2) Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, 266237, China
The quantitative real-time reverse transcription PCR (qRT-PCR) is a popularized technique for quantification of relative mRNA levels due to the advantage of its low cost, specificity, sensitivity and large dynamic range (Huggett et al., 2005). In qRT-PCR, the expression levels of target genes are usually normalized to the endogenous controls, also called reference genes. The purpose of using reference genes is to remove or reduce the variations in the quantity and quality of total RNA among samples, thus making it possible for accurate comparison of transcript abundance of a gene of interest among various samples (Dheda et al., 2004; Zhu et al., 2008). Selecting an ideal reference gene that showing constant expression across all samples is an essential step for normalization and quantitative analysis in qRT-PCR. Though a suitable reference gene is very important, many studies are using commonly used reference genes without validation in qRT-PCR as-says. Some commonly used reference genes were confirmed to be unsuitable in some conditions (Zhu et al., 2008). For example, GAPDH, a widely used reference gene in human, has been reported to be an unsuitable reference gene in some other species (Long et al., 2010; Ghani et al., 2013), and so are many other traditionally reference genes, such as 18S rRNA and ACTB (Olsvik et al., 2005).
There have been many emerging studies to identify and validate reliable reference genes with various methods following the development of biotechnology. For example, serial analysis of gene expression, microarray analysis, and expressed tag sequences have been widely used for reference genes identification in human and other mammals (Velculescu et al., 1999; Zhu et al., 2008; Eisenberg and Levanon 2013); however, few similar studies have been conducted in other species. In recent years, RNA-seq has become a useful experimental tool to identify the genes with stable expression patterns. Many studies were carried out utilizing RNA-seq for reference genes selection not only in model organisms, but also in many non-model organisms, like kiwi, seaweed, and scallop (Gao et al., 2018; Liu et al., 2018; Li et al., 2019).
The black rockfish (S. schlegelii, hereafter denoted as 'rockfish') is a good model of teleost to study the adaption to viviparity (He et al., 2019). Some studies have been carried out focusing on the genes related to development, growth, immunity, and reproduction in this group of fish (Ma et al., 2014; Kugapreethan et al., 2017; Kim and Cho, 2019). Genes 18S rRNA, ACTB, and GADPH are usually adopted as the endogenous control of qRT-PCR in rockfish studies. A previous study of our lab compared the stabilities of several commonly used reference genes (18S rRNA, ACTB, GAPDH, TUBA, RPL17, EF1A, HPRT, and B2M) and defined RPL17, EF1A and ACTB as the most reliable three ones (Liman et al., 2013). However, some inconsistent results are still observed even we used the most stable reference gene in rockfish studies. The most stable reference gene usually is selected by the results in adult tissue, it is not confirmed whether it is still stable during early developmental stages. It is the best way to study as many samples as possible to select the reference genes. The availability of a chromosome-level genome and a large set of transcriptomes of rockfish (He et al., 2019) provide valuable resources to conduct the transcriptomewide analysis of suitable reference genes selection.
In this study, we analyzed 89 transcriptome datasets, and 121 candidate reference genes were identified based on four criteria. The expression stabilities of eight candidates and four commonly used genes were validated by qRT-PCR and four statistical methods. The results revealed that candidates we recommended are more stable than traditionally used reference genes. This will benefit further gene quantification analysis of the rockfish and other teleost species.
2 Materials and Methods 2.1 Sample CollectionAll adult rockfish in this study were obtained from the fish hatchery of Qingdao Beibao Aquatic Co., Ltd., Shandong, China. They were transported to our laboratory and maintained in aerated seawater for acclimation. Then, they were sacrificed and the following tissues were dissected: heart, liver, spleen, kidney, brain, gill, intestine, testis and ovary. All samples were frozen in liquid nitrogen and stored at −80℃ for further RNA extraction. Each tissue was collected in triplicate or more.
Embryos at six different stages (1-cell, 32-cell, blastula, gastrula, somite and pre-hatching) were collected as described in our previous study (He et al., 2019). Then, they were stored at −80℃ for further analyses.
2.2 RNA-Seq DatasetsTotally 89 RNA-libraries, including 63 from different tissues and 26 from six developmental stages (1-cell, 32-cell, blastula, gastrula, somite and pre-hatching), were selected with three biological replicates or more. In each gene 100 bp from each end was sequenced using BGI-500 platform. Then, the transcriptome data were processed and the raw counts were converted into TPM (Transcripts per million) values using the RNA-seq by Expectation Maximization software package (Li and Dewey, 2011).
2.3 Identification of Reference Genes Using Transcriptome DataModified methods as described by Li et al. (2019) from the method of Eisenberg and Levanon (2013) were used to select reference genes among all tissues and at different early developmental stages of rockfish. Briefly, four criteria utilized to screen reference genes were as follows: 1) expression could be detected in all tissues and developmental stages of embryo; 2) low dispersion degree over all tissues and developmental stages by requiring log2 (TPM) < 1; 3) no log2 (TPM) differed from the mean log2 (TPM) by two or more in each tissue and developmental stage regarding expression level; 4) mean log2 (TPM) > 6, where 6 was the median of log2 (TPM) values of all genes in all tissues and developmental stages. As described in Li et al. (2019), the stability of reference genes was further evaluated according to the CV value (stdev/mean). Based on the criteria mentioned above, mean log2 (TPM) > 6 and dispersion degree log2 (TPM) < 1, the CV values of reference genes we identified are always less than 0.17.
2.4 RNA Extraction and cDNA SynthesisTotal RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer's protocol. After RNA purification, the integrity and concentration of RNA extracted was assessed via 1.5% agarose gel electrophoresis and determined by a Nano photometer. The cDNA was synthesized with Reverse Transcriptase M-MLV kit (Takara) and the final volume was 20 μL. The cDNA products were stored at −20℃ for further analyses.
2.5 Primer Design and qRT-PCRIn total, we selected ten candidate reference genes from transcriptome data, and four commonly used reference genes for further qRT-PCR validation. Primers of these genes were designed by Primer 5.0 software. The criteria of primer design were as follows: amplicon lengths of 80-200 bp, annealing temperature of 60-63℃, primer lengths of 18-25 bp, and GC content of 40%-60%. The information of primers is listed in Table 1. All primers amplification efficiencies were determined by the standard curve generated from two-fold dilution series of cDNA concentration (0.625-20 ng μL−1).
|
|
Table 1 List of primers used for qRT-PCR analysis |
qRT-PCR was conducted in a 384-well plate using a Light Cycler 480 RT-PCR system (Roche Molecular Biochemical, Mannheim, Germany). Each reaction contained 2 μL cDNA (5 ng μL−1), 10 μL Light Cycler 480 SYBR Green I Master, 0.5 μL of each primer and 7 μL ddH2O. Three biological replicates were required for each sample, and three technical replicates were performed for each biological replicate. RT-PCR cycling conditions were 95℃ for 5 min, followed by 45 cycles at 95℃ for 15 s, and 61℃ for 45 s. The specificity of each primer was determined by melting curve and gel electrophoresis.
2.6 qRT-PCR Data AnalysisThe stabilities of reference genes determined in our study were evaluated by ReFinder (https://www.heartcure.com.au/reffinder/) (Liu et al., 2014), which integrates four commonly used statistical approaches: geNorm, NormFinder, Bestkeeper, and comparative delt-Ct method (Vandesompele et al., 2002; Andersen et al., 2004; Silver et al., 2006). Then the geometric mean was calculated to guarantee a comprehensive analysis (Chen et al., 2011).
3 Results 3.1 Selection of the Reference Genes from the Transcriptome Data of RockfishWe identified rockfish candidate reference genes among 24094 transcripts from 89 RNA-seq datasets, which are available at CNSA (CNGB Nucleotide Sequence Archive) under the accession ID CNP0000222. Firstly, we obtained 8357 (34.7%), 439 (2%), 242 (1%) genes respectively, according to the first three criteria (Fig. 1). Then, we figured out the median log2 (TPM) value among genes that met the first three criteria was approximately six. Finally, the fourth criteria were applied to screen genes with an average log2 (TPM) > 6, resulting in 121 candidate reference genes for all tissues and early developmental stages in rockfish (Fig. 1). As shown in Table 2, the 15 most stable genes have mean log2 (TPM) values ranging from 6.33 to 11.00, and the lowest CV values vary from 0.069 to 0.093.
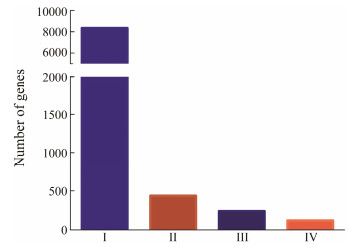
|
Fig. 1 The number of genes that met our fourth criteria. (Ⅰ) TPM > 0; (Ⅱ) standard-deviation log2 (TPM) < 1; (Ⅲ) no log2 (TPM) differed from the mean log2 (TPM) by two or more; (Ⅳ) mean log2 (TPM) > 6. |
|
|
Table 2 The top 15 candidate reference genes identified from the 89 transcriptome datasets of S. schlegeli |
To validate the candidate reference genes we identified, eight genes were chosen from the top 15 candidates and four commonly used were selected to compare their expression profiles. The eight candidates include METAP2, EIF5A1, BTF3L4, TPT1, UBC, RAB10, DLD and PAIRB, the four commonly used reference genes include TUBA, ACTB, RPL17 and GAPDH (Table 3).
|
|
Table 3 Detailed information on the 8 selected candidate reference genes and the 4 commonly used reference genes |
Considering log2 (TPM) values, candidate reference genes have lower variances than commonly used reference genes as shown in Fig. 2. For example, GAPDH is a widely used reference genes in mammals, while its log2 (TPM) values (ranging from 2 to 14) exhibit high variances in rockfish tissues and developmental stages. Additionally, RPL17 and ACTB also show relatively high variances. In contrast to these commonly used reference genes, candidates identified from the transcriptome datasets have smaller variances in log2 (TPM) values among different tissues and developmental stages. Our transcriptome datasets suggest the candidate reference genes identified are more stable than commonly used reference genes in rockfish tissues and early developmental stages.
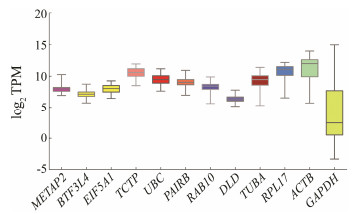
|
Fig. 2 Evaluation of the reference gene candidates and commonly used reference genes based on RNA-seq analysis. A boxplot showing the log2 (TPM) values of the 8 candidate reference genes and 4 commonly used reference genes among tissues and developmental stages. |
To validate the results from our transcriptome datasets, we conducted the qRT-PCR of twelve genes and four methods (comparative delta-Ct, geNorm, Bestkeeper, and Norm-Finder) were used for further expression stability evaluation. We used the Pearson coefficient to determine the correlation between Ct values and log2 (TPM) values, and there is a negative correlation between these two values (R = −0.636, P < 0.01).
As shown in Fig. 3, the Ct values of each gene in different tissues and developmental stages were determined. Among commonly used reference genes, GAPDH displays the highest variation of Ct values (17-32), and RPL17 (Ct 21-29) also expresses variably in different tissues and developmental stages. TUBA and ACTB have relative stable Ct values. In contrast to commonly used genes, most candidates have smaller variances than commonly used reference genes. For example, METAP2 (Ct 22-25), RAB10 (25-27), EIF5A1 (24-27) display very low expression variations. According to the results of qRT-PCR, Ct values of most candidates are relatively more stable than commonly used reference genes, consisting with our transcriptome data.
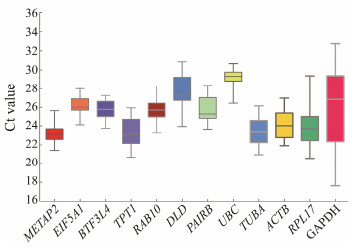
|
Fig. 3 Evaluation of the reference genes based on qRT-PCR analysis. A boxplot shows the Ct values of the 8 candidate reference genes and 4 commonly used reference genes in different tissues and at different early developmental stages. |
After having summarized the qRT-PCR results, it's necessary to compare the expression stability between the candidates and commonly used reference genes by the statistical methods for determining the optimal reference genes in further normalization. Here, we applied the four statistical methods for stability analysis: delta-Ct, Best-Keeper, geNorm and NormFinder (Fig. 4).
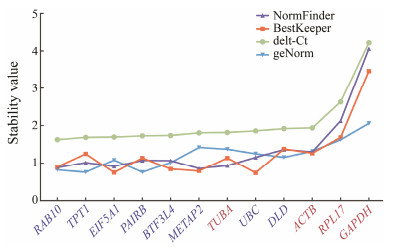
|
Fig. 4 Expression stability of the twelve genes based on qRT-PCR experiments. The stability was evaluated based on geNorm, NormFinder and comparative delta-Ct analyses of the qRT-PCR data. |
According to the delta-Ct method, RAB10 was identified as the most stable reference gene with the highest ranking value. UBC, METAP2, TPT1/PAIRB were most stable genes by the BestKeeper, NormFinder, geNorm methods respectively. In consistent with our observation of CV values and Ct Values, GAPDH and RPL17 showed the lowest ranking values among twelve genes by four methods. Although the stability rankings of these genes are variable by different methods, the candidates selected from transcriptome data tend to have higher ranking values compared to the commonly used reference genes. The result of stability analysis with different statistical analysis methods further confirmed the reliability of candidates selected based on transcriptome data.
4 DiscussionThe inappropriate reference genes for normalizing transcription levels can cause systematic errors in qRT-PCR
analysis, which can cause wrong interpretation of results. Thus selecting optimal reference genes is very important in gene quantitative analysis under different experimental conditions. The same reference genes can have different expression stability in different species, especially between mammals and lower vertebrates like fish. For example, GAPDH has been widely identified and used as a housekeeping gene in mammals (Sun et al., 2012; Niu et al., 2016), but it has a significant expression variation in Atlantic halibut and zebrafish (Tang et al., 2007; Fernandes et al., 2008). However, most reference genes for qRT-PCR in fishes are selected from commonly used reference genes in mammals, which can lead to unnecessary errors in gene quantitative analysis. With the wide application of omics in molecular biology, transcriptomic data have been becoming a reliable source for the selection of optimal reference genes for gene quantitative analysis (Yang et al., 2014; Fu et al., 2015; Gao et al., 2018).
In this study, we modified a method developed by Eisenberg and Levanon (2013) to select suitable reference genes in rockfish. The reasonability and advantage of the method were introduced in detail in a previous study (Li et al., 2019). We aimed to screen reference genes with high stability among different tissues and early developmental stages, so we integrated the transcriptome data of different adult tissues and developmental stages. With four criteria, only 121 genes with CV values < 1 were selected from all 24094 genes, suggesting the method we used is very stringent. Furthermore, we analyzed the transcriptome data of tissues and early developmental stages respectively by four criteria. Interestingly, 375 and 752 genes were obtained respectively, suggesting a significant difference in gene expression patterns between tissues and developmental stages, which is similar to some other studies (Carlyle et al., 1996; Jorgensen et al., 2006; Liu et al., 2014). After comparing two gene sets obtained from tissues and developmental stages, we obtained a core set of 122 genes, and most of them are overlapped with the candidate reference genes from 121 gene sets, confirming the stringency and reliability of the methods we applied.
Top 15 candidate reference genes with the lowest CV values are all annotated proteins, and most of them have been widely studied in model organisms. For example, UBC has been identified as a reliable reference gene in some organisms (Mosley and HogenEsch, 2017; Wang et al., 2017; Zhao et al., 2018). From these 15 genes, 8 genes were selected for further study to detect the reliability of our method.
The results of RNA-seq analysis of eight top candidates and four commonly used reference genes were validated by qRT-PCR, and four statistical methods were used to evaluate their expression stabilities. The stability ranking values of most reference genes obtained from log2 (TPM) values and Ct values are consistent in samples used for qRT-PCR and RNA-seq. Furthermore, candidate reference genes showed higher stabilities than those commonly used reference genes. ACTB and GAPDH are the least stable genes among twelve genes, which is consistent with a previous study in rockfish, and they have been widely described to be unsuitable for gene quantitative analysis in some species (Jorgensen et al., 2006; De Jonge et al., 2007; Du et al., 2013).
Intriguingly, TUBA gets a medium ranking value by qRT-PCR, showing it is more stable than the other three commonly used reference genes. However, in the process of screening reference genes based on four criteria, TUBA was filtered out during the second step for screening genes with low dispersion degree over all tissues and developmental stages (log2 (TPM) < 1) (The standard deviation value < 1). The CV of TUBA is 0.131. Considering these two results, we don't think TUBA is a perfect reference gene in rockfish, since it is not like the candidates we recommended, such as RAB10, EIF5A1, PAIRB and BTF3L4, which show the high stability ranking values evaluated both from RNA-seq and qRT-PCR. In non-model organisms, especially in teleosts, the reference genes used for normalization in qRT-PCR experiments usually are from mammals, like GAPDH, ACTB and TUBA. Their unstable expressions under different conditions may cause misleading results during normalization. The reference genes identified in this study can be widely applied in teleost studies. The results of this study are from the black rockfish cultured under general conditions. When the fish are cultured in an environment with different stresses, whether the expressions of these genes are still stable need to be studied carefully.
5 ConclusionsIn this study, 89 transcriptome datasets of rockfish were employed to identify reference genes, and 121 candidate reference genes were selected from the transcriptomes based on four criteria. Combing RNA-seq and qRT-PCR assays, the expression stabilities of candidates we identified were higher than commonly used reference genes. RAB10, EIF5A1, PAIRB and BTF3L4 might be the best reference genes in rockfish. Our studies provide valuable resources for further gene expression analysis in teleost.
AbbreviationsMETAP2, Methionine aminopeptidase 2; BTF3L4, Transcription factor BTF3 homolog 4; EIF5A1, Eukaryotic translation initiation factor 5A-1; TCTP, Translationally-controlled tumor protein; UBC, Ubiquitin C; PAIRB, Plasminogen activator inhibitor 1 RNA-binding protein; RAB10, Ras-related protein Rab-10; DLD, Dihydrolipoamide dehydrogenase; TUBA, α-Tubulin; RPL17, Ribosomal protein L17; ACTB, β-actin; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; qRT-PCR, Quantitative reverse transcription PCR; TPM, Transcripts per million.
AcknowledgementThis study was supported by the National Key R & D Program of China (No. 2018YFD0900101).
Andersen, C. L., Jensen, J. L. and Ørntoft, T. F., 2004. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64(15): 5245-5250. DOI:10.1158/0008-5472.CAN-04-0496 (  0) 0) |
Carlyle, W. C., Cynthia, A. T., Jonathan, R. V., Kenneth, M. M., David, C. H. and Jay, N. C., 1996. Changes in β-actin mRNA expression in remodeling canine myocardium. Journal of Molecular and Cellular Cardiology, 28(1): 53-63. DOI:10.1006/jmcc.1996.0006 (  0) 0) |
Chen, D., Pan, X., Peng, X., Mary, A. F. and Zhang, B., 2011. Evaluation and identification of reliable reference genes for pharmacogenomics, toxicogenomics, and small RNA expression analysis. Journal of Cellular Physiology, 226(10): 2469-2477. DOI:10.1002/jcp.22725 (  0) 0) |
de Jonge, H. J., Fehrmann, R. S., de Bont, E. S., Hofstra, R. M., Gerbens, F., Kamps, W. A., de Vries, E. G., van der Zee, A. G., te Meerman, G. J. and ter Elst, A., 2007. Evidence based selection of housekeeping genes. PLoS One, 2(9): e898. DOI:10.1371/journal.pone.0000898 (  0) 0) |
Dheda, K., Huggett, J. F., Bustin, S. A., Johnson, M. A., Rook, G. and Zumla, A., 2004. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques, 37(1): 112-119. DOI:10.2144/04371RR03 (  0) 0) |
Du, Y., Zhang, L., Xu, F., Huang, B., Zhang, G. and Li, L., 2013. Validation of housekeeping genes as internal controls for studying gene expression during Pacific oyster (Crassostrea gigas) development by quantitative real-time PCR. Fish & Shell-fish Immunology, 34(3): 939-945. (  0) 0) |
Eisenberg, E. and Levanon, E. Y., 2013. Human housekeeping genes, revisited. TRENDS in Genetics, 29(10): 569-574. DOI:10.1016/j.tig.2013.05.010 (  0) 0) |
Fernandes, J. M., Mommens, M., Hagen, O., Babiak, I. and Solberg, C., 2008. Selection of suitable reference genes for realtime PCR studies of Atlantic halibut development. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 150(1): 23-32. DOI:10.1016/j.cbpb.2008.01.003 (  0) 0) |
Fu, Y., He, W., Wang, L. and Wei, Y., 2015. Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Frontiers in Plant Science, 6: 475. (  0) 0) |
Gao, D., Kong, F., Sun, P., Bi, G. and Mao, Y., 2018. Transcrip-tome-wide identification of optimal reference genes for expression analysis of Pyropia yezoensis responses to abiotic stress. BMC Genomics, 19(1): 251. DOI:10.1186/s12864-018-4643-8 (  0) 0) |
Ghani, M., Sato, C. and Rogaeva, E., 2013. Segmental duplications in genome-wide significant loci and housekeeping genes; Warning for GAPDH and ACTB. Neurobiology of Aging, 34(6): 1710. (  0) 0) |
He, Y., Chang, Y., Bao, L., Yu, M., Li, R., Niu, J., Fan, G., Song, W., Seim, I. and Qin, Y., 2019. A chromosome-level genome of black rockfish, Sebastes schlegelii, provides insights into the evolution of live birth. Molecular Ecology Resources, 19: 1309-1321. DOI:10.1111/1755-0998.13034 (  0) 0) |
Huggett, J. F., Dheda, K., Bustin, S. A. and Zumla, A., 2005. Realtime RT-PCR normalisation; strategies and considerations. Genes and Immunity, 6(4): 279. DOI:10.1038/sj.gene.6364190 (  0) 0) |
Jorgensen, S. M., Kleveland, E. J., Grimholt, U. and Gjoen, T., 2006. Validation of reference genes for real-time polymerase chain reaction studies in Atlantic salmon. Marine Biotechnology, 8(4): 398-408. DOI:10.1007/s10126-005-5164-4 (  0) 0) |
Kim, H. S. and Cho, S. H., 2019. Dietary inclusion effect of feed ingredients showing high feeding attractiveness to rockfish (Sebastes schlegeli Hilgendorf 1880) on the growth performance, feed utilization, condition factor and whole body composition of fish (Ⅱ). Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 231(2019): 66-73. (  0) 0) |
Kugapreethan, R., Umasuthan, N., Wan, Q., Thulasitha, W. S., Kim, C. and Lee, J., 2017. Comparative analysis of two thioredoxin-like genes in black rockfish Sebastes schlegelii and their possible involvement in redox homeostasis and innate immune responses. Developmental & Comparative Immunology, 67: 43-56. (  0) 0) |
Li, B. and Dewey, C. N., 2011. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12(1): 323. DOI:10.1186/1471-2105-12-323 (  0) 0) |
Li, Y., Zhang, L., Li, R., Zhang, M., Li, Y., Wang, H., Wang, S. and Bao, Z., 2019. Systematic identification and validation of the reference genes from 60 RNA-Seq libraries in the scallop Mizuhopecten yessoensis. BMC Genomics, 20(1): 288. DOI:10.1186/s12864-019-5661-x (  0) 0) |
Ma, L., Wang, W., Liu, C., Yu, H., Wang, Z., Wang, X., Qi, J. and Zhang, Q., 2013. Selection of reference genes for reverse transcription quantitative real-time PCR normalization in black rockfish (Sebastes schlegeli). Marine Genomics, 11: 67-73. DOI:10.1016/j.margen.2013.08.002 (  0) 0) |
Lin, F., Jiang, L., Liu, Y., Lv, Y., Dai, H. and Zhao, H., 2014. Genome-wide identification of housekeeping genes in maize. Plant Molecular Biology, 86(4-5): 543-554. DOI:10.1007/s11103-014-0246-1 (  0) 0) |
Liu, C., Xin, N., Zhai, Y., Jiang, L., Zhai, M., Zhang, Q. and Qi, J., 2014. Reference gene selection for quantitative real-time RT-PCR normalization in the half-smooth tongue sole (Cynoglossus semilaevis) at different developmental stages, in various tissue types and on exposure to chemicals. PLoS One, 9(3): e91715. DOI:10.1371/journal.pone.0091715 (  0) 0) |
Liu, J., Huang, S., Niu, X., Chen, D., Chen, Q., Tian, L., Xiao, F. and Liu, Y., 2018. Genome-wide identification and validation of new reference genes for transcript normalization in developmental and post-harvested fruits of Actinidia chinensis. Gene, 645: 1-6. DOI:10.1016/j.gene.2017.12.012 (  0) 0) |
Long, X., Wang, J., Ouellet, T., Rocheleau, H., Wei, Y., Pu, Z., Jiang, T., Lan, X. and Zheng, Y., 2010. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Molecular Biology, 74(3): 307-311. DOI:10.1007/s11103-010-9666-8 (  0) 0) |
Ma, L., Wang, W., Yang, X., Jiang, J., Song, H., Jiang, H., Zhang, Q. and Qi, J., 2014. Characterization of the Dmrt1 gene in the black rockfish Sebastes schlegeli revealed a remarkable sex-dimorphic expression. Fish Physiology and Biochemistry, 40(4): 1263-1274. (  0) 0) |
Mosley, Y. C. and HogenEsch, H., 2017. Selection of a suitable reference gene for quantitative gene expression in mouse lymph nodes after vaccination. BMC Research Notes, 10(1): 689. DOI:10.1186/s13104-017-3005-y (  0) 0) |
Niu, G., Yang, Y., Zhang, Y., Hua, C., Wang, Z., Tang, Z. and Li, K., 2016. Identifying suitable reference genes for gene expression analysis in developing skeletal muscle in pigs. PeerJ, 4: e2428. DOI:10.7717/peerj.2428 (  0) 0) |
Olsvik, P. A., Lie, K. K., Jordal, A. E., Nilsen, T. O. and Hordvik, I., 2005. Evaluation of potential reference genes in realtime RT-PCR studies of Atlantic salmon. BMC Molecular Biology, 6(1): 21. DOI:10.1186/1471-2199-6-21 (  0) 0) |
She, X., Rohl, C. A., Castle, J. C., Kulkarni, A. V., Johnson, J. M. and Chen, R., 2009. Definition, conservation and epigenetics of housekeeping and tissue-enriched genes. BMC Genomics, 10(1): 269. DOI:10.1186/1471-2164-10-269 (  0) 0) |
Silver, N., Best, S., Jiang, J. and Thein, S. L., 2006. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology, 7(1): 33. DOI:10.1186/1471-2199-7-33 (  0) 0) |
Sun, Y., Li, Y., Luo, D. and Liao, D. J., 2012. Pseudogenes as weaknesses of ACTB (Actb) and GAPDH (Gapdh) used as reference genes in reverse transcription and polymerase chain reactions. PLoS One, 7(8): e41659. DOI:10.1371/journal.pone.0041659 (  0) 0) |
Tang, R., Dodd, A., Lai, D., McNabb, W. C. and Love, D. R., 2007. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochimica et Biophysica Sinica, 39(5): 384-390. DOI:10.1111/j.1745-7270.2007.00283.x (  0) 0) |
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A. and Speleman, F., 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7): research0034.1. (  0) 0) |
Velculescu, V. E., Madden, S. L., Zhang, L., Lash, A. E., Yu, J., Rago, C., Lal, A., Wang, C. J., Beaudry, G. A. and Ciriello, K. M., 1999. Analysis of human transcriptomes. Nature Genetics, 23(4): 387. (  0) 0) |
Wang, M., Li, Q., Xin, H., Chen, X., Zhu, X. and Li, X., 2017. Reliable reference genes for normalization of gene expression data in tea plants (Camellia sinensis) exposed to metal stresses. PLoS One, 12(4): e0175863. DOI:10.1371/journal.pone.0175863 (  0) 0) |
Yang, H., Liu, J., Huang, S., Guo, T., Deng, L. and Hua, W., 2014. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene, 538(1): 113-122. DOI:10.1016/j.gene.2013.12.057 (  0) 0) |
Zeng, J., Liu, S., Zhao, Y., Tan, X., Aljohi, H. A., Liu, W. and Hu, S., 2016. Identification and analysis of house-keeping and tissue-specific genes based on RNA-seq data sets across 15 mouse tissues. Gene, 576(1): 560-570. DOI:10.1016/j.gene.2015.11.003 (  0) 0) |
Zhao, X., Yang, H., Chen, M., Song, X., Yu, C., Zhao, Y. and Wu, Y., 2018. Reference gene selection for quantitative realtime PCR of mycelia from Lentinula edodes under high-temperature stress. BioMed Research International. DOI:10.1155/2018/1670328 (  0) 0) |
Zhu, J., He, F., Song, S., Wang, J. and Yu, J., 2008. How many human genes can be defined as housekeeping with current expression data?. BMC Genomics, 9(1): 172. DOI:10.1186/1471-2164-9-172 (  0) 0) |
 2021, Vol. 20
2021, Vol. 20


