2) Collaborative Innovation Center for Zhejiang Marine High-Efficiency and Healthy Aquaculture, Ningbo University, Ningbo 315211, China;
3) Third Institute of Oceanography, State Oceanic Administration, Xiamen 361001, China
DNA methylation has been proposed to be a predominant epigenetic mechanism underlining the response of organisms to environmental change (Bird, 2002). In eukaryotes, methylation is confined to cytosine and has been described in CG, CHG and CHH (H = A, T or C). DNA methylation may render genes inaccessible, and inactivate their transcription (Doerfler, 1983). Methylation is essential for development (Smith and Meissner, 2013) and associates with a number of key processes such as genomic imprinting and polyploidization (Li et al., 1993; Jiang et al., 2016), X-chromosome inactivation (Csankovszki et al., 2001), aging (Maegawa et al., 2010) and environmental adaptation (Christensen et al., 2009). Environmental stimuli may cause heritable phenotypic variation through methylation (Angers et al., 2010). DNA methylation plays an important role in adapting to environmental stresses in, for example, cold (Shan et al., 2013), heavy metals (Ou et al., 2012), temperature (Navarromartín et al., 2011) and salinity (Wang et al., 2014). Osmotic-stressed plants hold higher level demethylated DNA than the non-stressed (Zhang et al., 2013). In addition, DNA methylation level of Cynoglossus semilaevis significantly decreased at salinity 15 (Li et al., 2017).
The salinity of coastal aquaculture area is sensitive to climate, especially rainfall and typhoon (Feng et al., 2005), which may significantly affect the survival and growth of marine animals. Swimming crab (Portunus trituberculatus) widely inhabiting the coasts of China, Japan and Korea is one of the most important commercial species in China (Chen et al., 2006). It can adapt to low salinity environments (Ruscoe et al., 2004; Pan et al., 2007); however, its survival and growth are significantly affected by low salinity.
Methylation-sensitive amplified polymorphisms (MSAP) is effective for describing genome methylation status (Xiong et al., 1999). MSAP is a modification of amplified fragment length polymorphism (AFLP). Osmoregulation has been well documented in crustaceans; however DNA methylation of crustacean is scarcely studied. The present study was carried out to investigate the change of DNA methylation pattern in the muscle, hepatopancreas and gill of P. trituberculatus at low salinities in order to understand the potential role of DNA methylation when P. trituberculatus is low salinity stressed.
2 Materials and Methods 2.1 Crab Rearing and SamplingThe crab individuals used in this study were obtained from XinYi Aquatic Products Limited Company, Zhejiang, China. The crab individuals were acclimated at 25℃ for a week, during which the salinity of the aerated seawater was maintained at 24. During the entire experiment, crab individuals were fed with fresh fish with the uneaten removed, and one third of the water was changed daily. The low salinity of the seawater was adjusted by adding aerated tap water.
One hundred twenty individuals with an average weight of 25.32 g ± 3.12 g were placed in six pools (8 m × 2 m × 1 m), 20 each. The pools were divided into the control (salinity 24) and experimental (salinity 12) groups, three pools each. The crab was reared for 30 days with gill, muscle and hepatopancreas sampled for DNA extraction with a standard phenol/chloroform protocol.
2.2 MSAP Procedure and Fragment IsolationMSAP was performed following the method of Xiong et al. (1999). DNA (200 ng) was double-digested with one of the methylation-sensitive enzymes, either Hpa Ⅱ or Msp Ⅰ, and the methylation-insensitive EcoR Ⅰ. Hpa Ⅱ and Msp Ⅰ recognize tetranucleotide sequence, 5'-CCGG-3', differentially. Hpa Ⅱ is inactive when two cytosines are methylated but cuts the sequence when any one of two cytocines is methylated (hemimethylated), whereas Msp Ⅰ cuts 5'-C5mCGG-3' but not 5'-5mCCGG-3'. Therefore, comparisons of EcoR Ⅰ/Hpa Ⅱ and EcoR Ⅰ/Msp Ⅰ digestion profiles allows us to detect the methylation status at this site.
Genomic DNA (200 ng) was digested with 20 U Hpa Ⅱ or Msp Ⅰ and 10 U EcoR Ⅰ in 20 μL volume at 37℃ for 6 h. The digestion was stopped at 65℃ for 10 min. The digested DNA was ligated with EcoR Ⅰ and Hpa Ⅱ/Msp Ⅰ adaptors for 12 h. Pre-amplification was performed in a 20 μL volume containing 2 μL ligated products, 0.4 μL 10 mmol L-1 dNTP (each), 2 μL 10×Buffer, 0.2 μL 5 U μL-1 Taq DNA polymerase, 0.5 μL 10 μmol L-1 HM adaptor and 0.5 μL 10 μmol L-1 EcoR Ⅰ adaptor. PCR was performed on a master-cycler gradient thermal cycler (Eppendorf) by predenaturing at 94℃ for 2 min followed by 30 cycles of denaturing at 94℃ for 30 s, annealing at 56℃ for 30 s, and extending at 72℃ for 60 s, and a final extension at 72℃ for 10 min. The PCR product was used as the template for the second PCR by denaturing at 94℃ for 2 min, followed by 11 cycles of denaturing at 94℃ for 30 s, annealing at 65℃ for 30 s and extending at 72℃ for 60 s (the annealing temperature was decreased to 55℃, one degree each cycle, touchdown PCR), 25 cycles of denaturing at 94℃ for 30 s, annealing at 65℃ for 30 s and extending at 72℃ for 60 s and a final extension at 72℃ for 10 min. Primers and adaptors are listed in Table 1.
|
|
Table 1 Sequences of adaptors and primers used for MSAP |
Amplification product was resolved via 8% denaturing polyacrylamide gel, and visualized by silver-staining. A 10-bp DNA ladder (Invitrogen Inc.) was used as markers of allele size.
2.3 Data AnalysisOnly unambiguous and reproducible fragments were scored. The different types of bands were counted three times to minimize artificial error. Based on presence (1) or absence (0) of EcoR Ⅰ/Hpa Ⅱ and EcoR Ⅰ/Msp Ⅰ bands, respectively, four types of band patterns were identified, A, present in both (1-1), non-methylation; B, present in only EcoR Ⅰ/Hpa Ⅱ (1-0) and hemi-methylation; C, present in only EcoR Ⅰ/Msp Ⅰ (0-1) and full methylation; and D, absent in both (0-0), hyper-methylation or uninformative because of the absence of a fragment. The analysis of DNA methylation fragments was conducted using a chisquare test in SPSS19.0 (Fig. 1).
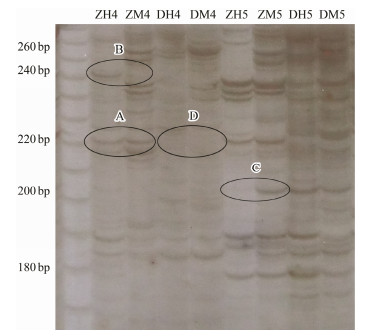
|
Fig. 1 DNA methylation patterns detected by MSAP assay with PAGE electrophoresis. ZH, DNA digested by EcoR Ⅰ and Hpa Ⅱ, control; ZM, DNA digested by EcoR Ⅰ and Msp Ⅰ, control; DH, DNA digested by EcoR Ⅰ and Hpa Ⅱ, experimental group; DM, DNA digested by EcoR Ⅰ and Msp Ⅰ, experimental group. A, no methylation site; B, hemi-methylation site; C, full-methylation site; and D, hyper-methylation or uninformative site. |
The statistical analysis of the methylation epiloci in experimental and control groups are presented in Table 2. The methylation ratio of control ranged from 47.31% (muscle) to 22.94% (hepatopancreas) to 17.69% (gill). The ratio of experimental group varied similarly from 39.61% (muscle) to 25.19% (hepatopancreas) to 17.26% (gill). In comparison with control, the methylation ratio decreased in muscle (by 7.70%, P < 0.01) but increased in hepatopancreas (by 2.25%, P < 0.05).
|
|
Table 2 Statistical analysis of methylation epiloci |
The change of DNA methylation pattern under low salinity stress is listed in Table 3. Three changes of methylation epiloci were identified including 1) unchanged (A1-A3), 2) demethylated (B1-B6); and 3) methylated (C1-C6). The percentage of methylation pattern change is shown in Fig. 2. At low salinity, most of the epiloci remained unchanged, but demethylation and methylation occurred in three tissues simultaneously. The largest proportion of demethylation was detected in muscle, and the methylation ratio of these three tissues was highly similar.
|
|
Table 3 The different methylation patterns |
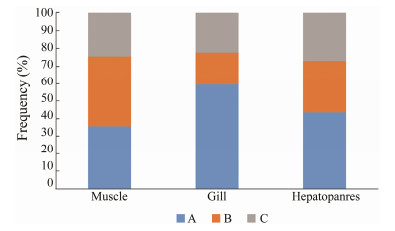
|
Fig. 2 The percentage of the methylation change pattern after low salinity stressing. A, unchanged epiloci; B, demethylated epiloci; C, methylated epiloci. |
DNA methylation is an epigenetic process which helps to control gene transcription. It is important through the whole life in eukaryotes. Although various cell types have the same genome, these cells have different methylomes at different times and in different spaces. In rice (Oryza sativa L.), the DNA methylation in leaf and root was 16.43% and 22.13% (Wang et al., 2011). In chicken, the methylation level was about 29.7% in muscle, 27.5% in liver, 27.5% in heart, and 26.1% in kidney (Xu et al., 2011). In grass carp (Ctenopharyngodon idella), a high level of methylation was detected (75.9%) (Cao et al., 2007). DNA methylation has been well-studied in vertebrates and plants but less studied in most invertebrates. In the present study, the genomic baseline of DNA methylation of P. trituberculatus was 17.69% in gill, 22.94% in hepatopancreas, and 47.31% in muscle, being consistent with the reported in other marine invertebrates varing between 20% and 50% (Jiang et al., 2014; Sun et al., 2014; Zhao et al., 2015; Kong et al., 2017). The different DNA methylation levels in different tissues suggest a tissue-specific epigenetic regulation of gene expression in differentiation (Zhao et al., 2015). In our study, the higher methylation rate in the muscle (47.31%) may be due to the relatively low metabolic rate and low expression of genes in this tissue.
Salinity is one of the most important abiotic factors that influence not only the distribution and abundance of crustaceans, but also their general physiology and well being (Romano and Zeng, 2012). DNA methylation plays an important role in resisting abiotic stress (Bird, 2002). The changes of DNA methylation under low salinity stress have been documented recently in Cynoglossus semilaevis and Crassostrea gigas (Li et al., 2017; Zhang et al., 2017), both showed a downward trend in methylation in gill at earlier low-salinity stage (7 days and 24 h, respectively); however, they recovered to the original and control level at long term (60 days and 5 days, respectively). In the present study, when the crab subjected to low salinity stress for as long as 30 days, the methylation ratio was not significantly affected in gill (decreased by 0.43%, P > 0.05). Gill was reported as the direct regulation tissue for osmoregulation (Mcnamara and Faria, 2012). The relatively stable methylation level in gill reflects its greater adaptability to environmental salinity stress. Despite the lack of change in overall methylation ratio in gill of P. trituberculatus after low salinity stress, changes in the methylation pattern of gill tissue could still be observed (Table 3), indicating that DNA methylation may act as a dynamic balance to regulate the low salinity-resistance in the gill of P. trituberculatus.
Difference of methylation during low salinity stress mainly occurred in muscle (7.70%, P < 0.01) and hepatopancreas (2.25%, P < 0.05), both are tissue-specific. Muscle and hepatopancreas have been reported to suffer a significant alteration of amino acid composition and deformation of microstructure (Han et al., 2014; Koyama et al., 2018), which presumably link with a strong suppression or activation of transcription and translation under low salinity stress.
DNA methylation often associated with the change of gene expression; however, the relationship between DNA methylation and gene expression is more complicated than the initially predicted. The effect of DNA methylation on gene expression varies in different tissues, sequences and genome regions (Wang et al., 2014). To further understand the relationship between methylation and expression of a specific gene, the gene expression profiles and methylation sites should be carefully examined.
5 ConclusionsThe results showed that the baseline DNA methylation is different in muscle, gill and hepatopancreas of P. trituberculatus (17.69% to 47.31%), demonstrating the important role of DNA methylation in tissue-specific epiloci. Furthermore, DNA methylation patterns of three tissues changed after P. trituberculatus was cultured at salinity 12 for 30 days, which suggests that DNA methylation may have an important role in salinity adaptation. The results provided a basis for further research on the potential role of DNA methylation in P. trituberculatus during adapting to low salinity.
AcknowledgementsThis study was supported by the grants from the National Natural Science Foundation of China (No. 4147 6124), the Natural Science Foundation of Zhejiang Province (No. LY17C190005), the Major Agriculture Program of Ningbo (No. 2017C110007), the Ningbo Science and Technology Project (No. 2016C10037), the Open Fund of Ningbo University (No. xkzsc1505) and K C Wong Magana Fund in Ningbo University. We would like to thank Dr. Sarah-Louise Counter Selly (University of Stirling, UK) for her linguistic assistance during the preparation of this manuscript.
Angers, B., Castonguay, E. and Massicotte, R., 2010. Environmentally induced phenotypes and DNA methylation: How to deal with unpredictable conditions until the next generation and after. Molecular Ecology, 19(7): 7-1283. (  0) 0) |
Bird, A., 2002. DNA methylation patterns and epigenetic memory. Genes and Development, 16(1): 1-6. DOI:10.1101/gad.964002 (  0) 0) |
Cao, Z., Ding, W., Yu, J., Cao, L. and Wu, T., 2007. Differences in methylated loci among different grass carp individuals from one pair of parents. Acta Zoologica Sinica, 6: 1083-1088. (  0) 0) |
Chen, G. Q., Jiang, M. M., Chen, B., Yang, Z. E. and Lin, C., 2006. Emergy analysis of Chinese agriculture. Agriculture Ecosystems and Environment, 115(1): 161-173. (  0) 0) |
Christensen, B. C., Houseman, E. A., Marsit, C. J., Zheng, S., Wrensch, M. R., Wiemels, J. L., Nelson, H. H., Karagas, M. R., Padbury, J. E., Bueno, R., Sugarbaker, D. J., Yeh, R. E., Wiencke, J. K. and Kelsey, K. E., 2009. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon cpg island context. PLoS Genetics, 5(8): e1000602. DOI:10.1371/journal.pgen.1000602 (  0) 0) |
Csankovszki, G., Nagy, A. and Jaenisch, R., 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. The Journal of Cell Biology, 153(4): 4-773. (  0) 0) |
Doerfler, W., 1983. DNA methylation and gene activity. Cell, 52(53): 93. (  0) 0) |
Feng, X. D., Hong, L. Z., Hua, X. X. and Ping, X. I., 2005. Ehe influence of typhoon on the sea surface salinity in the warm pool of the western pacific. Acta Oceanologica Sinica, 27(6): 6-343. (  0) 0) |
Han, X. K., Gao, B. Q., Wang, H. F., Liu, P., Chen, P. and Li, H., 2014. Effects of low salinity stress on microstructure of gill and hepatopancreas and family survival rate of Portunus trituberculatus. Progress in Fishery Sciences, 35(1): 1-104 (in Chinese with English abstract). (  0) 0) |
Jiang, Q., Li, Q., Yu, H. and Kong, L., 2016. Inheritance and variation of genomic DNA methylation in diploid and triploid Pacific oyster (Crassostrea gigas). Marine Biotechnology, 18: 124-132. DOI:10.1007/s10126-015-9674-4 (  0) 0) |
Jiang, Q., Yu, H., Kong, L. F. and Li, Q., 2014. Analysis of DNA methylation in different tissues of the Pacific oyster (Crassostrea gigas) with the fluorescence-labeled methylation-sensitive amplified polymorphism (F-MSAP). Journal of Fishery Sciences of China, 21(4): 4-676 (in Chinese with English abstract). (  0) 0) |
Kong, N., Liu, X., Li, J. Y., Mu, W. D., Lian, J. W., Xue, Y. J. and Li, Q., 2017. Effects of temperature and salinity on survival, growth and DNA methylation of juvenile Pacific abalone, Haliotis discus hannai Ⅰno. Chinese Journal of Oceanology and Limnology, 35(5): 5-1248. (  0) 0) |
Koyama, H., Mizusawa, N., Hoashi, M., Tan, E., Yasumoto, K., Jimbo, M., Ikeda, D., Yokoyama, T., Asakawa, S., Piyapattanakorn, S. and Watabe, S., 2018. Changes in free amino acid concentrations and associated gene expression profiles in the abdominal muscle of kuruma shrimp (Marsupenaeus japonicus) acclimated at different salinities. Journal of Experimental Biology, 221: jebl68997. (  0) 0) |
Li, E., Beard, C. and Jaenisch, R., 1993. Role for DNA methylation in genomic imprinting. Nature, 366(6453): 6453-362. (  0) 0) |
Li, S., He, F., Wen, H., Li, J., Si, Y., Liu, M., He, H. and Huang, Z., 2017. Analysis of DNA methylation level by methylation-sensitive amplification polymorphism in half smooth tongue sole (Cynoglossus semilaevis) subjected to salinity stress. Journal of Ocean University China, 16(2): 2-269. (  0) 0) |
Maegawa, S., Hinkal, G., Kim, H. S., Shen, F., Li, Z., Zhang, J., Zhang, N., Liang, S., Donehower, L. A. and Issa, J. P., 2010. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Research, 20(3): 3-332. (  0) 0) |
Mcnamara, J. C. and Faria, S. C., 2012. Evolution of osmoregulatory patterns and gill ion transport mechanisms in the decapod Crustacea: A review. Journal of Comparative Physiology B, 182(8): 8-997. (  0) 0) |
Navarromartin, L., Vinas, J., Ribas, L., Diaz, N., Gutierrez, A., Croce, L. D. and Piferrer, F., 2011. DNA methylation of the gonadal aromatase (cypl9a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genetics, 7(12): e1002447. DOI:10.1371/journal.pgen.1002447 |
Ou, X., Zhang, Y., Xu, C., Lin, X., Zang, Q., Zhuang, T., Jiang, L., Wettstein, D. and Liu, B., 2012. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L). PLoS One, 7(9): e41143. DOI:10.1371/journal.pone.0041143 (  0) 0) |
Pan, L. Q., Zhang, L. J. and Liu, H. Y., 2007. Effects of salinity and pH on ion-transport enzyme activities, survival and growth of litopenaeus vannamei postlarvae. Aquaculture, 273(4): 4-711. (  0) 0) |
Romano, N. and Zeng, C. S., 2012. Osmoregulation in decapod crustaceans: Implications to aquaculture productivity, methods for potential improvement and interactions with elevated ammonia exposure. Aquaculture, 334: 12-23. (  0) 0) |
Ruscoe, I. M., Shelley, C. C. and Williams, G. R., 2004. The combined effects of temperature and salinity on growth and survival of juvenile mud crabs (Scylla serrata Forskal). Aquaculture, 238(1-4): 239-247. DOI:10.1016/j.aquaculture.2004.05.030 (  0) 0) |
Shan, X., Wang, X., Yang, G., Wu, Y., Su, S., Li, S., Liu, H. and Yuan, Y., 2013. Analysis of the DNA methylation of maize (Zea mays L.) in response to cold stress based on methyla-tion-sensitive amplified polymorphisms. Journal of Plant Biology, 56(1): 1-32. (  0) 0) |
Smith, Z. D. and Meissner, A., 2013. DNA methylation: Roles in mammalian development. Nature Reviews Genetics, 14(3): 3-204. (  0) 0) |
Sun, Y., Hou, R., Fu, X., Sun, C., Wang, S., Wang, C., Li, N., Zhang, L. and Bao, Z., 2014. Genomewide analysis of DNA methylation in five tissues of Zhikong scallop, Chlamys farreri. PLoS One, 9(1): e86232. DOI:10.1371/journal.pone.0086232 (  0) 0) |
Xiong, L. Z., Xu, C. G., Maroof, M. A. S. and Zhang, Q., 1999. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Molecular and General Genetics Mgg, 261(3): 3-439. (  0) 0) |
Xu, Q., Zhang, Y., Sun, D. X., Wang, Y. C., Tang, S. Q. and Zhao, M., 2011. Analysis of DNA methylation in different chicken tissues with MSAP. Hereditas, 33(6): 620. (  0) 0) |
Zhang, C. Y., Wang, N. N., Zhang, Y. H., Feng, Q. Z., Yang, C. W. and Liu, B., 2013. DNA methylation involved in proline accumulation in response to osmotic stress in rice (Oryza sativa). Genetics and Molecular Research, 12(2): 2-1269. (  0) 0) |
Zhang, X., Li, Q., Kong, L. F. and Yu, H., 2017. DNA methylation changes detected by methylation-sensitive amplified polymorphism in the Pacific oyster (Crassostrea gigas) in response to salinity stress. Genes & Genomics, 39: 1173-1181. (  0) 0) |
Zhao, Y., Chen, M., Storey, K. B., Sun, K. and Yang, H., 2015. DNA methylation levels analysis in four tissues of sea cucumber Apostichopus japonicus based on fluorescence-labeled methylation-sensitive amplified polymorphism (F-MSAP) during aestivation. Comparative Biochemistry and Physiology B, 181: 26-32. DOI:10.1016/j.cbpb.2014.11.001 (  0) 0) |
Wang, J., Marowsky, N. C. and Fan, C., 2014. Divergence of gene body DNA methylation and evolution of plant duplicate genes. PLoS One, 9(10): e110357. DOI:10.1371/journal.pone.0110357 (  0) 0) |
Wang, M., Qin, F., Xie, C., Li, W., Yuan, J., Kong, F., Yu, W., Xia, G. and Liu, S., 2014. Induced and constitutive DNA methylation in a salinity-tolerant wheat introgression line. Plant and Cell Physiology, 55(7): 7-1354. (  0) 0) |
Wang, W. S., Pan, Y. J., Zhao, X. Q., Dwivedi, D., Zhu, L. H., Ali, J., Fu, B. Y. and Li, Z. K., 2011. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). Journal of Experimental Botany, 62(6): 6-1951. (  0) 0) |
 2019, Vol. 18
2019, Vol. 18


