, respectively. There was a significant decrease for NO2−-N, NO3−-N, PO43−-P between shellfish and macro-algae areas (P < 0.05). The removal of COD in the shellfish area was significant (P < 0.05), reaching a minimum level of 1.28 mg L−1.
|
|
Table 1 Nutrient removal efficiency rate in IAWBS |
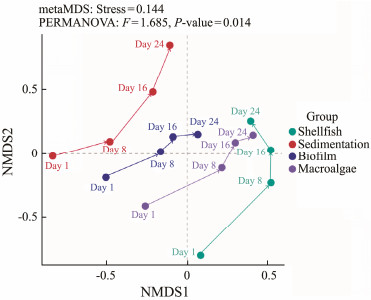
There was considerable overlap among the BCs within same area and among different areas (Fig.1). Most BC of the same treated area grouped together. However, overlaps of BC among the treated areas were also observed. Conversely, when considering different units, there was a distinct pattern where the BC in sedimentation did not overlap with those from the other three units (biofilm, shellfish, and macro-algae). It was further observed that the BC of macro-algae largely overlapped with shellfish and biofilm, while the communities of shellfish and biofilm did not interact with each other (Fig.1). These results indicated that BC was not affected by time interval but was impacted by the different units in the system.
|
Fig. 1 Nonmetric multidimensional scaling (NMDS) ordination indicating the dissimilarities in the bacterioplankton community composition in the system based on Bray-Curtis matrix. |
A subsequent PERMANOVA analysis highlighted significant disparities in BC. Specifically, BC dynamics in the treated units were significant (P < 0.001; Table 2). Additionally, BC was notably influenced by the time interval (P < 0.001; Table 2). Remarkably, there was a meaningful interaction with time interval and treated units affecting the BC composition (P < 0.001; Table 2).
|
|
Table 2 Quantitative effects of time interval sampling and treated units on the difference in bacterioplankton community structure (BC) using PERMANOVA with adonis function (BC: symbolizes the bacterioplankton community) |
This study confirms that bacterioplankton assemblages are predominantly affected by both spatial regions and the timing of sampling. Moreover, a significant disparity in BC (P < 0.001; Table 2) was associated with nutrient content, indicating that changes in BC are contingent upon variations in nutrient concentrations. This suggests that specific sampling times and the regions influence the abundance and dynamics of BC. Furthermore, alpha diversity metrics underscored a significant difference between sedimentation and the other units (macro-algae, shellfish, and biofilm) in terms of richness, Chao1, and the Shannon index (Table 3). In the sedimentation unit, the Chao1 index increased from day 1 to day 8 and then significantly decreased on subsequent days (P < 0.05). A similar pattern was observed for richness and Shannon indices, which increased significantly and then began to decrease (P < 0.05). In the biofilm unit, the Chao1index significantly decreased from day 1 to day 8 but increased on days 16 and 24 (P < 0.05). The richness and Shannon indices followed the same pattern, increasing and then decreasing. In the shellfish area, there was a significant decrease on day 8, followed by significant increases on days 16 and 24 (P < 0.05). The richness and Shannon indices also followed the pattern of Chao1. In the macro-algae unit, there was a significant increase in Chao1 index (P < 0.05), but overall, Chao1 decreased compared to the initial levels in the sedimentation unit (P < 0.05).
|
|
Table 3 The α diversity of the bacterioplankton community (mean ± SD) over treated units at different sampling time |
|
|
Table 4 Difference of bacterioplankton community structure in different sampling time interval in the treatment areas |
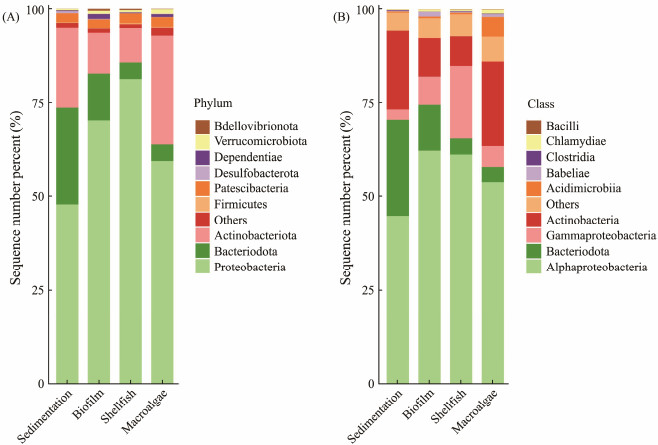
From the identified phyla, the ten most plentiful phyla were selected for interpretation, at the phylum level, the predominant phyla included Proteobacteria, Bacteriodota, and Actinobacteria. When distributions across units were examined, Proteobacteria was most abundant in the shellfish region, with its lowest presence observed in the sedimentation unit. Actinobacteria, in contrast, had its highest abundance in the macro-algae region, with minimal presence in the shellfish unit. Bacteriodota dominated the sedimentation region but was least prevalent in the macro-algae unit (Fig.2A). Delving into class-level analysis, the most abundant class was Alphaproteobacteria (belonging to phylum Proteobacteria), followed by Bacteroidia. Then Gammaproteobacteria and Actinobacteria classes were present in the same pattern as their phyla across the units (Fig.2B).
|
Fig. 2 Relative abundances of bacterio-plankton communities. (A), at phylum level (top 10); (B), at class level (top 10). |
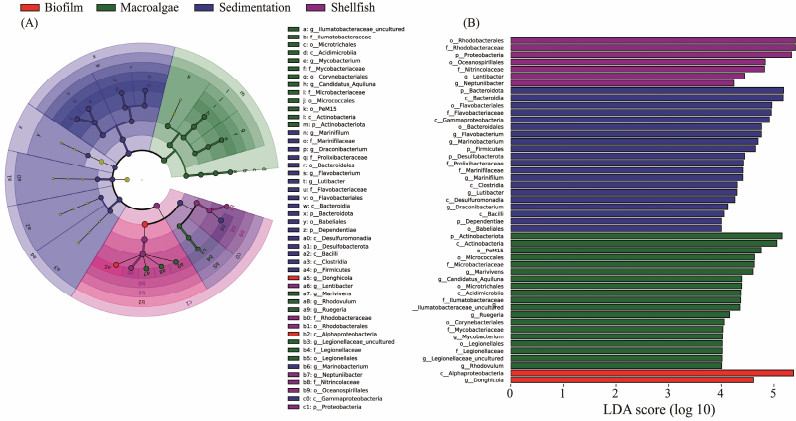
The potential biomarkers which were revealed by the phylum Actinobacteria are present in the macro-algae unit while in the shellfish unit, the proteobacteria were most abundant phylum (P < 0.05; Fig.3A). And in the biofilm unit genus Donghicola was the most dominant biomarkers (P < 0.05; Fig.3A). Desulfobacterota was the most dominant phyla present in the sedimentation area. The genera Marinobacterium, Marinifilum and Flavobacterium were potential biomarkers (Fig.3A). BC composition was further portrayed especially focusing on the genus level (Fig.3B). The LDA was structured based on different units. Unit-wise analysis of LDA scores revealed the significant dominant genus diversity in the shellfish region. Key findings included: Shellfish unit: Dominated by the Marinobacterium genus. Sedimentation unit: Flavobacterium emerged as the most prevalent. Macroalgae region: Genus Ruegeria was predominantly found. Biofilm unit: The Donghicola genus was the most abundant (P < 0.05; Fig.3B). The macro-algae and sedimentation area showed a significantly altered BC composition (P < 0.05; Fig.3).
|
Fig. 3 (A) LEfSe cladogram representing the distribution of statistically significant taxa (biomarkers) from phylum to genus level under four treatment areas. Each circle is related to the relative abundance of the given taxa. The biomarkers of each treated areas are symbolized in color (red indicate biofilm area, green indicate macro-algae, blue indicate sedimentation and purple indicate shellfish). The circles symbolize the taxonomic levels from phylum inside to genus out. (B) LDA scores histogram identifies which clades among the detected taxa with a statistical and biological difference between the communities at a threshold value of 4. |
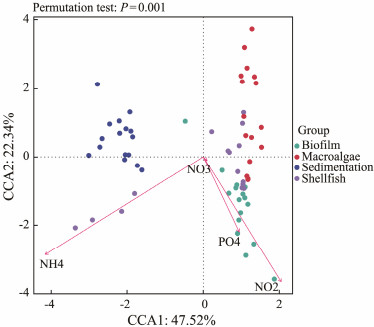
The association between BC and abiotic parameters was delineated in Fig.4 and Table 1. Notably, NO3−-N, NO2−-N, NH4+-N, and PO43−-N emerged as the most significant variables associated with BC, as indicated in Fig.4 (P < 0.001). Negative correlations were discerned between BC and NO2−-N, NH4+-N. CCA bi-plot showed the clear partitioning among the four groups associated with the nutrients. NO2−-N and PO43−-N were positively correlated with the BC of the biofilm and while the NH4+-N was negatively correlated with the shellfish area. NO3−-N acted as partitioned line. The BC in macro-algae and sedimentation units differed completely from biofilm, while shellfish overlapped slightly with biofilm, and shellfish BC related to the macro-algae units.
|
Fig. 4 Canonical correspondence analysis (CCA) indicating the correlation between bacterioplankton communities and abiotic and biotic parameters that were selected as the most parsimonious set of explanatory variables (P < 0.05) in the system. |
Bioremediation operates as a natural self-cleaning mechanism where microorganisms either undergo aerobic or anaerobic transformations, converting organic matter into biomass. Recent researches have shown that holistic bioremediation strategies offer more efficient solutions for mitigating eutrophication (Wang et al., 2018; Lukwambe et al., 2019). Our study assessed an integrated system comprising sedimentation, biofilm, shellfish, and macro-algae, which acted as bio-filters. This system was moderately successful in modulating the BC composition, controlling infective bacteria, and transforming nitrogen-rich effluents into valuable biomass. Such a configuration enhances the biodegradability of particulate organic matter via microbial processes like ammonification, nitrification, and denitrification, thereby optimizing nutrient removal the concentrations of all nutrients significantly decreased, such as Wang et al. (2021) found that the ammonia oxidation performance of aerobic granular sludge was improved. In our study, the macro-algae area has demonstrated good inorganic nutrient removal efficiency, achieved a removal rate of over 85% for NO2−-N, NO3−-N, and PO43−-P (Table 1). Additionally, transitioning from the shellfish area to the macro-algae area, there was a significant decrease in all nutrient concentrations, while the CODMn has reached its lowest level in the shellfish, indicating that shellfish might be more effective in reducing CODMn of the system (Table 1). Bacillus subtilis showcased impressive purification capabilities, achieving removal rates of 81% for NH4+-N, 87% for NO3−-N, 91% for TN, and 52% for NO2−-N (Shao et al., 2021). Our findings showed the same pattern (Table 1). The transition of NH4+ -N to NO3−-N principally relies on the nitrification process. Autotrophic bacteria, primarily ammonia-oxidizing bacteria (AOB) and nitrite oxidizing bacteria (NOB), orchestrate this transformation, with many of these bacteria falling under the Proteobacteria phylum (Nicholaus et al., 2019; Lukwambe et al., 2020). Notably, macro-algae regions exhibited a pronounced self-purification effect when juxtaposed with other bio-filtration zones. This is attributed to the assimilative capabilities of emergent macro-algae, which play a pivotal role in nutrient circulation and transformation. Globally acknowledged for their nutrient assimilation efficacy, macro-algae have been instrumental in ecological restoration endeavors, ameliorating water quality and fostering long-term ecosystem stability (Wang et al., 2009; Søndergaard et al., 2010). They also fortify the sediment-water interface, promote bacterial mineralization of organic substances, and establish a favorable milieu for bacterial mineralization (Rehman et al., 2017).
Sedimentation zones recorded peak concentrations of NO3−-N and NO2−-N, likely due to water not being treated with PSB. However, in the bio-filter units, there was a marked decrease in NO3−-N and NO2−-N contents, hinting at the uptake of these nutrients by the integrated constituents of biofilm, shellfish, macro-algae, and introduced PSB. Several studies have postulated the influence of TN on the functional diversity of BC in various ecosystems (Wang et al., 2016). Still, it's noteworthy that the nutrient removal efficiency in the shellfish and biofilm zones was not as high as in the macro-algae region (Table 1). Potential explanations include insufficient stocking durations or sub optimal macro-benthos growth, both of which could undermine removal rates. Additionally, bioturbation by clams may have elevated nutrient release from sediments (Nicholaus et al., 2019).
4.2 Dynamics of Bacterial CommunitiesIAWBSs are extensively employed in aquaculture wastewater treatment. Their superiority, as compared to standalone biological or physicochemical treatments, arises from the integration of diverse treatments that enhance the wastewater purification performance (Ahmad et al., 2022). The phylogenetic diversity and consistency of the bacterial community expanded, peaking at the growth stage (Jiang et al., 2019). The bioremediation system for wastewater is mostly influenced by biological processes, which as a result are affected by changes in the BC structure within the system. The ANOSIM analysis approved that both unit and time interval significantly influenced BC. Moreover, the NMDS analysis (Fig.1) highlighted a deficit in BC from day 1 to day 24 in the sedimentation unit, and shellfish unit devoid of PSB. Conversely, in treated units, there was an evident augmentation in BC, as depicted in (Fig.1). This suggests that the treated units have relatively stable BC compared to sedimentation, and that shrimp effluent release possibly alters BC coupling in the system (Drury et al., 2013). Besides solidity, a necessarily dynamic BC (able to adapt to environmental changes) is crucial for proper system function (Ayala-del-Río et al., 2004).
For a bioremediation system to function optimally, a bacterial community must possess the dynamism to adapt to environmental shifts (Ayala-del-Río et al., 2004). Taxonomic biomarkers such as Marivivens, Candidatus Aquiluna and Mycobacterium were observed in the sedimentation unit (Fig.3B), although their comparative abundance was not considerable. Prior research has spotlighted pathogenic bacterial taxa, most notably Vibrio, Aeromonas, Acinetobacter, Arcobacter, Flavobacterium, and Pseudoalteromonas (Xiong et al., 2015; Lamb et al., 2017; Wang et al., 2018). In contrast, potential bacteriolytic taxa characterized by Actinobacteria (family Nitriliruptoraceae) and Firmicutes (family Bacillaceae; order Bacillales) were substantially distributed in macro-algae and mud treatment units (Fig.2). These taxa might play significant roles in suppressing unwanted organisms. In summation, the longevity and efficiency of aquaculture bioremediation systems are contingent upon a robust and stable microbial community. It's imperative that within this ecosystem, antagonistic or detrimental bacterial species remain subdued.
4.3 Role of PSB and Microbes in Nutrient RemovalEffective wastewater treatment is fundamentally anchored in the robustness of the microbial community. Fluctuations in microbial diversity can destabilize the ecological balance of the system, thereby influencing nutrient removal mechanisms (Wang et al., 2011). Microbes transform nitrogen into different shapes through nitrification, ammonification, and denitrification. This study revealed negligible nitrate (NO3−-N) in biofilter units. However, increased overall removal rates (Table 3) were possibly related to emergent macro-algae releasing oxygen to stimulate microbial ammonium (NH4+-N) nitrification (Van Hulle et al., 2010; Rehman et al., 2017). Under these situations, autotrophic (anammox) nitrifying bacteria oxidize ammonium aerobically to nitrate. Subsequently, under anoxic conditions, heterotrophic denitrifying bacteria transform nitrate into nitrogen gas or nitrous oxide (Chen et al., 2019).
Furthermore, the introduction of clams augments the nitrifying bacterial population within the sediment, fostering the oxidation of nitrite to nitrate (Lukwambe et al., 2019). Notably, this effect is orchestrated predominantly by autotrophic bacteria, specifically AOB and NOB (Rud et al., 2017). Denitrifying bacteria such as Actinobacteria, Flavobacteriaceae, and Delfusobacteraea, proficient in nitrate-to-nitrite conversion, demonstrated significant diversity and abundance across the system (Fig.2B). This affirms the role of bio-filters in promoting the proliferation of autotrophic nitrifying bacteria and heterotrophic denitrifying bacteria, which underpin the nitrification and denitrification cycles. Most of these bacteria align with the phylum Proteobacteria. In our investigation, Alphaproteobacteria emerged as a dominant subset, with a notably dispersed bacterial composition within the aquatic milieu.
In our study, Alphaproteobacteria of Proteobacteria dominated all units, with bacterial composition in water being dispersed (Fig.2B). Additionally, Chlamydia may exhibit salt acceptance in early stages and are concerned to nitrification and denitrification processes within the treatment system (Yan et al., 2016). The macro-algae region registered a surge in Actinobacteria, renowned for their pollutant removal capabilities (Alvarez et al., 2017). Diverse biomarker performances among treatment groups were observed. Alphaproteobacteria, crucial for biological nitrogen and phosphorus elimination in preliminary stages, was succeeded by Gammaproteobacteria, abundant with nitrogen and phosphorus-functional bacteria. Additionally, Cohaesibacter, with its biodegradability feature (Dai et al., 2015), became predominant midway through the experiment. As the research progressed, the PSB-associated Bacillus and Micrococcaceae emerged as potential biomarkers, signifying the influence of PSB on native bacteria.
Regarding spatial distribution, Proteobacteria predominated in the shellfish zone, Actinobacteria in the macro-algae region, and Bacteriodota in the sedimentation unit. In the biofilm territory, Bacteriodota and Actinobacteria appeared almost equally abundant (Fig.2A), with proteobacteria marginally trailing. Notably, these species, characterized as halophilic heterotrophic nitrifying bacteria, play pivotal roles in heterotrophic nitrification and denitrification, thus enhancing nutrient removal efficacy (Shitu et al., 2021). The bio-film saturated bio-filter unit displayed the presence of several potential biomarkers, crucial for purification. A plethora of algae-lysing bacteria, including Actinobacteria, were predominantly concentrated in the biofilm-bio-filter region (Fig.2A). This confirms the dual role of Actinobacteria and Bacillus sp. not only in pathogenic bacterial elimination but likewise in nutrient removal from the system. Aforementioned literature has underscored the importance of Bacillus sp., Roseobacter, and Actinobacteria strains as key probiotics essential for mitigating pathogenic algae-bacteria, promoting nitrogen elimination, and enhancing water quality (Ghai et al., 2014; Lukwambe et al., 2020).
Ultimately, bacterial composition greatly dictates the system's wastewater treatment efficacy. For instance, the underwhelming degradation of NO2−-N may arise from the dominance of AOB in the nitrification cycle, coupled with a scarcity of NOB, leading to delayed NO2−-N removal. The NH4+-N concentration influences bacterial dynamics, and their reciprocal interactions invariably shape the system's overall performance (Veillette et al., 2011). Bacterial metabolism is foundational for effective biological wastewater treatment, with species composition steering potential metabolic pathways. These pathways, in turn, determine the post-treatment wastewater quality (Cydzik-Kwiatkowska and Zielińska, 2016).
5 ConclusionsAn IAWBS has emerged as a sustainable method to improve water quality of aquaculture effluents and to remove possible environmental pollution. The current study aimed to use IAWBS with PSB (Rhodopseudomonas, Rhodobacteria, Rhodococcus) which was successfully employed as a bioremediation agent for the treatment of wastewater obtained from shrimp. The results showed that significant removal efficiencies of CODMn, NH4+-N, NO3−-N, NO2−-N, PO43−-P were 71.42%, 91.37%, 91.72%, 87.20%, and 88.80%, respectively. NMDS analysis showed that BC was not affected by the time interval but affected by different units. Notably, the PERMANOVA analysis highlighted a significant interaction between the time intervals and treated units, influencing the composition of the microbial community (R2 = 0.152; P = 0.001). This study provides a better understanding of the role of PSB in an IAWBS that can effectively treat culture tail-water, which is of great significance for the sustainable development of aquaculture. However, more studies are needed to justify the efficiency of this approach to improving IAWBS.
AcknowledgementsThis research was financially supported by the National Key R&D Program of China (No. 2020YFD0900201), the Ningbo Public Welfare Technology Application Research Project (No. 2022S164), and the K. C. Wong Magna Fund in Ningbo University.
Author Contributions
Muhammad Naeem Ramzan: methodology, data analysis, raw material preparation, writing-original draft, data curation writing-review and editing. Ding Shen, , Yingzhen Wei and Arslan Emmanuel: software and surveys. Wen Yang: raw material preparation. Jinyong Zhu: supervision and funding acquisition. Yangcai Wang: administration and supervision. Zhongming Zheng: conceptualization, investigation, data analysis, writing-review and editing, funding acquisition, supervision, project administration.
Data Availability
The data and references presented in this study are available from the corresponding author upon reasonable request.
Declarations
Ethics Approval and Consent to Participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for Publication
Informed consent for publication was obtained from all participants.
Conflict of Interests
The authors declare that they have no conflict of interests.
Ahmad, A. L., Chin, J. Y., Mohd Harun, M. H. Z., and Low, S. C., 2022. Environmental impacts and imperative technologies towards sustainable treatment of aquaculture wastewater: A review. Journal of Water Process Engineering, 46: 102553. DOI:10.1016/j.jwpe.2021.102553 (  0) 0) |
Aparicio, J. D., Saez, J. M., Raimondo, E. E., Benimeli, C. S., and Polti, M. A., 2018. Comparative study of single and mixed cultures of Actinobacteria for the bioremediation of co-contaminated matrices. Journal of Environmental Chemical Engineering, 6(2): 2310-2318. DOI:10.1016/j.jece.2018.03.030 (  0) 0) |
Ayala-del-Río, H. L., Callister, S. J., Criddle, C. S., and Tiedje, J. M., 2004. Correspondence between community structure and function during succession in phenol-and phenol-plus-trichloroethene-fed sequencing batch reactors. Applied and Environmental Microbiology, 70(8): 4950-4960. DOI:10.1128/AEM.70.8.4950-4960.2004 (  0) 0) |
Alvarez, A., Saez, J. M., Costa, J. S. D., Colin, V. L., Fuentes, M. S., Cuozzo, S. A., et al., 2017. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere, 166: 41-62. DOI:10.1016/j.chemosphere.2016.09.070 (  0) 0) |
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al., 2010. QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5): 335-336. DOI:10.1038/nmeth.f.303 (  0) 0) |
Chen, L., Tsui, M. M. P., Lam, J. C. W., Wang, Q., Hu, C., Wai, O. W. H., et al., 2019. Contamination by perfluoroalkyl substances and microbial community structure in Pearl River Delta sediments. Environmental Pollution, 245: 218-225. DOI:10.1016/j.envpol.2018.11.005 (  0) 0) |
Cydzik-Kwiatkowska, A., and Zielińska, M., 2016. Bacterial communities in full-scale wastewater treatment systems. World Journal of Microbiology and Biotechnology, 32: 1-8. DOI:10.1007/s11274-016-2012-9 (  0) 0) |
Dai, H. H., Gao, J. F., Wang, Z. Q., Zhao, Y. F., and Zhang, D., 2020. Behavior of nitrogen, phosphorus and antibiotic resistance genes under polyvinyl chloride microplastics pressures in an aerobic granular sludge system. Journal of Cleaner Production, 256: 120402. DOI:10.1016/j.jclepro.2020.120402 (  0) 0) |
Drury, B., Scott, J., Rosi-Marshall, E. J., and Kelly, J. J., 2013. Triclosan exposure increases triclosan resistance and influences taxonomic composition of benthic bacterial communities. Environmental Science & Technology, 47(15): 8923-8930. DOI:10.1021/es401919k (  0) 0) |
Dai, W., Zhang, J., Tu, Q., Deng, Y., Qiu, Q., and Xiong, J., 2017. Bacterioplankton assembly and interspecies interaction indicating increasing coastal eutrophication. Chemosphere, 177: 317-325. DOI:10.1016/j.chemosphere.2017.03.034 (  0) 0) |
Dai, Y., Jiang, Y., and Su, H., 2015. Influence of an aniline supplement on the stability of aerobic granular sludge. Journal of Environmental Management, 162: 115-122. DOI:10.1016/j.jenvman.2015.05.017 (  0) 0) |
Deng, Y., Zhang, P., Qin, Y., Tu, Q., Yang, Y., He, Z., et al., 2016. Network succession reveals the importance of competition in response to emulsified vegetable oil amendment for uranium bioremediation. Environmental Microbiology, 18: 205-218. DOI:10.1111/1462-2920.12981 (  0) 0) |
FAO (Food Agriculture Organization of the United Nations), 2018. The state of world fisheries and aquaculture 2018-meeting the sustainable development goals. CC BY-NC-SA 3.0 IGO.
(  0) 0) |
Ghai, R., Mizuno, C. M., Picazo, A., Camacho, A., and Rodriguez‐Valera, F., 2014. Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing. Molecular Ecology, 23: 6073-6090. DOI:10.1111/mec.12985 (  0) 0) |
Hosseini, S., Azadi, D., and Absalan, A., 2023. Bioremediation of phenol, sulfate sodium, and polycyclic aromatic hydrocarbons by Rhodococcus sp. first time isolated and molecular characterized from aquatic and terrestrial ecosystems. Water and Environment Journal, 37(3): 594-603. DOI:10.1111/wej.12862 (  0) 0) |
Ivshina, I., Bazhutin, G., and Tyumina, E., 2022. Rhodococcus strains as a good biotool for neutralizing pharmaceutical pollutants and obtaining therapeutically valuable products: Through the past into the future. Frontiers in Microbiology, 13: 967127. DOI:10.3389/fmicb.2022.967127 (  0) 0) |
Jamoneau, A., Passy, S. I., Soininen, J., Leboucher, T., and Tison‐Rosebery, J., 2018. Beta diversity of diatom species and ecological guilds: Response to environmental and spatial mechanisms along the stream watercourse. Freshwater Biology, 63: 62-73. DOI:10.1111/fwb.12980 (  0) 0) |
Jiang, W., Tian, X., Li, L., Dong, S., Zhao, K., Li, H., et al., 2019. Temporal bacterial community succession during the start-up process of biofilters in a cold-freshwater recirculating aquaculture system. Bioresource Technology, 28: 121441. DOI:10.1016/j.biortech.2019.121441 (  0) 0) |
Jones, A. B., Dennison, W. C., and Preston, N. P., 2001. Integrated treatment of shrimp effluent by sedimentation, oyster filtration and macroalgal absorption: A laboratory scale study. Aquaculture, 193(1-2): 155-178. DOI:10.1016/S0044-8486(00)00486-5 (  0) 0) |
Kuyukina, M. S., and Ivshina, I. B., 2019. Bioremediation of contaminated environments using Rhodococcus. Biology of Rhodococcus, 13: 231-270. DOI:10.1007/978-3-030-11461-9_9 (  0) 0) |
Labbate, M., Seymour, J. R., Lauro, F., and Brown, M. V., 2016. Anthropogenic impacts on the microbial ecology and function of aquatic environments. Frontiers in Microbiology, 7: 210082. DOI:10.3389/fmicb.2016.01044 (  0) 0) |
Lamb, J. B., Van De Water, J. A. J. M., Bourne, D. G., Altier, C., Hein, M. Y., Fiorenza, E. A., et al., 2017. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science, 355: 731-733. DOI:10.1126/science.aal1956 (  0) 0) |
Li, X. N., Song, H. L., Li, W., Lu, X. W., and Nishimura, O., 2010. An integrated ecological floating bed employing plant, freshwater clam and biofilm carrier for purification of eutrophic water. Ecological Engineering, 36: 382-390. DOI:10.1016/j.ecoleng.2009.11.004 (  0) 0) |
Liu, S., Wang, C., Wang, P., Chen, J., Wang, X., and Yuan, Q., 2018. Variation of bacterioplankton community along an urban river impacted by touristic city: With a focus on pathogen. Ecotoxicology and Environmental Safety, 165: 573-581. DOI:10.1016/j.ecoenv.2018.09.006 (  0) 0) |
Lukwambe, B., Nicholaus, R., Zhang, D., Yang, W., Zhu, J., and Zheng, Z., 2019. Successional changes of microalgae community in response to commercial probiotics in the intensive shrimp (Litopenaeus vannamei Boone) culture systems. Aquaculture, 511: 734257. DOI:10.1016/j.aquacul-ture.2019.734257 (  0) 0) |
Lukwambe, B., Nicholaus, R., Zhao, L., Yang, W., Zhu, J., and Zheng, Z., 2020. Microbial community and interspecies interaction during grazing of ark shell bivalve (Scapharca subcrenata) in a full-scale bioremediation system of mariculture effluents. Marine Environmental Research, 158: 104956. DOI:10.1016/j.marenvres.2020.104956 (  0) 0) |
Martínez-Córdova, L. R., Robles-Porchas, G. R., Vargas-Albores, F., Porchas-Cornejo, M. A., and Martínez-Porchas, M., 2022. Microbial bioremediation of aquaculture effluents. Microbial Biodegradation and Bioremediation, 46: 409-417. DOI:10.1016/B978-0-323-85455-9.00009-6 (  0) 0) |
Mishra, B., and Tiwari, A., 2022. Sustainable aquaculture wastewater remediation through diatom and biomass valorization. Biomass, Biofuels, Biochemicals, 38: 181-202. (  0) 0) |
Nicholaus, R., Lukwambe, B., Zhao, L., Yang, W., Zhu, J., and Zheng, Z., 2019. Bioturbation of blood clam Tegillarca granosa on benthic nutrient fluxes and microbial community in an aquaculture wastewater treatment system. International Biodeterioration & Biodegradation, 142: 73-82. DOI:10.1016/j.ibiod.2019.05.004 (  0) 0) |
Nixon, S. W., Granger, S. L., and Nowicki, B. L., 1995. An assessment of the annual mass balance of carbon, nitrogen, and phosphorus in Narragansett Bay. Biogeochemistry, 77: 31. DOI:10.1007/BF00000805 (  0) 0) |
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'hara, R., et al., 2013. Community Ecology Package. R Package Version, 2: 321-326, http://cran.r-project.org/package¼vegan.
(  0) 0) |
Oyebamiji, O. O., Boeing, W. J., Holguin, F. O., Ilori, O., and Amund, O., 2019. Green microalgae cultured in textile wastewater for biomass generation and biodetoxification of heavy metals and chromogenic substances. Bioresource Technology Reports, 7: 100247. DOI:10.1016/j.biteb.2019.100247 (  0) 0) |
Rehman, F., Pervez, A., Khattak, B. N., and Ahmad, R., 2017. Constructed wetlands: Perspectives of the oxygen released in the rhizosphere of macrophytes. Clean Soil Air Water, 45: clen.201600054. DOI:10.1002/clen.201600054 (  0) 0) |
Rud, I., Kolarevic, J., Holan, A. B., Berget, I., Calabrese, S., and Terjesen, B. F., 2017. Deep-sequencing of the bacterial microbiota in commercial-scale recirculating and semi-closed aquaculture systems for Atlantic salmon post-smolt production. Aquacultural Engineering, 78: 50-62. DOI:10.1016/j.aquaeng.2016.10.003 (  0) 0) |
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al., 2011. Metagenomic biomarker discovery and explanation. Genome Biology, 12: 1-18. DOI:10.1186/gb-2011-12-6-r60 (  0) 0) |
Shao, Y., Zhong, H., Wang, L., and Elbashier, M. M., 2021. Use of Bacillus subtilis D9 to purify coastal aquaculture wastewater and improve grass carp resistance to vibrio infection. Aquaculture Environment Interactions, 13: 249-258. DOI:10.3354/aei00404 (  0) 0) |
Shitu, A., Liu, G., Zhang, Y., Ye, Z., Zhao, J., Zhu, S., et al., 2021. Enhancement of mariculture wastewater treatment using moving bed biofilm reactors filled with modified biocarriers: Characterisation, process performance and microbial community evaluation. Journal of Environmental Management, 291: 112724. DOI:10.1016/j.jenvman.2021.112724 (  0) 0) |
Søndergaard, M., Johansson, L. S., Lauridsen, T. L., Jørgensen, T. B., Liboriussen, L., and Jeppesen, E., 2010. Submerged macrophytes as indicators of the ecological quality of lakes. Freshwater Biology, 55: 893-908. DOI:10.1111/j.1365-2427.2009.02331.x (  0) 0) |
Tarabas, O. V., Hnatush, S. O., Moroz, O. M., and Kovalchuk, M. M., 2019. Wastewater bioremediation with using of phototrophic non-sulfur bacteria Rhodopseudomonas yavorovii IMV B-7620. Ecology and Noospherology, 30(2): 63-67. DOI:10.15421/031911 (  0) 0) |
Van Hulle, S. W. H., Vandeweyer, H. J. P., Meesschaert, B. D., Vanrolleghem, P. A., Dejans, P., and Dumoulin, A., 2010. Engineering aspects and practical application of autotrophic nitrogen removal from nitrogen rich streams. Chemical Engineering Journal, 162: 1-20. DOI:10.1016/j.cej.2010.05.037 (  0) 0) |
Vergani, L., Mapelli, F., Suman, J., Cajthaml, T., Uhlik, O., and Borin, S., 2019. Novel PCB-degrading Rhodococcus strains able to promote plant growth for assisted rhizoremediation of historically polluted soils. PLoS One, 14(8): e0221253. DOI:10.1371/journal.pone.0221253 (  0) 0) |
Veillette, M., Viens, P., Ramirez, A. A., Brzezinski, R., and Heitz, M., 2011. Effect of ammonium concentration on microbial population and performance of a biofilter treating air polluted with methane. Chemical Engineering Journal, 171: 1114-1123. DOI:10.1016/j.cej.2011.05.008 (  0) 0) |
Verdegem, M. C. J., 2013. Nutrient discharge from aquaculture operations in function of system design and production environment. Reviews in Aquaculture, 5: 158-171. DOI:10.1111/raq.12011 (  0) 0) |
Wang, L., Sun, J., Zheng, W., Huang, T., Zhang, Y., Wu, Z., et al., 2018. Effects of a combined biological restoration technology on nitrogen and phosphorus removal from eutrophic water. Polish Journal of Environmental Studies, 27(5): 77609. DOI:10.15244/pjoes/77609 (  0) 0) |
Wang, S., Jin, X., Jiao, L., and Wu, F., 2009. Response in root morphology and nutrient contents of Myriophyllum spicatum to sediment type. Ecological Engineering, 35: 1264-1270. DOI:10.1016/j.ecoleng.2009.05.012 (  0) 0) |
Wang, X., Wen, X., Yan, H., Ding, K., Zhao, F., and Hu, M., 2011. Bacterial community dynamics in a functionally stable pilot-scale wastewater treatment plant. Bioresource Technology, 102: 2352-2357. DOI:10.1016/j.biortech.2010.10.095 (  0) 0) |
Wang, Z., Gao, J., Dai, H., Zhao, Y., Li, D., Duan, W., et al., 2021. Microplastics affect the ammonia oxidation performance of aerobic granular sludge and enrich the intracellular and extracellular antibiotic resistance genes. Journal of Hazardous Materials, 409: 124981. DOI:10.1016/j.jhazmat.2020.124981 (  0) 0) |
Wang, Z., Li, W., Li, H., Zheng, W., and Guo, F., 2020. Phylogenomics of Rhodocyclales and its distribution in wastewater treatment systems. Scientific Reports, 10(1): 3883. DOI:10.1038/s41598-020-60723-x (  0) 0) |
Wei, Y., Shen, D., Lukwambe, B., Wang, Y., Yang, W., Zhu, J., et al., 2022. The exogenous compound bacteria alter microbial community and nutrients removal performance in the biofilm unit of the integrated aquaculture wastewater bioremediation systems. Aquaculture Reports, 27: 101414. DOI:10.1016/j.aqrep.2022.101414 (  0) 0) |
Wen, S., Liu, H., He, H., Luo, L., Li, X., Zeng, G., et al., 2016. Treatment of anaerobically digested swine wastewater by Rhodobacter blasticus and Rhodobacter capsulatus. Bioresource Technology, 222: 33-38. DOI:10.1016/j.bior-tech.2016.09.102 (  0) 0) |
Wu, P., Hu, Y., Wang, Y., Wu, Y., Li, N., Zhang, Y., et al., 2021. The regulation of the disease resistance, mTOR and NF-kB signaling pathway of Aristichthys nobilis using Rhodopseudomonas. Wastewater Treatment, 104: 103517. DOI:10.1016/j.dci.2020.103733 (  0) 0) |
Xiong, J., Chen, H., Hu, C., Ye, X., Kong, D., and Zhang, D., 2015. Evidence of bacterioplankton community adaptation in response to long-term mariculture disturbance. Scientific Reports, 5(1): 15274. DOI:10.1038/srep15274 (  0) 0) |
Xu, C. B., Sun, X. K., Li, Y. Y., Wang, Y. G., and Meng, X. L., 2011. Study on optimization of the culture conditions for four Rhodopseudomonas. Advanced Materials Research, 393-395: 976-979. DOI:10.4028/www.scientific.net/amr.393-395.976 (  0) 0) |
Yang, Y. Y., and Toor, G. S., 2016. δ15N and δ18O reveal the sources of nitrate-nitrogen in urban residential stormwater runoff. Environmental Science & Technology, 50(6): 2881-2889. DOI:10.1021/acs.est.5b05353 (  0) 0) |
Yan, L., Zhang, S., Hao, G., Zhang, X., Ren, Y., Wen, Y., et al., 2016. Simultaneous nitrification and denitrification by EPSs in aerobic granular sludge enhanced nitrogen removal of ammonium-nitrogen-rich wastewater. Bioresource Technology, 202: 101-106. DOI:10.1016/j.biortech.2015.11 (  0) 0) |
 2025, Vol. 24
2025, Vol. 24


