2) College of Life Sciences, Yantai University, Yantai 264005, China
Gametogenesis, including spermatogenesis and oogenesis, is a complex process that is regulated by internal and external factors (Zhang et al., 2014). Internal factors, including the regulation of spatial and temporal genes and the formation of an RNA-induced silencing complex, play major roles in vertebrate gametogenesis (Schultz et al., 2003). The Argonaute family of proteins forms a typical element of the RNA-induced silencing complex (RISC) and plays a crucial role regulating small RNA in gene silencing. The Argonaute family proteins are divided into the Ago- and Piwi subfamilies (Carmell et al., 2002). The Ago-subfamily proteins exist in all types of cells. They can combine with small interfering RNAs and micro-RNAs (He and Hannon, 2004), and have crucial role in RNA interference. The Piwi-subfamily proteins are usually abundant in germ cells. They tend to bind with PIWI-interacting RNAs (piRNAs), and inhibit the expression of transposable elements (TEs) further ensuring the stability of genome (Aravin et al., 2008).
Piwi genes, which belong to the Piwi-subfamily, are crucial components of the RISC involved in RNA interference and play an essential regulatory role in gametogenesis and gonadal development (Hammond et al., 2001). Piwi genes are conserved and characterized by sequences encoding two domains: a PAZ (Piwi-Argonaute-Zwille)-encoding sequence in the center of the nucleotide sequence and a C-terminus PIWI domain-encoding sequence (Filipowicz, 2005; Pan et al., 2012). Both of these domains are responsible for the interaction between Piwi proteins and piRNA.
Piwi genes are enriched in germ cells, and mutations in piwi genes can cause failure of germline cell division. After combining with specific piRNAs, Piwi proteins firstly affect the expression at the post-transcriptional level and then regulate the transcriptional silencing (Pandey et al., 2018). The functions of piwi genes in gametogenesis have been well investigated, particularly in some model organisms. For example, three homologs (Miwi, Miwi2, and Mili) have been identified in mice, all of which are enriched in germ cells (Taborska et al., 2019). Additionally, Miwi, Mili2, and Mili mutations lead to obvious defects in spermatogenesis that cause infertility (Manakov et al., 2015). In some lower vertebrates and fishes, such as zebrafish, ziwi (piwil1) and zili (piwil2) are highly expressed in the germline (Houwing et al., 2007). A ziwi mutation causes failure of germ cell maintenance, while a lack of zili affects differentiation of germ cells (Houwing et al., 2008). Studies involving piwi homologous genes in teleosts, such as Scophthalmus maximus, Oreochromis niloticus, and Cyprinus carpio, have reported that piwi genes are involved in the reproductive cycle and influence sex differentiation (Zhou et al., 2012; Xiao et al., 2013; Wang et al., 2017).
The hypothalamic-pituitary-gonadal (HPG) axis regulates the Piwi-piRNA pathway and affects reproductive activities (Wang et al., 2018). Multiple hormones in the HPG axis interact to regulate individual reproductive activities. Gonadotropin-releasing hormone, which is synthesized and secreted by the hypothalamus, stimulates pituitary gonadotropic cells to release gonadotropins, such as follicle-stimulating hormone and luteinizing hormone (LH), which regulate the secretion of steroids (García-Lopez et al., 2010). Exogenously administered human chorionic gonadotropin (hCG), which is similar to LH, promotes maturation of the gonads and the ovulation during the spawning period in teleosts (Zhou et al., 2015; Zhao et al., 2018). As steroids, 17α-methyltestosterone (17α-MT) regulates sex determination and differentiation in males (Lee et al., 2017), and estradiol-17β (E2) is a critical hormone stimulated by gonadotropins that controls growth and maturation of oocytes (Gauthier-Clerc et al., 2006). Previous studies have shown that steroids negatively affected piwi in mice (Pan et al., 2012), suggesting that piwi genes might be important in the pathways that the hormones are included in regulating the reproductive development. In teleosts, the HPG axis hormones suppress piwi expression in carp (Zhou et al., 2014), but the detailed regulatory functions of piwi in other teleosts have rarely been studied.
Paralichthys olivaceus is an important cultured fish species in northern China, Korea, and Japan (Sun et al., 2013). The sex ratio of P. olivaceus is close to 1:1 in the natural environment. Considering that females sexually mature earlier and are substantially larger than males, the culture of primarily females would help optimize production. Increasing the female sex ratio has become a hot topic in the field of breeding (Yamamoto 1999; Tian et al., 2009). Genetic manipulation and hormonal treatment are major methods for regulating sex differentiation in fish (Zhang et al., 2010). As piwi is an important gene regulating gonadal development and gametogenesis, clarifying the molecular mechanisms of piwi will help to understand its important role in the regulation of reproduction and gender differentiation. Furthermore, exploring the mechanism of hormone interaction with piwi genes is important for culturing primarily females and thereby improving the economic value of the aquaculture.
In this study, we cloned the piwil2 gene in P. olivaceus to elucidate its molecular and genetic features. Then, we identified the localization of piwil2 during gametogenesis and its transcriptional activity in adult tissues and during embryonic development. The transcriptional activity of the piwil2 promoter regions was analyzed to investigate the transcriptional regulatory elements. Furthermore, we analyzed the effects of hormones including hCG, 17α-MT, and E2 on piwil2, to understand the regulatory mechanisms of piwil2 during gonadal development. In general, we analyzed the gene structure and evolutionary status of the piwil2 gene, and further explored its expression pattern and regulatory factors. The results will provide a foundation for further study of gonadal development in P. olivaceus.
2 Materials and Methods 2.1 Fish and Embryo CollectionExperimental fish were provided by a commercial hatchery in Haiyang, Shandong Province, China. Animal experiments were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals (State Science and Technology Commission of China for No. 2, October 31, 1988. http://www.gov.cn/gongbao/content/2011/content_1860757.htm). Six 1.5-year-old adult fish (three females and three males) were obtained for tissue distribution studies, which included specimens from the heart, liver, spleen, kidney, brain, gills, muscle, intestines, testes, and ovaries. Body lengths were 310-420 mm and body weights were 520-720 g. Gonads for in situ hybridization (ISH) were collected and fully measured. In addition, ten different larval stages, including unfertilized eggs, 1-cell, 8-cell, 64-cell, blastula, gastrula, neurula, heart-beating, hatching, and 1-day post-hatch (dph) embryos and larvae were gathered for experiments. All tissues and larvae were frozen in liquid nitrogen and stored at −80℃ for further experiments.
2.2 RNA Extraction and cDNA SynthesisTotal RNA from different P. olivaceus tissues or embryos was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. Then, RNase-free DNase Ⅰ (TaKaRa, Dalian, China) was used to dislodge any remaining DNA. The concentration and quality of the DNA were determined by 1.5% agarose gel electrophoresis and spectrophotometry, respectively. cDNA was synthesized with total RNA and random hexamer primers using the Reverse Transcriptase M-MLV Kit (TaKaRa). β-actin was selected as reference gene to verify the success of reverse transcription and the quality of the cDNA.
2.3 Gene piwil2 Molecular CloningPrimers (Piwil2-core-Fw/Rv, Table 1) designed based on conserved sequences of other teleosts were used to obtain the core cDNA fragment of piwil2. Next, agarose gel electrophoresis and a Zymoclean Gel DNA Recovery Kit (Zymo Research, Orange, CA, USA) were used to separate and purify the amplified polymerase chain reaction (PCR) products. Then, the PCR products were ligated into the pMD-19T vector for sequencing (TaKaRa). The P. olivaceus transcriptome which was previously sequenced by our laboratory (Wang et al., 2014) was used to ensure sequence accuracy.
|
|
Table 1 Primers used to analyze the Paralichthys olivaceus piwil2 gene |
The BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/) was used to carry out the homology search for the piwil2 nucleotide sequence. The highly conserved piwil2 protein domains, including the PAZ and PIWI amino acid (aa) sequences, were deduced through the SMART program (http://smart.embl-heidelberg.de/). The neighborjoining (NJ) method was applied to establish the phylogenetic tree according to the alignment of piwil2 amino acid sequences using MEGA 6.0 software (Tamura et al., 2013). The scale bar is a numerical scale representing the differences between the amino acid sequences of different species. The branch support statistic was 500.
2.5 Quantitative Real-Time PCR (qRT-PCR)qRT-PCR was used to detect the expression profiles of piwil2 in different tissues, embryonic developmental stages, and hormone treatments. β-actin was selected as a reference gene to measure piwil2 mRNA expression levels. All primers, including piwil2-qPCR-Fw/Rv and β-actin-Fw/Rv (Table 1), were designed by the Integrated DNA Technologies website (http://sg.idtdna.com/Primerquest/Home/Index) in non-conserved domains. The pre-experimental amplification was performed to verify primer specificity. qRT-PCR was performed with 2× SYBR Green qPCR MasterMix (Novoprotein, Shanghai, China) using a LightCycler 480 (Roche, Forrentrasse, Switzerland). The reaction volume was 20 μL, including 10 μL of 2× SYBR qPCR SuperMix, 0.4 μL of each primer (10 μmol L−1), 1 μL of cDNA (20 ng μL−1), and 8.2 μL of nuclease-free water. The PCR reaction conditions were 95℃ (5 min) for pre-incubation followed by 40 cycles at 95℃ (15 s), 60℃ (15 s) and 72℃ (45 s). At least three templates were analyzed to guarantee the accuracy of the experimental results. All data were subjected to the 2−ΔΔCt method.
2-6 In situ Hybridization (ISH)ISH was performed to localize the piwil2 mRNA in gonadal tissues as described previously. The specific primers (Piwil2-ISH-Fw and Piwil2-ISH-Rv, Table 1) were designed according to piwil2 whole-genome sequencing. The sense and anti-sense probes were synthesized using a DIG RNA Labeling Kit (SP6/T7) (Roche, Mannheim, Germany), following the manufacturer's instructions. The paraffin sections were stained and combined with the piwil2 antisense probe according to a previous study (Gao et al., 2013).
2.7 Construction of Deletion Vectors and the Luciferase AssayTo reveal the piwil2 promoter's transcriptional regulatory mechanisms, deleted vectors were constructed by ligating the piwil2 promoter fragments with different sizes, including pGL3-M1 (2083 bp), pGL3-M2 (1541 bp), pGL3-M3 (1117 bp), pGL3-M4 (798 bp), and pGL3-M5 (300 bp), into the luciferase reporter vector pGL3-Basic. The primer sequences are shown in Table 1. The human 293T cell line was used in the luciferase assay considering the transfection efficiency of cells. The human 293T cell line was donated by the Key Laboratory of Marine Genetics and Breeding, Ministry of Education, College of Marine Life Sciences, Ocean University of China. LipoGene™ 2000 PLus Transfection Reagent (US Everbright Inc., Suzhou, China) was used for the transient transfection experiments. The Dual-Luciferase Reporter Assay System (Promega, Mannheim, Germany) was used to perform the dual-luciferase reporter assays after 24 h of transfection according to the manufacturer's instructions. All samples in the dualluciferase reporter assay were assessed with three replicates.
2.8 In vivo Hormone TreatmentThe exogenous hormone hCG is highly similar to HPG axis hormone LH, and has been widely used to manipulate for spawn in production because of the abundant raw materials. In the same way, 17α-MT and E2 are also readily available and have similar function to the steroids in HPG axis. Thus, hCG, 17α-MT, and E2 (Sigma-Aldrich, St. Louis, MO, USA) were selected for in vivo experiments to examine the effects of hormones on piwil2 expression. For the hCG experiment, the male and female fish were divided into three groups (n = 3), respectively, including a control group (injected with saline), a low concentration group (injected with 600 IU kg−1 bodyweight), and a high concentration group (injected with 3000 IU kg−1 bodyweight). For the 17α-MT experiment, males were divided into three groups (n = 3), including a control group (injected with saline), a low concentration group (injected with 1.5 μg kg−1 bodyweight), and a high concentration group (injected with 50 μg kg−1 bodyweight). For the E2 experiment, males were divided into two groups (n = 3), including a control group (injected with saline) and an E2 group (injected with 3 μg kg−1 bodyweight). The hormones were injected intraperitoneally. The same hormone treatment was administered for the following 5 days and the treated tissue samples were obtained on day 6. Previous research on other teleosts provided the references for the treatment concentrations used in this study (Xiao et al., 2013; Li et al., 2018; Wang et al., 2018). Each group was maintained in a stable condition for 5 days. All gonads were collected on day 6 and were stored at −80℃ for further study.
2.9 In vitro Hormone TreatmentThe gonads of three male fish were dissected and washed three times in phosphate-buffered saline (PBS) containing 1% penicillin-streptomycin solution. The tissues were minced into small pieces (mean length = 1.5 cm) in a Petri dish. Then the isometric segments were cultured in a 12-well culture plate for further in vitro hormone treatments.
Four different concentrations of 17α-MT and E2, such as 10−6, 10−8, 10−10, and 10−12 mol L−1, were added to the treatment groups, respectively. The tissues from each well were collected after 4 h of treatment, and stored for further qRT-PCR to determine piwil2 expression.
2.10 Statistical AnalysisData are expressed as mean ± standard error (SE) of at least three independent experiments, and were analyzed by one-way analysis of variance. All qRT-PCR data were analyzed using SPSS 20.0 software (IBM, Armonk, NY, USA). P-values < 0.05 were considered significant.
3 Results 3.1 piwil2 Cloning and Sequence AnalysisThe open reading frame (ORF) of the piwil2 gene was obtained from the P. olivaceus testes by PCR. The full-length ORF was 3192 bp and encoded 1063 amino acids. An analysis of the protein structure identified PAZ and PIWI as conserved domains in piwil2 (Fig. 1A). The PAZ domain was located at positions 477–616 at the N-terminal and contained 140 amino acids. The PIWI domain, which contained 292 amino acids, was located at the C-terminal at positions 758–1049. The PAZ (Fig. 1B) and PIWI domains (Fig. 1C) in P. olivaceus were highly conserved after a comparison with other teleosts, suggesting that these two domains may play an essential role maintaining functions of the piwil2 gene among different species.

|
Fig. 1 Amino acid sequence and structural domain analysis of the piwil2 gene. (A) The amino acid sequence analysis of the piwil2 gene. The green square represents the PAZ domain and the blue square represents the PIWI domain. The amino acids positions of the structural domain are respectively marked in the picture. (B) Alignment of the piwil2 PAZ domain among different species. Dark shadowed regions represent the same amino acid residues and the grey shadowed regions represent similar amino acids residues. (C) Alignment of the piwil2 PIWI domain among different species. The selected species include Paralichthys olivaceus (Po, XM_020094094.1); Oryctolagus cuniculus (Oc, XM_008249876.1); Monopterus albus (Ma, XM_020590301.1); Oreochromis niloticus (On, XM_003445662.4); Mus musculus (Mm, NM_001364321.1); Homo sapiens (Hs, NM_018068.5); Xenopus tropicalis (Xt, XM_018091922.1); and Scophthalmus maximus (Sm, KY_ 123251.1). The amino acid sequences are from the GenBank accession numbers in brackets. |
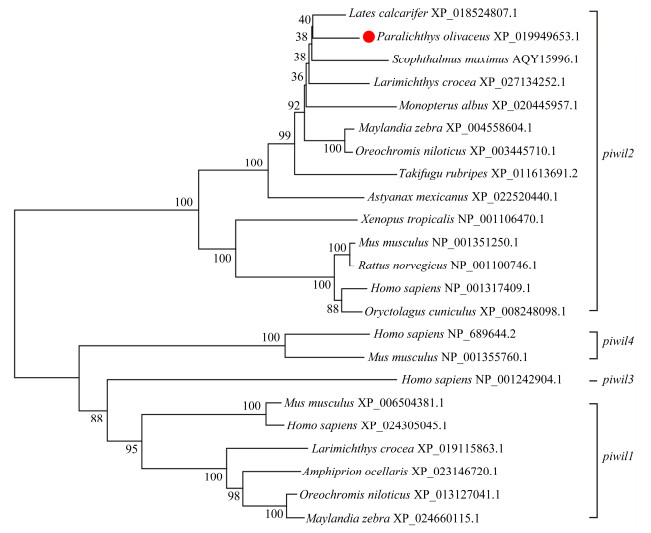
A phylogenetic tree was established using the amino acid sequences and the NJ method to demonstrate the evolutionary relationships between P. olivaceus and other vertebrates (Fig. 2). As expected, the two piwi homologs can be branched into two main clades, called the piwil1 and piwil2 clades in the phylogenetic tree. Only a few mammals, such as humans and mice, possess the piwil3 and piwil4 clades. P. olivaceus piwil2 was clearly clustered with other teleosts and belonged to the teleost subgroup, while the others formed a tetrapod subgroup.
|
Fig. 2 piwil2 phylogenetic tree in Paralichthys olivaceus with other representative vertebrates, predicted by the amino acid sequences. The species and GenBank accession numbers are shown. |
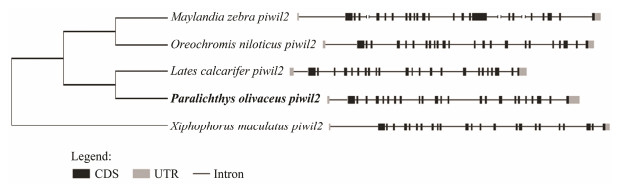
The gene structure was highly conserved between P. olivaceus and other species, as the structures had the same number of exons and introns (Fig. 3). The exon and intron data were uploaded to the supplementary file. As the untranslated region (UTR) sequences of various species were incomplete, only the ORF sequences were the focus of our analysis. Twenty-three exons and 23 introns were found in P. olivaceus piwil2 after comparing piwil2 cDNA and genomic DNA. For most species, there was an intron inserted in the 5'UTR of piwil2, and the 5'UTR sequence is always part of one-to-a-few first exons. The similarity of the genomic organization revealed the conservative of piwil2 genes among teleosts during the evolution.
|
Fig. 3 Comparison of piwil2 genomic structure between Paralichthys olivaceus and other teleosts. The black areas represent exons, the lines represent introns, and the gray areas represent the untranslated region (UTR) area. |
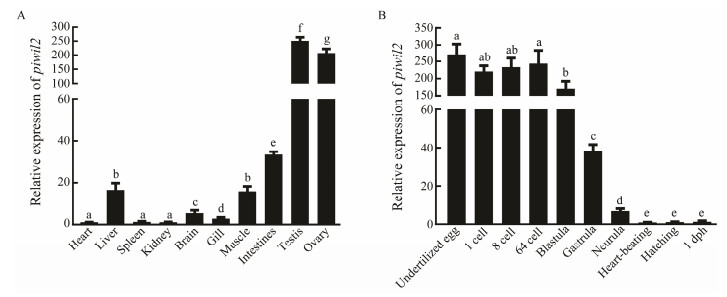
The piwil2 mRNA expression levels in different tissues, including heart, brain, liver, intestines, kidney, spleen, muscle, gills, testes, and ovaries were analyzed by qRT-PCR (Fig. 4A). The results showed that piwil2 was particularly strongly expressed in the testes and ovaries, and showed a sexually dimorphic expression pattern. Furthermore, piwil2 was also expressed in other somatic tissues at low levels.
|
Fig. 4 The relative expression levels of piwil2 in different tissues and stages of embryonic development. (A) The piwil2 expression patterns in various tissues of males and females. (B) The piwil2 expression pattern during different stages of embryonic development. All data are expressed as mean ± SEM. from three separate individuals (n = 3). Different superscripts indicate that P < 0.05. |
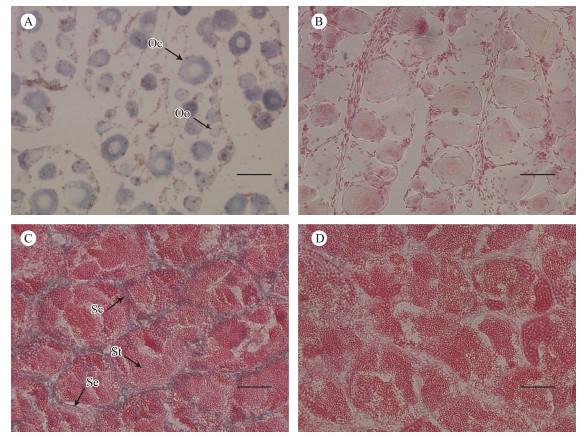
ISH was used to further explore the tissue-specific expression pattern of piwil2 (Fig. 5). Strong positive signals were detected throughout the cytoplasm of oogonia and oocytes in ovarian sections. The signals were concentrated in the Sertoli cells and spermatocytes of testicular sections, with no signal in spermatids. These results indicate that the piwil2 gene might play a crucial role in gametogenesis.
|
Fig. 5 The mRNA distribution of piwil2 in gonadal cells. The antisense probe labeled with DIG was stained blue (A and C). The sense probe is the negative control and the hybridization was unstained (B and D). Se, Sertoli cells; Sc, spermatocytes; St, spermatid; Oo, oogonia; Oc, oocytes. Scale bars = 50 µm. |
To determine the piwil2 expression pattern during embryogenesis, piwil2 mRNA levels were analyzed at different embryonic development stages by qRT-PCR (Fig. 4B). Piwil2 was maternally inherited as it was highly expressed from the 1-cell to the blastula stage. The expression levels suddenly decreased from the beginning of the gastrula stage and continued to decrease until birth. Considering the difference in piwil2 expression levels at different embryogenesis stages, we speculate that piwil2 might perform different functions at these stages.
3.5 The piwil2 Core Promoter Region and Related Transcriptional Regulatory ElementsFive different deletion fragments of the piwil2 promoter region were obtained and connected to upstream of the luciferase reporter gene to detect the core promoter region (Fig. 6). The transcriptional initiation site (ATG) for the promoter deletion assay was designated as +1 and the piwil2 upstream 5'-flanking sequence from −2143 bp to −60 bp was analyzed after removing all introns. piwil2 expression increased significantly in the M5 fragment. The M4 fragment included M5, but the expression level of piwil2 in M4 decreased slightly. Expression increased in the M3 fragment, which was similar to that in M5. Subsequently, piwil2 expression in the M2 region decreased rapidly and then recovered in the M1 fragment. Therefore, the deletion of the fragment between −360 bp to −60 bp seemed to be the core promoter region of piwil2.
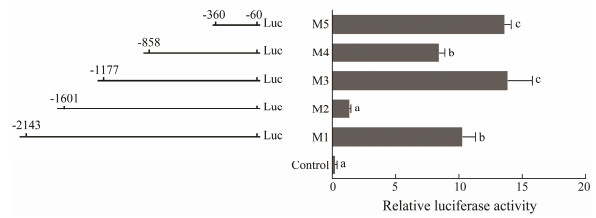
|
Fig. 6 Transcriptional activities of deleted vectors measured by the luciferase assay. Left, A series of deletion fragments linked with the luciferase gene in the pGL3 vector. Right, Relative activities of the deleted vectors pGL3 M1-M5 determined by the luciferase assay. Different superscripts indicate that P < 0.05. |
Then, the transcription elements of these five different deletion fragments were analyzed. Some transcription elements, such as SMAD, NKX6, CAAT, and NF-1F, were detected in the core promoter region (M5). M2 and M4 highly inhibited piwil2 expression, in which GATA, CART, IRFF, and HOMF were found. BRAC, NF-1F, HNF, and AP1R were detected in the M1 and M3 regions, neutralizing the inhibitory effect of the previously deleted fragments. All of these transcriptional elements may be closely related to regulation of piwil2 expression and may further affect gametogenesis and gonadal development.
3.6 Effect of in vivo and in vitro Hormone Treatments on piwil2The piwil2 expression levels were analyzed under the hormone treatments to understand the effects on the piwil2 gene. As the result, hCG suppressed piwil2 expression in males (Fig. 7A). In females, the high concentration of hCG had an inhibitory effect on piwil2, while the low concentration promoted piwil2 expression (Fig. 7B). For 17α-MT, both the high and low concentrations increased piwil2 expression in males (Fig. 7C). Additionally, piwil2 transcription was promoted in response to E2 (Fig. 7D).
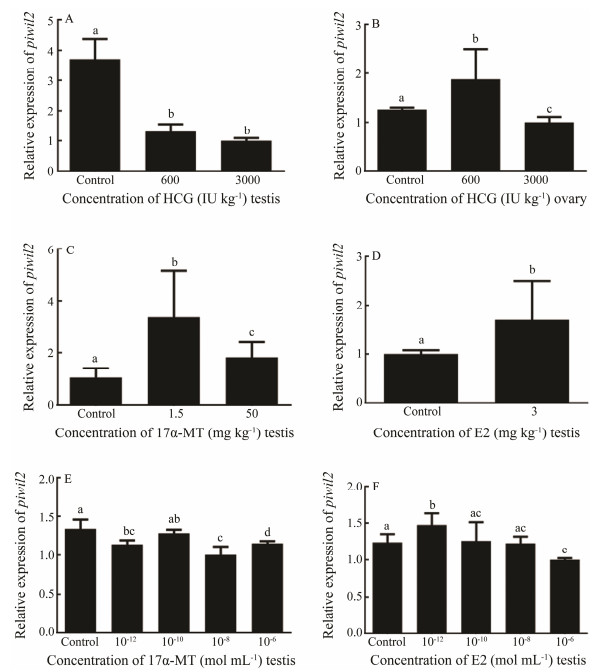
|
Fig. 7 The relative expression levels of piwil2 after the hormone treatments. (A) In vivo hCG treatment of testis. (B) In vivo hCG treatment of ovary. (C) In vivo 17α-MT treatment of testis. (D) In vivo E2 treatment of testis. (E) In vitro 17α-MT treatment of testicular cells. (F) In vitro E2 treatment of testicular cells. All data are shown as mean ± SEM (n = 3 in every group). Different superscripts indicate that P < 0.05. |
Different concentrations of sex hormones were used for the in vitro treatment to determine the piwil2 mRNA levels. As shown in Fig. 7E, all 17α-MT-treated groups showed a slight decrease in piwil2 mRNA levels compared with the control group, particularly when the concentration was 10−8 mol L−1. However, piwil2 expression declined slightly when the E2 concentration was 10−6 mol L−1 and increased when the E2 concentration was 10−12 mol L−1 (Fig. 7F).
4 DiscussionThe piwi gene is involved in many complex processes, such as spermatogenesis, egg activation, and fertilization (Wang et al., 2018). Piwi proteins play a crucial role during gonadal development and gametogenesis (Qiao et al., 2002), and when combined with piRNA, they participate in the Ping-Pong cycle to regulate genomic integrity and retrotransposon repression (Aravin et al., 2008). The piwi gene has been extensively researched in vertebrates and invertebrates, with some studies employing model organisms, including mice and zebrafish (Kuramochi-Miyagawa et al., 2001; Thomson and Lin, 2009). However, information regarding piwil2 in teleosts is still relatively scarce. Therefore, in this study, we identified and explored the fundamental functions of piwil2 in P. olivaceus.
4.1 Gene piwil2 Is Highly Conserved in VertebratesGene piwil2 from P. olivaceus was cloned to investigate its potential functions. The piwil2 from P. olivaceus contained the same PAZ and PIWI domains as in other teleost species, indicating that piwil2 plays a conserved role in different species. Evidence gathered from a phylogenetic tree showed that P. olivaceus piwil2 was more consistent with the teleost branch and clearly separate from tetrapods. Therefore, it was concluded that piwil2 is highly conserved and might participate in the same regulatory mechanisms in different species.
4.2 Piwil2 Transcripts Are More Abundant in the Testes than in the OvariesPiwil2 is a germline-specific gene that is enriched in gonadal tissues (Wen et al., 2018). Previous studies have shown that piwil2 is expressed in the testes and ovaries and that its expression is higher in testes than in ovaries (Mishima et al., 2008). In addition, piwil2 is highly expressed in the cytoplasm of oocytes and oogonia of Oryzias latipes (Zhao et al., 2012). In this study, the expression pattern of piwil2 in P. olivaceus was similar with that described in previous studies with other species, demonstrating that piwil2 has a crucial function in germline stem cell maintenance and gametogenesis. In addition, piwil2 expression was relatively lower in somatic tissues, indicating that piwil2 has other essential functions beyond reproduction, which need to be further explored.
4.3 Piwil2 May Be Involved in the Regulation of EmbryogenesisSome studies have reported that PIWI and piRNA regulate gene expression at the post-transcriptional level and participate in the regulation of embryonic development, gender determination, and other events (Schwager et al., 2015). It has been inferred that the piRNA pathway is highly active during early embryonic development, as piwil2 is expressed at high levels at this time (Rouget et al., 2010). The genome recombination and chromatin remodeling processes also occur during early embryogenesis (Sunanaga et al., 2010; Zhang et al., 2010), suggesting that the high piwil2 expression levels during this period regulate transcription and ensure stability of the genome. The piwil2 expression pattern during embryogenesis in P. olivaceus was similar to those in other species, such as S. maximus and X. tropicalis (Zhang et al., 2010; Wang et al., 2017). Therefore, the present study provides strong evidence that the piwil2-encoded protein participates in embryogenesis and has conserved functions (Li et al., 2011).
4.4 Transcription Elements Regulate piwil2 ExpressionDefining the core promoter region of the piwil2 gene has importance for revealing the transcriptional regulatory mechanisms and for further regulating piwil2 expression (Chang et al., 2015). In this study, the −360 to −60 bp region in the piwil2 5'-UTR contained some important transcriptional elements (SMAD, NKX6, CAAT, and NF-1F) that significantly enhanced promoter activity and was, therefore, suggested to be the core promoter region of the piwil2 gene. The −858 to −360 bp and the −1601 to −1177 bp sections in the 5'-flanking region were regions where piwil2 expression was suppressed. As some studies have demonstrated that piwi is inhibited by transcriptional elements (Sohn et al., 2014; Chang et al., 2015), the transcriptional elements in this fragment (GATA, CART, IRFF, and HOMF) may downregulate the activation of the promoter in P. olivaceus. The transcriptional regulatory mechanisms present in the −1177 to −858 bp and −2143 to −1607 bp regions played a positive role, thereby counteracting the inhibitory effects of the previous regions. BRAC, NF-1F, HNF, and AP1R in these regions potentially promoted piwil2 expression. Although these results are based on a bioanalytical level of analysis and prediction, these transcriptional elements may play an important role regulating piwil2 expression, and their detailed mechanisms in P. olivaceus need to be further studied.
4.5 The HPG Axis Affects piwil2 ExpressionGametogenesis and gonadal development in teleosts are regulated by a series of complex systems. Previous studies have shown that the HPG axis is an important neuroendocrine regulatory system (Fuqua and Rogol, 2013). Therefore, in this study, hCG, 17α-MT, and E2 were selected to verify their effects on piwil2 expression. They all play a crucial role in gametogenesis by regulating the piRNA pathway (Scholz and Gutzeit, 2000; Zhang et al., 2010; Miura et al., 2013; Zhou et al., 2015). As piwil2 is a key gene in the piRNA pathway, piwil2 may be involved in the regulation of HPG axis hormones during gametogenesis.
The expression of piwil2 decreased in males treated with hCG. The high hCG concentration inhibited piwil2 expression in females, whereas the low concentration promoted expression. These results are similar to those previously reported in Nile tilapia (Xiao et al., 2013). Additionally, 17α-MT and E2 promoted piwil2 expression in males. As 17α-MT masculinizes the gonads and promotes spermatogenesis and a lack of E2 disrupts early spermatogenesis (Lee et al., 1986; Murata et al., 2002), we hypothesized that sex hormones regulate the formation of spermatogenesis, leading to a change of piwil2 expression (Hansson et al., 1976).
The in vitro treatment was designed to explore the effect of complex regulatory system including cytokines and other factors on piwil2 expression at the cellular level after the hormone treatments. 17α-MT decreased piwil2 expression in testes in vitro, which was opposite to the in vivo result. We inferred that this result may be due to the complex and interrelated regulatory systems in individuals. The HPG axis, an important hormone regulatory pathway, is affected by many feedback loops (Rosvall et al., 2016). In the in vivo treatment, a high 17α-MT concentration not only promoted spermatogenesis, but also regulated the secretion of gonadotropins through negative feedback control. All of these mechanisms masked the true level of piwil2 responding to 17α-MT in vivo. A low E2 concentration increased piwil2 expression, which was consistent with the in vivo experiment. A higher E2 concentration had a negative effect on cell activities, leading to the inhibition of piwil2 expression. These results suggest that piwil2 was affected by the HPG axis and participates in gametogenesis (Bohórquez et al., 2017).
5 ConclusionsIn this study, we investigated the basic functions and regulatory factors of piwil2 in P. olivaceus. The piwil2 cDNA sequence from the testes of P. olivaceus was identified. A genetic structural analysis showed that piwil2 was conserved in different species. The tissue expression pattern analysis demonstrated that the piwil2 expression level was higher in testes than in other tissues. The tissue localization results suggested that piwil2 was highly expressed in the cytoplasm of oogonia and oocytes in the ovaries, as well as in Sertoli cells and spermatocytes in the testes. Moreover, the core promoter region was detected, and some major transcriptional elements were identified to explore the piwil2 transcriptional regulatory mechanism. In addition, piwil2 was maternally inherited and determined to play a vital role in the mechanism underlying hormonal regulation. Taken together, these results provide a theoretical basis for further exploring the role of hormones in the regulation of the expression of piwil2 gene.
AcknowledgementsThis study was supported by the National Natural Science Foundation of China (No. 31672646) and the Natural Science Foundation of Shandong Province (No. ZR 2017MC072).
Aravin, A.A., Sachidanandam, R., Bourc'his, D., Schaefer, C., Pezic, D., Toth, K.F., Bestor, T. and Hannon, G.J., 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular Cell, 31(6): 785-799. DOI:10.1016/j.molcel.2008.09.003 (  0) 0) |
Bohórquez, M.O.T., Mechaly, A.S., Elisio, M., Chalde, T., Canosa, L.F., Miranda, L.A. and Somoza, G.M., 2017. Kiss-peptins and their receptors in the brain-pituitary-gonadal axis of Odonthestes bonariensis:Their relationship with gameto-genesis along the reproductive cycle. General and Compa-rative Endocrinology, 252: 209-218. DOI:10.1016/j.ygcen.2017.06.028 (  0) 0) |
Carmell, M.A., Xuan, Z., Zhang, M.Q. and Hannon, G.J., 2002. The Argonaute family:Tentacles that reach into RNAi, deve-lopmental control, stem cell maintenance, and tumorigenesis. Genes and Development, 16(21): 2733-2742. DOI:10.1101/gad.1026102 (  0) 0) |
Chang, G., Chen, R., Xu, L., Ma, T., Wang, H., Chen, J., Zhang, Y., Li, Z., Wan, F., Guo, X., Xu, Q., Zhao, M. and Chen, G., 2015. DNA methylation and NF-Y regulate Piwil1 expression during chicken spermatogenesis. Animal Reproduction Science, 162: 95-103. DOI:10.1016/j.anireprosci.2015.09.016 (  0) 0) |
Filipowicz, W., 2005. RNAi:The nuts and bolts of the RISC ma-chine. Cell, 122(1): 17-20. DOI:10.1016/j.cell.2005.06.023 (  0) 0) |
Fuqua, J.S. and Rogol, A.D., 2013. Neuroendocrine alterations in the exercising human:Implications for energy homeostasis. Metabolism, 62(7): 911-921. DOI:10.1016/j.cell.2005.06.023 (  0) 0) |
Gao, J., Wang, J., Jiang, J., Fan, L., Wang, W., Liu, J., Zhang, Q. and Wang, X., 2013. Identification and characterization of a nanog homolog in Japanese flounder (Paralichthys olivaceus). Gene, 531(2): 411-421. DOI:10.1016/j.gene.2013.08.030 (  0) 0) |
García-Lopez, A., de Jonge, H., Nobrega, R.H., de Waal, P.P., van Dijk, W., Hemrika, W., Taranger, G.L., Bogerd, J. and Schulz, R.W., 2010. Studies in zebrafish reveal unusual cellu-lar expression patterns of gonadotropin receptor messenger ribonucleic acids in the testis and unexpected functional dif-ferentiation of the gonadotropins. Endocrinology, 151(5): 2349-2360. DOI:10.1210/en.2009-1227 (  0) 0) |
Gauthier-Clerc, S., Pellerin, J. and Amiard, J., 2006. Estradiol-17β and testosterone concentrations in male and female Mya arenaria(Mollusca bivalvia) during the reproductive cycle. General and Comparative Endocrinology, 145(2): 133-139. DOI:10.1016/j.ygcen.2005.08.004 (  0) 0) |
Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R. and Hannon, G.J., 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293(5532): 1146-1150. DOI:10.1126/science.1064023 (  0) 0) |
Hansson, V., Calandra, R., Purvis, K., Ritzen, M. and French, F.S., 1976. Hormonal regulation of spermatogenesis. Vitam Horm, 34: 187-214. DOI:10.1016/S0083-6729(08)60076-X (  0) 0) |
He, L. and Hannon, G.J., 2004. MicroRNAs:Small RNAs with a big role in gene regulation. Nature Reviews Genetics, 5(7): 522. DOI:10.1038/nrg1379 (  0) 0) |
Houwing, S., Berezikov, E. and Ketting, R.F., 2008. Zili is re-quired for germ cell differentiation and meiosis in zebrafish. The EMBO Journal, 27(20): 2702-2711. DOI:10.1038/emboj.2008.204 (  0) 0) |
Houwing, S., Kamminga, L.M., Berezikov, E., Cronembold, D., Girard, A., Van, H.D.E., Filippov, D.V., Blaser, H., Raz, E., Moens, C.B., Plasterk, R.H.A., Hannon, G.J., Draper, B.W. and Ketting, R.F., 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell, 129(1): 69-82. DOI:10.1016/j.cell.2007.03.026 (  0) 0) |
Kuramochi-Miyagawa, S., Kimura, T., Yomogida, K., Kuroiwa, A., Tadokoro, Y., Fujita, Y., Sato, M., Matsuda, Y. and Nakano, T., 2001. Two mouse piwi-related genes:miwi and mili. Mecha-nisms of Development, 108(1-2): 121-133. DOI:10.1016/s0925-4773(01)00499-3 (  0) 0) |
Lee, C.S., Tamaru, C., Banno, J. and Kelley, C., 1986. Inf-luence of chronic administration of LHRH-analogue and/or 17α-methyltestosterone on maturation in milkfish, Chanos cha-nos. Aquaculture, 59(2): 147-159. DOI:10.1016/0044-8486(86)90127-4 (  0) 0) |
Lee, S.L.J., Horsfield, J.A., Black, M.A., Rutherford, K., Fisher, A. and Gemmell, N.J., 2017. Histological and transcriptomic effects of 17α-methyltestosterone on zebrafish gonad develop-ment. BMC Genomics, 18(1): 557. DOI:10.1186/s12864-017-3915-z (  0) 0) |
Li, D., Sun, H., Deng, W., Tao, D., Liu, Y. and Ma, Y., 2011. Zili antagonizes Bmp signaling to regulate dorsal-ventral pattern-ing during zebrafish early embryogenesis. Zoological Science, 28(6): 397-402. DOI:10.2108/zsj.28.397 (  0) 0) |
Li, X., Yu, H., Wang, Y., Liu, X., Liu, Y., Qu, J. and Wang, X., 2018. Roles of two Sox9 genes during gonadal development in Japanese flounder:Sex differentiation, spermatogenesis and gonadal function maintenance. International Journal of Mole-cular Sciences, 19(2): 512. DOI:10.3390/ijms19020512 (  0) 0) |
Manakov, S.A., Pezic, D., Marinov, G.K., Pastor, W.A., Sa-chidanandam, R. and Aravin, A.A., 2015. miwi2 and mili have differential effects on piRNA biogenesis and DNA methyla-tion. Cell Reports, 12(8): 1234-1243. DOI:10.1016/j.celrep.2015.07.036 (  0) 0) |
Mishima, T., Takizawa, T., Luo, S.S., Ishibashi, O., Kawa-higashi, Y., Mizuguchi, Y., Ishikawa, T., Mori, M., Kanda, T., Goto, T. and Takizawa, T., 2008. MicroRNA (miRNA) clo-ning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction, 136(6): 811-822. DOI:10.1530/REP-08-0349 (  0) 0) |
Miura, S., Kobayashi, Y., Bhandari, R.K. and Nakamura, M., 2013. Estrogen favors the differentiation of ovarian tissues in the ambisexual gonads of anemonefish Amphiprion clarkii. Journal of Experimental Zoology Part A:Ecological Genetics and Physiology, 319(10): 560-568. DOI:10.1002/jez.1818 (  0) 0) |
Murata, Y., Robertson, K., Jones, M. and Simpson, E., 2002. Ef-fect of estrogen deficiency in the male:The ArKO mouse mo-del. Molecular and Cellular Endocrinology, 193(1-2): 7-12. DOI:10.1016/s0303-7207(02)00090-4 (  0) 0) |
Pan, Y., Hu, M., Liang, H., Wang, J. and Tang, L., 2012. The ex-pression of the PIWI family members miwi and mili in mice testis is negatively affected by estrogen. Cell and Tissue Re-search, 350(1): 177-181. DOI:10.1007/s00441-012-1447-z (  0) 0) |
Pandey, R., Homolka, D., Olotu, O., Sachidanandam, R., Kotaja, N. and Pillai, R.S., 2018. Exonuclease domain-containing 1 enhances MIWI2 piRNA biogenesis via its interaction with TDRD12. Cell Reports, 24(13): 3423-3432. DOI:10.1016/j.celrep.2018.08.087 (  0) 0) |
Qiao, D., Zeeman, A., Deng, W., Looijenga, L. and Lin, H., 2002. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to se-minomas. Oncogene, 21(25): 3988-3999. DOI:10.1038/sj.onc.1205505 (  0) 0) |
Rosvall, K.A., Bergeon Burns, C.M., Jayaratna, S.P., Dossey, E.K. and Ketterson, E.D., 2016. Gonads and the evolution of hormonal phenotypes. Integrative and Comparative Biol-ogy, 56(2): 225-234. DOI:10.1093/icb/icw050 (  0) 0) |
Rouget, C., Papin, C., Boureux, A., Meunier, A., Franco, B., Ro-bine, N., Lai, E.C., Pelisson, A. and Simonelig, M., 2010. Maternal mRNA deadenylation and decay by the piRNA path-way in the early Drosophila embryo. Nature, 467(7319): 1128-1132. DOI:10.1038/nature09465 (  0) 0) |
Scholz, S. and Gutzeit, H., 2000. 17-α-ethinylestradiol affects reproduction, sexual differentiation and aromatase gene ex-pression of the medaka (Oryzias latipes). Aquatic Toxicology, 50(4): 363-373. DOI:10.1016/s0166-445x(00)00090-4 (  0) 0) |
Schultz, N., Hamra, F.K. and Garbers, D.L., 2003. A multitude of genes expressed solely in meiotic or postmeiotic sperma-togenic cells offers a myriad of contraceptive targets. Pro-ceedings of the National Academy of Sciences, 100(21): 12201-12206. DOI:10.1073/pnas.1635054100 (  0) 0) |
Schwager, E.E., Meng, Y. and Extavour, C.G., 2015. vasa and piwi are required for mitotic integrity in early embryogenesis in the spider Parasteatoda tepidariorum. Developmental Biol-ogy, 402(2): 276-290. DOI:10.1016/j.ydbio.2014.08.032 (  0) 0) |
Sun, P., You, F., Ma, D., Li, J. and Zhang, P., 2013. Sex steroid changes during temperature-induced gonadal differentiation in Paralichthys olivaceus (Temminck & Schegel, 1846). Jour-nal of Applied Ichthyology, 29(4): 886-890. DOI:10.1111/jai.12128 (  0) 0) |
Sunanaga, T., Inubushi, H. and Kawamura, K., 2010. Piwi-expressing hemoblasts serve as germline stem cells during post-embryonic germ cell specification in colonial ascidian, Botry-llus primigenus. Development, Growth & Differentiation, 52(7): 603-614. DOI:10.1111/j.1440-169X.2010.01196.x (  0) 0) |
Taborska, E., Pasulka, J., Malik, R., Horvat, F., Jenickova, I., Matosevic, Z.J., and Svoboda, B., 2019.Restricted and non-essential redundancy of RNAi and piRNA pathways in mouse oocytes.BioRxiv: 678177, DOI: 10.1371/journal.pgen.1008261.
(  0) 0) |
Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S., 2013. MEGA6:Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2725-2729. DOI:10.1093/molbev/mst197 (  0) 0) |
Thomson, T. and Lin, H., 2009. The biogenesis and function of PIWI proteins and piRNAs:Progress and prospect. Annual Review of Cell and Developmental, 25: 355-376. DOI:10.1146/annurev.cellbio.24.110707.175327 (  0) 0) |
Tian, Y., Chen, S., Xu, T., Deng, H. and Ding, H., 2009. The com-parison of growth performances of Japanese flounder (Para-lichthys olivaceus) families and selection of parents with good trait. Journal of Fisheries of China, 33(6): 901-911. (  0) 0) |
Wang, H., Wang, B., Liu, J., Li, A., Zhu, H., Wang, X. and Zhang, Q., 2018. Piwil1 gene is regulated by hypothalamic-pituitary-gonadal axis in turbot (Scophthalmus maximus):A different effect in ovaries and testes. Gene, 658: 86-95. DOI:10.1016/j.gene.2018.03.016 (  0) 0) |
Wang, H., Wang, B., Liu, X., Liu, Y., Du, X., Zhang, Q. and Wang, X., 2017. Identification and expression of piwil2 in turbot Scophthalmus maximus, with implications of the in-volvement in embryonic and gonadal development. Compa-rative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 208: 84-93. DOI:10.1016/j.cbpb.2017.04.007 (  0) 0) |
Wang, W., Wang, J., You, F., Ma, L., Yang, X., Gao, J., He, Y., Qi, J., Yu, H., Wang, Z., Wang, X., Wu, Z. and Zhang, Q., 2014. Detection of alternative splice and gene duplication by RNA sequencing in Japanese flounder, Paralichthys olivaceus. G3:Genes, Genomes, Genetics, 4(12): 2419-2424. DOI:10.1534/g3.114.012138 (  0) 0) |
Wen, X., Wang, D., Li, X., Zhao, C., Wang, T., Qian, X. and Yin, S., 2018. Differential expression of two Piwil orthologs during embryonic and gonadal development in pufferfish, Ta-kifugu fasciatus. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 219: 44-51. DOI:10.1016/j.cbpb.2018.03.005 (  0) 0) |
Xiao, J., Zhou, Y., Luo, Y., Zhong, H., Huang, Y., Zhang, Y., Luo, Z., Ling, Z., Zhang, M. and Gan, X., 2013. Suppression effect of LHRH-A and hCG on Piwi expression in testis of Nile tilapia Oreochromis niloticus. General and Comparative Endocrinology, 189: 43-50. DOI:10.1016/j.ygcen.2013.04.021 (  0) 0) |
Yamamoto, E., 1999. Studies on sex-manipulation and produc-tion of cloned populations in hirame, Paralichthys olivaceus(Temminck et Schlegel). Aquaculture, 173(1-4): 235-246. DOI:10.1016/S0044-8486(98)00448-7 (  0) 0) |
Zhang, D., Duarte-Guterman, P., Langlois, V.S. and Trudeau, V.L., 2010. Temporal expression and steroidal regulation of piRNA pathway genes (mael, piwi, vasa) during Silurana (Xenopus) tropicalis embryogenesis and early larval development. Com-parative Biochemistry and Physiology Part C:Toxicology & Pharmacology, 152(2): 202-206. DOI:10.1016/j.cbpc.2010.04.005 (  0) 0) |
Zhang, L., Liu, W., Shao, C., Ning, Z., Li, H., Liu, K., Dong, Z., Qi, Q., Zhao, W. and Chen, S., 2014. Cloning, expression and methylation analysis of piwil2 in half-smooth tongue sole (Cy-noglossus semilaevis). Marine Genomics, 18: 45-54. DOI:10.1016/j.margen.2014.04.004 (  0) 0) |
Zhao, C., Zhu, W., Yin, S., Cao, Q., Zhang, H., Wen, X., Zhang, G., Xie, W. and Chen, S., 2018. Molecular characterization and expression of Piwil1 and Piwil2 during gonadal develop-ment and treatment with HCG and LHRH-A2 in Odontobutis potamophila. Gene, 647: 181-191. DOI:10.1016/j.gene.2018.01.038 (  0) 0) |
Zhao, H., Duan, J., Cheng, N. and Nagahama, Y., 2012. Speci-fic expression of Olpiwi1 and Olpiwi2 in medaka (Oryzias la-tipes) germ cells. Biochemical and Biophysical Research Com-munications, 418(4): 592-597. DOI:10.1016/j.bbrc.2011.12.062 (  0) 0) |
Zhou, Y., Hao, G., Zhong, H., Wu, Q., Lu, S., Zhao, Q. and Liu, Z., 2015. Human chorionic gonadotropin promotes expression of protein absorption factors in the intestine of goldfish (Ca-rassius auratus). Genetics and Molecular Research, 14(3): 8306-8313. DOI:10.4238/2015.July.27.19 (  0) 0) |
Zhou, Y., Wang, F., Liu, S., Zhong, H., Liu, Z., Tao, M., Zhang, C. and Liu, Y., 2012. Human chorionic gonadotropin sup-presses expression of Piwis in common carp (Cyprinus carpio) ovaries. General and Comparative Endocrinology, 176(2): 126-131. DOI:10.1016/j.ygcen.2011.11.044 (  0) 0) |
Zhou, Y., Zhong, H., Liu, S., Yu, F., Hu, J., Zhang, C., Tao, M. and Liu, Y., 2014. Elevated expression of Piwi and piRNAs in ovaries of triploid crucian carp. Molecular and Cellular En-docrinology, 383(1-2): 1-9. DOI:10.1016/j.mce.2013.11.019 (  0) 0) |
 2020, Vol. 19
2020, Vol. 19


