2) School of Marine Science and Engineering, Qingdao Agricultural University, Qingdao 266109, China;
3) Shandong Provincial Key Laboratory of Biochemical Engineering, College of Marine Science and Biological Engineering, Qingdao University of Science and Technology, Qingdao 266042, China;
4) Wuqiong Food Co., Ltd., Raoping 515726, China
The immune system plays vital roles in vertebrates which can response and defend against the invasion of pathogens. nNOS is an important molecule in immune system as the mainly producer of NO which can prove to be important role in the function for neuromodulation, molecule signaling deliver or immune function (Lee et al., 1994; Gomes et al., 2020). Especially in the process of immunization, many reports have confirmed that NO involved in the pathogenesis and control of infectious diseases, tumors, autoimmune processes and chronic degenerative diseases. However, as the producer of NO, the research of nNOS in teleost was still very scarce.
Black rockfish (Sebastes schlegelii) is a common economic fish species in the coast of China, Korea and Japan. Compared to terrestrial animals, fishes inhabit in the microbial rich environment, seawater, thus had more pressure for their immune system to fight against pathogens. So that, the living environment of black rockfish was vulnerable that many bacterial pathogens can affect their growth and economic value, such as Edwardsiella tarda (Xu and Zhang, 2014), Vibrio anguillarum (Eguchi et al., 2000) and other pathogenic bacteria. With the rapid development of aquaculture, retarded E. tarda and V. anguillarum have been recognized as the major pathogens in freshwater and mariculture fish worldwide. Infection with E. tarda and V. anguillarum leads to the development of a systemic disorder in fishes which characterized by ascites, hernias, proptosis, and severe damage to the brain and internal organs (Mohanty and Sahoo, 2007; Fran et al., 2011).
Meanwhile, the immune functions of NOS especially for nNOS and iNOS in teleost were verified in previous research. Among the three types of NOS, the most widely type that found to be effective in the antibacterial immune response of teleost was iNOS. It had been identified with immunological efficacy in channel catfish (Ictalurus punctatus) (Yao et al., 2014), turbot (Scophthalmus maximus L.) (Losada et al., 2012) and other fish species. Compared with iNOS, the function of nNOS was often reflected in the response of the nervous system (Förstermann and Sessa, 2012). Actually, except for function in nerve cells, nNOS also can mediate the activation of some immune- related cells by producing NO like T-cell mast cells and neutrophils which perform critical roles in adaptive immune response (Kamimura et al., 2003; McCauley et al., 2007; Sadaf et al., 2021). For example, in turbot (S. maximus L.), the nNOS could be significantly induced in mucosal surfaces after V. anguillarum infection (Dong et al., 2016). At present, some anti-inflammatory factors have been identified and functionally analyzed in black rockfish to against these two pathogens infection like CC chemokines 25 (CCL25) (Wang et al., 2020), Nod-like receptor (NLR) (Cao et al., 2021), but there was still a lack of systematic research on the expression patterns of nNOS under the stimulation. Therefore, we tried to identify the nNOS gene in black rockfish in gene and protein level. Firstly, the characteristic of nNOS gene was obtained or predicted. The phylogenetic tree was constructed to analyze the conservation of nNOS gene among species. Finally, the expression pattern of nNOS gene and content in response to pathogen stimulation was analyzed by quantitative Real-time PCR (qRT-PCR) and Elisa assay.
2 Materials and Methods 2.1 Genome-Wide Identification of nNOS Gene in Black RockfishIn order to identify the nNOS gene of black rockfish, the nNOS database were first constructed containing nNOS genes of several vertebrates and a variety of fish (including Homo sapiens, Mus musculus, Gallus gallus, Chrysemys picta, Xenopus tropicalis, Danio rerio, Cyprinus carpio, Perca flavescens) which downloaded from NCBI. After that, the black rockfish nNOS gene sequence (the e-value ≤1e-5) were obtained from the existing S. schlegelii database through the local blast.
2.2 Analysis of Characteristics of nNOS Genes in Black RockfishFor further identification of nNOS, the sequence of nNOS was extracted from black rockfish genome. After that, the structure of gene was visualized in GSDS2.0 (http://gsds.gao-lab.org/) and the molecular weight (MW) and isoelectric point (PI) were calculated by ProtParam analysis program in ExPASy (https://web.expasy.org/protparam/). Moreover, the subcellular localizations of black rockfish nNOS were predicted by neural nets method with ProtComp 9.0 on Softberry (http://www.softberry.com) and the protein domain were visualized by SMART (http://smart.embl-heidelberg.de/). A multiple alignment of the nNOS amino acid sequences were generated by MEGA6 program firstly. And then made it visible by using DNAman program. In addition, the STRING (http://string-db.org/) was used for clearly finding the immune-related protein which interacted with nNOS in black rockfish. At last, the secondary structure of protein was predicted by Phyre2 online tool (Kelley et al., 2015).
2.3 Phylogenetic Analysis of nNOS Gene in Multiple SpeciesFor deducing the phylogenetic relationship between the black rockfish and other species, the nNOS gene of black rockfish was aligned with NOS sequences from various other classes of vertebrates (contained H. sapiens, M. musculus, G. gallus, Alligator mississippiensis, X. tropicalis), as well as a variety of fish species (contained Tachysurus fulvidraco, Pangasianodon hypophthalmus, Electrophorus electricus, Colossoma macropomum, Pygocentrus nattereri, Carassius auratus, D. rerio, Cuplea harengus, Denticeps clupeoides, Sparus aurata, Acanthopagrus latus, Seriola dumerili, M. albus, Myripristis murdjan, Megalops cyprinoides). The phylogenetic analysis was processed in MEGA6 software (Tamura et al., 2013). And the NOS protein sequences of species required for phylogenetic analysis were all downloaded from the NCBI database. During construction of the phylogenetic, the protein sequences were aligned by MU SCLE and the ProtTest was used to determine the G+I+T evolutionary model (Edgar, 2004; Abascal et al., 2005).
2.4 Sampling Collection and Bacteria Bath-ChallengeFor exploring the nNOS gene expression level in healthy tissues of black rockfish (including blood, liver, spleen, gill, skin, intestine, kidney and brain), the experimental fish which had total average body length about 15 cm were fed in flowing seawater system firstly. When the healthy tissues were extracted from fishes, these tissue samples were extracted with total three replicates from random 6 fishes individuals. The extracted samples were summarized in control group which were divided into two parts, one part was kept in −20℃ freezer for nNOS Elisa experiments, and one part was kept in −80℃ ultra-low temperature freezer for isolating RNA. Meanwhile, to investigate the expression changes of nNOS after infection with pathogenic bacteria, a partial of healthy fish was selected for bath-challenge. Before challenge, for determining the accuracy of the challenge bacteria, the E. tarda and V. anguillarum were isolated from symptomatic fish and the purity of them were demonstrated by sequencing. After that, cultivate them overnight with the optimal conditions respectively. E. tarda was cultured in LB medium at 28℃ at 180 r min−1 and V. anguillarum was cultured in same type medium at 28℃ at 180 r min−1. Before infection, 100 μL of 10-fold serial dilutions bacteria was plated onto plates and then calculated their concentration using colony forming unit (CFU) per mL. The bath-challenge fish were divided into two large groups by two individual bacteria E. tarda and V. anguillarum. The final E. tarda concentration was 1×107 CFU mL−1 and V. anguillarum concentration was 5×107 CFU mL−1 (Wang et al., 2018; Cao et al., 2021). And one large group fish (contained 72 fishes) could be divided into 4 groups (one group contained 18 fishes) with different time point after infection including 2, 6, 12 and 24 h, which four time points. Previous related studies had shown that the 2, 6, 12 and 24 h four time points were important immune response nodes in fish autoimmune after pathogen infection (Eguchi et al., 2000; Cao et al., 2021). Subsequently, the fish for challenge should be immersed 2 h with two pathogenic bacteria which reached the challenge concentration separately.
2.5 qRT-PCR Expression Fold Analysis of SsnNOS for Healthy TissuesTo clarify the nNOS gene expression fold in black rockfish, the total RNA of healthy tissues (contained gill, intestine, skin, liver, brain, kidney, spleen, muscle) and the infected tissues (important autoimmune tissues in fish contained gill, intestine, skin, brain, kidney, spleen) were extracted by RNAiso reagent (TAKARA, China). After that, the total RNA was reversely transcribed into cDNA by PrimeScriptTMRT reagent Kit (TAKARA, China) with manufacture's study. The ribosomal protein L17 (RPL17) was selected as the internal reference primer. Meanwhile, the primer of nNOS and RPL17 were designed by using PrimerQuest Tool (https://www.idtdna.com/pages/tools/primerquest). As for experiment, the solution consists of 10 μL of SYBR® Premix Ex TaqTM II (TliRNaseH Plus), 0.6 μL of each primer, 2 μL of the 10-fold dilution cDNA and nuclease-free water 6.8 μL. The qRT-PCR program was set as follows: 5 min at 95℃ firstly, followed by 35 cycles of 95℃ for 5 s, 56℃ for 30 s and 72℃ for 30 s, finally up to 95℃ with a rate of 0.1℃ s−1 increment. In calculate program, the 2−ΔΔCt method was used to compute the expression fold change of nNOS gene among the healthy and infection tissues (Ma et al., 2013).
2.6 Elisa Assay for nNOS in Black Rockfish TissuesThe Fish Neuronal Nitric Oxide Synthase (nNOS) Elisa Kit (Jiancheng Bioengineering Institute, Nanjing, Jiangsu) was used for detecting the content of nNOS in black rockfish tissues. Firstly, the tissues sample were ground and centrifuged to enrich nNOS in the supernatant and each sample contained three parallel controls. After that, adding the sample and standards with Bio-Ab in 37℃ water bath 30 min when preparing reagents, supernatants and standards. Then, the HRP conjugate reagent was set in 96 wells plant which had washed 5 times and incubation at 37℃ 30 min. After that, the plate was washed five times again, and chromogen solution A and B were added in system incubation for ten minutes at 37℃. Finally, adding the stop solution into mixture solution and measuring the OD value under 450 nm wavelength within 10 min. Logistic was the model to calculate the fitted curves. Finally, the nNOS concentration of each sample was obtained by the standard curve method.
3 Results 3.1 Identification and Characterization of nNOS Genes in Black RockfishA nNOS gene was identified from the black rockfish genome named SsnNOS by blastn with databases which consisting of other species nNOS genes. In detail, the identified nNOS gene had 3780 bp full-length and located at Chr6 with a total of 27 segments of CDs distributed throughout itself (showed in Table 1 and Fig.1). And the nNOS protein which had 1424 acids was predicted that it had a molecular weight (MW) of 160546.71 and an isoelectric point (PI) at 6.38. Moreover, the subcellular localization showed the SsnNOS mainly existed in nuclear than other position in cells. As for structure domain, the SsnNOS protein beginning with the PDZ domain and ending with the NAD binding 1 domain, which inturn connected the NO synthase, Flaodoxin 1 and FAD binding 1 domains. According the results of the secondary structure prediction, 16 alpha-helices and 6 beta-sheets were found and the location of them also be clearlyshowed in Fig.2. In addition, the protein association network showed the SsnNOS could be associated with akt 1-3, asmtl, asl, gatm and other proteins. And the five important domains of SsnNOS were marked on Fig.3 of the alignment acid amino sequences. From the amino acid sequence alignment results, it can be seen that amino acid sequences in teleost were relatively conserved. The sequence similarity of paired align sequences was all higher than 80%. Among them, the black rockfish amino acid sequence had the highest sequence similarity with Seriola dumerili which value is 93.4 %.
|
|
Table 1 Characteristics of nNOS gene in black rockfish |

|
Fig. 1 Chromosome location and gene structure of nNOS in black rockfish. For whole-length SsnNOS gene, the UTR, CDs and intron were distinguished by different colors. And the location of gene was marked on figure. |

|
Fig. 2 Protein domain (A), two-dimensional structure (B) and association network (C) of SsnNOS. The 5 different domains in SsnNOS were showed in 5 colors. And the α-helix β-sheet random coil in protein two-dimensional structure of SsnNOS were demonstrated in red, yellow and green partially. The different interactions between the proteins were represented by different colored lines and listed in the bottom right of the figure. |

|
Fig. 3 Multiple sequence alignment results of teleost nNOS amino acid sequence. As for this figure, sequence similarity values were indicated by different colors, and the numerical values corresponding to specific colors were annotated to the right. |
The Fig.4 showed these NOS genes could be divided into three clades and the clustering of the three types of NOS genes was evident. The identified SsnNOS gene can be correctly placed in the nNOS subfamilies. Overall, NOS genes clustered significantly in species with higher degrees of evolution. The NOS gene sequences of high- grade organisms had a higher similarity and they had a distinct sequence divergence from the NOS genes of teleost, leading to their respective distribution in the phylogenetic tree. Meanwhile, NOS genes from teleost clustered into a single large clade compared with species with a higher degree of evolution. Among them, the nNOS gene which was the closest relative to the SsnNOS was from swamp eel (M. albus).

|
Fig. 4 Phylogenetic relationship of nNOS gene in black rockfish and other vertebrates. Using the software MEGA6 with G+I+T model to construct the phylogenetic tree. The parameters were set as follows: The proportion of invariable sites 0.32 and Gamma shape parameter 0.56, 1000 bootstrap. The different types of NOS genes were marked by color strips, and the NOS genes in black rockfish were highlighted in bule colors on genes name in iTOL. |
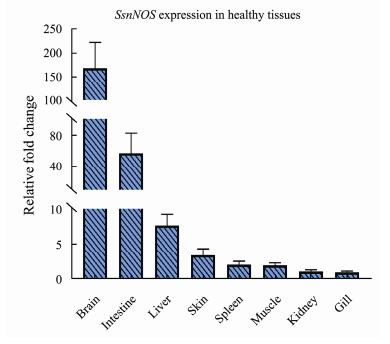
The expression analysis of SsnNOS in eight healthy tissues of black rockfish was certified by using qRT-PCR program. The SsnNOS was highly expression in brain and intestine (Fig.5). In detail, compared with gill which tissue with the lowest SsnNOS, the expression of brain was 168.51-fold and the intestine was 56.97-fold. In addition, the tissues which expressed below 5-fold change compared with gill were muscle (2.02-fold), spleen (2.13- fold), kidney (1.15-fold) and skin (3.46-fold). Besides, compared with other tissues, the liver had the moderate expression levels, about 7.69-fold. From the results, two tissues include brain and intestine with change fold significantly higher than the other tissues will be the key research subjects for following research which was the possible immunologic function of nNOS.
|
Fig. 5 Expression patterns of SsnNOS gene in eight healthy tissues. RPL17 was used as the reference gene. Expression levels were calibrated against tissue which had the lowest expression level. Each sample was mixed by 6 fishes and one tissue contained 3 three parallel sample. The results were presented as mean ± SE of fold changes. |
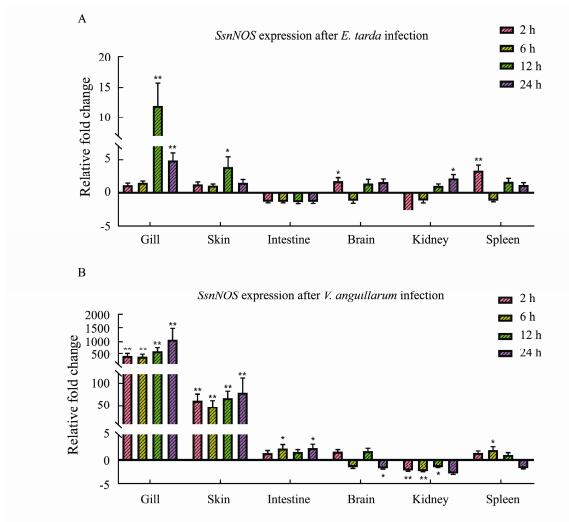
In order to obtain the expression pattern of SsnNOS when the 6 tissues (contained intestine, gill, spleen, brain, skin, kidney) of black rockfish were attacked by E. tarda or V. anguillarum, the qRT-PCR experiment was carried out. As the E. tarda infection group, the highest fold change of SsnNOS (12.00-fold) came from the gill tissue after 12 h infection. And the lowest expression level came from the kidney two hours after infection (−3.09-fold). From the trend of expression, the tissues where SsnNOS was up-regulated during 4 infection time were gill and skin. In contrast, the SsnNOS in intestine was down- regulated during the infection time. The expression of SsnNOS in the brain reached its highest value at 2 h after infection (1.72-fold) and its lowest value was obtained at 6 h (−1.16-fold). In the expression of spleen, the highest value (3.42-fold) and the lowest value (−1.16-fold) of SsnNOS were found when comparable to its expression level in brain.
In V. anguillarum infection group, the change fold of SsnNOS was also significantly different among tissues. For example, the SsnNOS expression in gill from 2 h to 6 h was a slight decrease (426.32-fold to 395.44-fold), but the expression showed the opposite upward trend from 6 h to 24 h (395.44-fold to 1065.58-fold). And the gill also had the highest fold change of SsnNOS among the six tissues. As for the second highest expression fold tissue skin, the SsnNOS expression change trend was consistent with that in gill, which had a lower expression fold interval relative to gill. The SsnNOS was expressed in the remaining four tissues intestine, brain, kidney and spleen with fold change absolute no higher than 3-fold. Among them, SsnNOS expression at 4 time points after intestinal infection with V. anguillarum was higher than the expression at healthy level, and the expression in kidney was just the opposite, all lower than the expression at healthy level. However, there was no regular trend in SsnNOS expression in the brain or spleen. The expression of SsnNOS in the brain was highest at 12 h (1.77-fold) and lowest at 24 h (-1.84-fold) after infection, whereas the expression in the spleen was highest at 6 h (1.98-fold) and lowest at 24 h (-1.81-fold) after infection (Fig.6). In summary, SsnNOS in different tissues with E. tarda or V. anguillarum infection diverged in their expression patterns at different time points.
|
Fig. 6 Expression patterns of SsnNOS genes after E. tarda (A) or V. anguillarum (B) infection in six tissues. RPL17 was used as the reference gene. The results were presented as mean ± SD of fold changes. The significantly differentially with the P-value range from 0.01– 0.05 was marked with * and the P-value less than 0.01 was marked with **. |
According to the Elisa assay analysis, concentration of SsnNOS protein was changed after infection with E. tarda or V. anguillarum in various tissues (Fig.7). In E. tarda infection group, the highest SsnNOS content was obtained from the gill tissue after 24 h of infection and the lowest was obtained from the brain tissue after 6 h of infection. From the perspective of the change trend, the SsnNOS content in the spleen has the most continuous change which decreased from uninfected to 24 h after infection. As for brain tissue, the SsnNOS was obviously lowest at 6 h time point and the pattern of expression was not evident at other time points. In addition, the concentration of SsnNOS in intestine was higher than that in brain tissues at all time points and the highest occurred at 12 h. Throughout, the gill had higher SsnNOS concentration than other tissues especially. Relatively, the SsnNOS content after V. anguillarum infection showed generally higher results which compared with SsnNOS after E. tarda infection. In brain, the SsnNOS activity had a decreasing trend from CON group until 12 h after infection, but the highest value was obtained at 24 h. Meanwhile, the change trend of SsnNOS activity in the intestine from 6 h to 24 h was similar to that in the brain. Interestingly, the change of SsnNOS activity was quite regular in kidney, it first decreasing from CON group to 2 h, and continuously increasing from 2 h to 24 h. However, the activity of SsnNOS in spleen again showed an alternating change of rising and falling, reaching a maximum at 12 h and a minimum at 6 h. Finally, the SsnNOS activity of gill in infection time were elevated compared to CON group, and maximum values were obtained 24 h post infection.

|
Fig. 7 Elisa assay of SsnNOS in five tissues after E. tarda (A) or V. anguillarum (B) infection. The results were presented as mean ± SD of SsnNOS concentration. The significantly differentially with the P-value range from 0.01 to 0.05 was marked with * and the P-value less than 0.01 was marked with **. |
NO is an immune transduction molecule which can be widely found in living organisms, possesses strong intraor inter-cellular conduction ability, and bind to various types of response receptors (Bogdan, 2001). As the main producer of NO, NOS has also been extensively studied. In NOS family, immune functions of iNOS have been strongly demonstrated. But there were few studies focus on the transduction of immune signals participants nNOS. As for research of SsnNOS in black rockfish, the length of its protein was about 1424 amino acids which similar to the NOS1 gene (1329 amino acids) in channel catfish and nNOS gene (1437 amino acids) in red drum (Zhou et al., 2009; Yao et al., 2014). The subcellular localization prediction showed the nNOS was produced and stored in nuclear. Moreover, the nNOS gene was located at number 6 chromosome in genome. As for the isoelectric point of nNOS, it was 6.38 which demonstrated that the nNOS protein contains more acidic amino acids. The domain of it was similar with the predicted domain of nNOS from previous studies for its protein structure (Alderton et al., 2001). It contained a PDZ domain that mediated synaptic association (Brenman et al., 1996) and three binding domains for transmitters, a FAD binding domain, a Flavodoxin domain which can bind one FMN molecule and a NAD binding domain. In addition, it also contained the major catalytic NO_synthase domain. In the prediction of protein interaction, SsnNOS showed the connection with akt protein. The phylogenetic tree constructed with SsnNOS sequences from different species provided strong support for ascription of SsnNOS gene. Obviously, the SsnNOS gene were orthologous to nNOS gene in other organisms. In particular, the nNOS gene from black rockfish clustered with the nNOS gene from swamp eel (M. albus) which showed the genetic relationship of nNOS gene between the two species. In addition, the iNOS and eNOS genes from other species were also clustered into one branch each respectively. The results of the phylogenetic tree illustrated that SsnNOS had high conservation among species, and the gene sequence did not change greatly during the evolution of species.
In qRT-PCR analysis, compared with other tissues, the SsnNOS was highest expressed in healthy brain. In addition, the fold change of SsnNOS was also highly in the important mucosal immune tissue intestine. Actually, the nNOS was an important gene expressed in central nervous system which can explain its highest expression in the brain (Förstermann and Sessa, 2012). Meanwhile, the results of nNOS expressed higher in intestine seem to support the unique function of transmission of immune signals black rockfish (Dong et al., 2016).
In qRT-PCR analysis after bath-challenge, the SsnNOS gene showed a differential fold change after infected by E. tarda or V. anguillarum in 5 tissues. For example, Ssn NOS expression was highest at 12 h after infection with E. tarda, but the highest expressed time point of SsnNOS was change to 24 h after infection with V. anguillarum. Moreover, the expression trend between gill and skin following infection of V. anguillarum was similar with high value which showed the strongest response to V. anguillarum infection. However, studies of nNOS expression in teleost following infection were still few now. But in previous study of channel catfish, the nNOS was induced in skin at early timepoints after Edwardsiella ictaluri (E. ictaluri) infection (Yao et al., 2014). It seemed to be because the gill and skin of teleost were important lymphoid tissues which were the first barrier to defend against pathogenic bacteria (Salinas, 2015). Interestingly, the expression levels of SsnNOS were reversed at various time points after infection with two different bacteria. But the relevant support in studies were still lack, it may be related to the different pathogenic mechanism of the two bacteria for teleost. Other study showed, the downregulated expression of iNOS gene in the intestine occurred after infection with Edwardsiella ictaluri in channel catfish (Yao et al., 2014). In black rockfish, the lack of iNOS and the significant expression differences after pathogen stimulation of SsnNOS most likely predict that the SsnNOS gene may have played a part in the role of iNOS gene in autoimmunity.
In protein level, the nNOS concentration in the five tissues was greatly affected by infection with different bacteria. Among the five tissues, SsnNOS has the highest concentration in gill which seems to corroborate the high expression of SsnNOS gene in gills after pathogen infection. It is obvious that the concentration of SsnNOS in gills increased more obviously after stimulation by V. anguillarum than infected by E. tarda. This phenomenon seems to be related to the different pathogenic mechanisms of the two bacteria. Interestingly, in the kidney and intestine which infected by different pathogens, there was a consistent tendency that the SsnNOS content first decreasing and then increasing. Probably because the pathogenic bacteria could first inhibit the innate immunity progress through secrete immunosuppressants like thioredoxin-like protein (Trxlp) (Yang et al., 2019). After that, the body responded to the stimulus by up-regulating SsnNOS to induce downstream immune response.
At the gene and protein level, the pattern expression of SsnNOS in response to pathogen infection showed that this gene had specificity expressed in various tissues, mainly in the brain, intestine, gill. The studies had confirmed the role of SsnNOS in the pathogen defense system in addition to it neurotransmitter functions, but the immune pathways and interacting immune molecules involved in SsnNOS required more experimental support at the protein level.
5 ConclusionsIn this study, the nNOS gene in black rockfish was identified based on genome information. According to the phylogenetic analysis, the SsnNOS in black rockfish showed the closest relationship with the M. albus nNOS. Meanwhile, qRT-PCR analysis showed the nNOS had the highest expression in healthy brain. Especially, SsnNOS gene expression was significantly upregulated in the gill and skin following pathogen stimulation, and the fold change in other tissues is dependent on the type of pathogen. In protein level, according to the Elisa assay for SsnNOS, the protein was significantly up-regulated in the gill tissue after V. anguillarum stimulation, and showed a first down- and then up-regulated expression pattern in some tissues like brain, intestine and kidney. It may provide a reference to explain the mechanism of immune escape of pathogenic bacteria. In a word, the work provided a systematic information about the SsnNOS gene. The evolutionary positions and potential response mechanisms of SsnNOS following bacteria challenge, which can be valuable for further studying their immune response mechanism.
AcknowledgementsThis study was supported by the Natural Science Foundation of Shandong Province (No. ZR2020QC214), the Young Experts of Taishan Scholars (No. tsqn201909130), the Science and Technology Support Plan for Youth Innovation of Colleges and Universities in Shandong Province (No. 2019KJF003), the 'First Class Fishery Discipline' Programme in Shandong Province, a special talent programme 'One Thing One Decision (YishiYiyi)' Programme in Shandong Province, China, and the Breeding Plan of Shandong Provincial Qingchuang Research Team (2019).
This study was supported by the Natural Science Foundation of Shandong Province (No. ZR2020QC214), the Young Experts of Taishan Scholars (No. tsqn201909130), the Science and Technology Support Plan for Youth Innovation of Colleges and Universities in Shandong Province (No. 2019KJF003), the 'First Class Fishery Discipline' Programme in Shandong Province, a special talent programme 'One Thing One Decision (YishiYiyi)' Programme in Shandong Province, China, and the Breeding Plan of Shandong Provincial Qingchuang Research Team (2019).
This study was supported by the Natural Science Foundation of Shandong Province (No. ZR2020QC214), the Young Experts of Taishan Scholars (No. tsqn201909130), the Science and Technology Support Plan for Youth Innovation of Colleges and Universities in Shandong Province (No. 2019KJF003), the 'First Class Fishery Discipline' Programme in Shandong Province, a special talent programme 'One Thing One Decision (YishiYiyi)' Programme in Shandong Province, China, and the Breeding Plan of Shandong Provincial Qingchuang Research Team (2019).
This study was supported by the Natural Science Foundation of Shandong Province (No. ZR2020QC214), the Young Experts of Taishan Scholars (No. tsqn201909130), the Science and Technology Support Plan for Youth Innovation of Colleges and Universities in Shandong Province (No. 2019KJF003), the 'First Class Fishery Discipline' Programme in Shandong Province, a special talent programme 'One Thing One Decision (YishiYiyi)' Programme in Shandong Province, China, and the Breeding Plan of Shandong Provincial Qingchuang Research Team (2019).
Abascal, F., Zardoya, R., and Posada, D., 2005. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics (Oxford, England), 21 (9): 2104-2105, https://doi.org/10.1093/bioinformatics/bti263.
(  0) 0) |
Alderton, W. K., Cooper, C. E., and Knowles, R. G., 2001. Nitric oxide synthases: Structure, function and inhibition. The Biochemical Journal, 357(3): 593-615. DOI:10.1042/0264-6021:3570593 (  0) 0) |
Bogdan, C., 2001. Nitric oxide and the immune response. Nature Immunology, 2(10): 907-916. DOI:10.1038/ni1001-907 (  0) 0) |
Brenman, J. E., Chao, D. S., Gee, S. H., McGee, A. W., Craven, S. E., Santillano, D. R., et al., 1996. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1 – Syntrophin mediated by PDZ domains. Cell, 84(5): 757-767. DOI:10.1016/s0092-8674(00)81053-3 (  0) 0) |
Cao, M., Yan, X., Li, Q., Fu, Q., Yang, N., Song, L., et al., 2021. Genome-wide identification and analysis of NOD-like receptors and their potential roles in response to Edwardsiella tarda infection in black rockfish (Sebastes schlegelii). Aquaculture, 30: 736803. DOI:10.1016/j.aquaculture.2021.736803 (  0) 0) |
(  0) 0) |
Edgar, R. C., 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5): 1792-1797. DOI:10.1093/nar/gkh340 (  0) 0) |
Eguchi, M., Fujiwara, E., and Miyamoto, N., 2000. Survival of Vibrio anguillarum in freshwater environments: Adaptation or debilitation? Journal of Infection and Chemotherapy: Official Journal of the Japan Society of Chemotherapy, 6 (2): 126-129, https://doi.org/10.1007/pl00012152.
(  0) 0) |
Förstermann, U., and Sessa, W. C., 2012. Nitric oxide synthases: Regulation and function. European Heart Journal, 33(7): 829-837. DOI:10.1093/eurheartj/ehr304 (  0) 0) |
Frans, I., Michiels, C. W., Bossier, P., Willems, K. A., Lievens, B., and Rediers, H., 2011. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. Journal of Fish Diseases, 34(9): 643-661. DOI:10.1111/j.1365-2761.2011.01279.x (  0) 0) |
Gomes, F., Cunha, F. Q., and Cunha, T. M., 2020. Peripheral nitric oxide signaling directly blocks inflammatory pain. Biochemical Pharmacology, 176: 113862. DOI:10.1016/j.bcp.2020.113862 (  0) 0) |
Kamimura, Y., Fujii, T., Kojima, H., Nagano, T., and Kawashima, K., 2003. Nitric oxide (NO) synthase mRNA expression and NO production via muscarinic acetylcholine receptor-mediated pathways in the CEM, human leukemic T-cell line. Life Sciences, 72(18-19): 2151-2154. DOI:10.1016/s0024-3205(03)00076-6 (  0) 0) |
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J., 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 10(6): 845-858. DOI:10.1038/nprot.2015.053 (  0) 0) |
Lee, K. H., Baek, M. Y., Moon, K. Y., Song, W. K., Chung, C. H., Ha, D. B., et al., 1994. Nitric oxide as a messenger molecule for myoblast fusion. The Journal of Biological Chemistry, 269(20): 14371-14374. (  0) 0) |
Losada, A. P., Bermúdez, R., Faílde, L. D., and Quiroga, M. I., 2012. Quantitative and qualitative evaluation of iNOS expression in turbot (Psetta maxima) infected with Enteromy- xum scophthalmi. Fish & Shellfish Immunology, 32(2): 243-248. DOI:10.1016/j.fsi.2011.11.007 (  0) 0) |
Ma, L. M., Wang, W. J., Liu, C. H., Yu, H. Y., Wang, Z. G., Wang, X. B., et al., 2013. Selection of reference genes for reverse transcription quantitative real-time PCR normalization in black rockfish (Sebastes schlegelii). Marine Genomics, 11: 67-73. DOI:10.1016/j.margen.2013.08.002 (  0) 0) |
McCauley, S. D., Gilchrist, M., and Befus, A. D., 2007. Regulation and function of the protein inhibitor of nitric oxide synthase (PIN)/dynein light chain 8 (LC8) in a human mast cell line. Life Sciences, 80(10): 959-964. DOI:10.1016/j.lfs.2006.11.025 (  0) 0) |
Mohanty, B. R., and Sahoo, P. K., 2007. Edward siellosis in fish: A brief review. Journal of Biosciences, 32(7): 1331-1344. DOI:10.1007/s12038-007-0143-8 (  0) 0) |
Sadaf, S., Nagarkoti, S., Awasthi, D., Singh, A. K., Srivastava, R. N., Kumar, S., et al., 2021. nNOS induction and NOSIP interaction impact granulopoiesis and neutrophil differentiation by modulating nitric oxide generation. Biochimica et Biophysica Acta Molecular Cell Research, 1868(7): 119018. DOI:10.1016/j.bbamcr.2021.119018 (  0) 0) |
Salinas, I., 2015. The mucosal immune system of teleost fish. Biology, 4(3): 525-539. DOI:10.3390/biology4030525 (  0) 0) |
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S.0., 2013. MEGA6: Molecular evolutionary genetics analysis version 6. Molecular Biology and Evolution, 30(12): 2725-2729. DOI:10.1093/molbev/mst197 (  0) 0) |
Wang, L., Jiang, L., Wu, C., and Lou, B., 2018. Molecular characterization and expression analysis of large yellow croaker (Larimichthys crocea) interleukin-12A, 16 and 34 after poly I: C and Vibrio anguillarum challenge. Fish & Shellfish Immunology, 74: 84-93. (  0) 0) |
Wang, J. J., Meng, Z. Q., Wang, G. H., Fu, Q., and Zhang, M., 2020. A CCL25 chemokine functions as a chemoattractant and an immunomodulator in black rockfish, Sebastes schle- gelii. Fish & Shellfish Immunology, 100: 161-170. DOI:10.1016/j.fsi.2020.02.063 (  0) 0) |
Xu, T. T., and Zhang, X. H., 2014. Edwardsiella tarda: An intriguing problem in aquaculture. Aquaculture, 431: 129-135. DOI:10.1016/j.aquaculture.2013.12.001 (  0) 0) |
Yang, D., Liu, X., Xu, W., Gu, Z., Yang, C., Zhang, L., et al., 2019. The Edwardsiella piscicida thioredoxin-like protein inhibits ASK1-MAPKs signaling cascades to promote pathogenesis during infection. PLoS Pathogens, 15(7): e1007917. DOI:10.1371/journal.ppat.1007917 (  0) 0) |
Yao, J., Li, C., Zhang, J. R., Liu, S. K., Feng, J. B., Wang, R. J., et al., 2014. Expression of nitric oxide synthase (NOS) genes in channel catfish is highly regulated and time dependent after bacterial challenges. Developmental and Comparative Immunology, 45(1): 74-86. DOI:10.1016/j.dci.2014.02.005 (  0) 0) |
Zhou, L. B., Bai, R., Tian, J. X., Liu, X. C., Lu, D. Q., Zhu, P., et al., 2009. Bioinformatic comparisons and tissue expression of the neuronal nitric oxide synthase (nNOS) gene from the red drum (Sciaenops ocellatus). Fish & Shellfish Immunology, 27(4): 577-584. DOI:10.1016/j.fsi.2009.07.010 (  0) 0) |
 2023, Vol. 22
2023, Vol. 22


