2) Laboratory for Marine Fisheries Science and Food Production Processes, Laoshan Laboratory, Qingdao 266237, China
Carotenoids are lipid-soluble, tetra-terpenoid organic pigments and are one of the most common pigments found in nature (Maoka, 2020). There is a wide variety of carotenoids, with about 1100 species reported, including 850 naturally occurring carotenoids (Yabuzaki, 2017; Maoka, 2020). Organisms capable of photosynthesis have the ability to synthesize carotenoids de novo. Together with chlorophyll, carotenoids play an indispensable role as pigments in photosynthetic organs. In animals, carotenoids cannot be synthesized de novo; therefore, they are either directly accumulated from diet or are partially modified by reactions of metabolism (Moran et al., 2010). Recent researches have demonstrated the essential functions of carotenoids in a range of physiological processes, encompassing gene expression, proliferation and differentiation, signaling and cell communication, antioxidant defense mechanisms as well as immune response. These functions are attributed to both the intrinsic properties of carotenoids themselves and the vitamin A-like metabolites generated during their metabolism (Karadas et al., 2005; Maoka, 2020).
Aquatic animals acquire carotenoids from sources such as algae and other organisms, and modify them through metabolic reactions (Maoka, 2020). Carotenoids are liberated from the food matrix upon ingestion by animals, incorporated into mixed microaggregates and absorbed in the intestine, subsequently undergoing transport, transformation, lysis or deposition within cells. These processes are affected by diverse exogenous elements, such as dietary, fluctuations in seasonality (Olsen et al., 2005; Chimsung et al., 2013) as well as photoperiod (Tan et al., 2021). Furthermore, inheritance is crucial in the metabolic mechanism of carotenoids (Toews et al., 2017).
MicroRNAs (miRNAs) perform pivotal functions in various physiological processes like pigmentation, immune response, and metabolism, across both plant and animal kingdoms. In the Pacific oyster Crassostrea gigas, miRNAs perform an essential regulatory function in pigmentation and are involved in the formation of shell color across various shellfish species (Feng et al., 2020). In Dunaliella salina, miRNAs are involved in the accumulation and metabolic processes of β-carotene. Currently, research on the digestion, absorption, transport, storage and metabolic transformation of shellfish β-carotene mainly focus on gene function (Li et al., 2010; Liu et al., 2015; Wan et al., 2022a, 2022b). Limited information is available regarding the function of miRNAs in β-carotene metabolism.
Bivalves are the second largest group of mollusks that live as filter feeders, and their diet consists of carotenoidrich microalgae in seawater (Maoka, 2009, 2011). Bivalves have the ability to absorb, accumulate and metabolize carotenoids and their derivatives from microalgae, while they can adapt to the complex and variable conditions of carotenoid supply in the environment (Maoka, 2020). Therefore, they serve as an excellent model for studying the mechanism underlying carotenoid metabolism. The Pacific oyster is the most productive bivalve shellfish with both significant ecological and economic values. Its soft body, especially the digestive gland, is a rich source of carotenoids that are beneficial for human health (Maoka, 2011). Therefore, a comprehensive investigation of the carotenoid metabolism mechanism of Pacific oyster can establish a theoretical foundation for further exploitation of its carotenoid-rich traits, which holds significant potential in enhancing the quality of Pacific oyster. Previous transcriptome sequencing has identified several genes related to carotenoid transport and storage such as apolipoproteins (Wan et al., 2022b). However, whether miRNAs are participating in regulating their differential expression is unclear.
In this study, we performed microRNA sequencing in order to investigate the regulatory mechanisms of carotenoid metabolism in Pacific oysters. Differences in miRNA profiles in the digestive glands of oysters fed with or without carotenoids were analyzed. Based on the available genomic information, both known and novel miRNAs were identified. The ceRNA networks were constructed to identify and analyze miRNAs related to carotenoid processing as well as their target genes by integrating miRNA and mRNA data. These results facilitate a better insight into the metabolic role of carotenoids in Pacific oysters.
2 Materials and Methods 2.1 AnimalsIn this study, the experimental component was consistent with previous research (Wan et al., 2022b). Briefly, the treatment groups of oysters were fed β-carotene (caro) while the control group (ctrl) did not receive any supplementation. After 30 days of treatment, digestive gland tissues were collected for total RNA extraction. The treatment and control groups consisted of three biological replicates and each was formed by mixing digestive gland tissues from three Pacific oysters.
2.2 miRNA Library Construction and SequencingTotal RNA was extracted from the digestive gland of Pacific oysters using the TRIzol (Invitrogen, USA). RNA purity and integrity were estimated using a 1% agarose gel and the concentration of RNA was measured by utilizing Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA).
Each sample was prepared with 3 μg of total RNA, and sequencing libraries were prepared using the NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, USA) following the manufacturer's instructions. PCR amplification was carried out utilizing LongAmp Taq 2× Master Mix, SR Primer for Illumina and index (X) primer. DNA fragments with a length of 140 – 160 bp (including the small noncoding RNA plus the 3' and 5' adaptors) were isolated and dissolved in 8 μL elution buffer. Single-end reads of 50 bp in length were generated using the Illumina Hiseq 2500/2000 platform.
2.3 Data ProcessingThe raw data were quality-checked using fastqc (0.12.1) and were subsequently filtered with trim_galore (0.6.10) software to remove adapter sequences and retain 18 – 30 bp fragments for downstream analyses. The sRNA reads were aligned to the reference (GCA_902806645.1) utilizing Bowtie (Langmead et al., 2009) without mismatch to analyze the expression and distribution on the reference.
2.4 Known miRNA AlignmentFirstly, mature miRNAs and precursor miRNAs from all animals in the miRBase 20.0 database were utilized as reference sequences for Pacific oyster miRNAs. Bowtie software was then used for map reads, which could align the Pacific oyster genome to the miRNA reference sequences to identify mature miRNA sequences specific to the Pacific oyster.
2.5 Novel miRNA PredictionPrediction of novel miRNA in Pacific oysters was carried out by utilizing mirdeep2 software with default parameters (Friedlander et al., 2012). The obtained alignments and annotations were summarized, taking into account the possibility of sRNA reads being aligned to multiple categories during the alignment and annotation process. To ensure each unique small RNA is mapped to a single annotation, we adhere to the following priority order: known miRNA > rRNA > tRNA > snRNA > snoRNA > repeat > exon > intron. The overall proportion of rRNA serves as an indicator of sample quality.
The quantifier.pl program from the mirdeep2 package was utilized for miRNA quantification, calculation of count value, and subsequent normalization. Standardization of miRNA expression levels were estimated by TPM (transcript per million) (Zhou et al., 2010).
miRanda (Enright et al., 2003), RNAhybrid (Kruger et al., 2006) and targetscan (McGeary et al., 2019) were employed to predict the target genes of differentially expressed miRNAs (DE-miRNA). The parameters for miRanda were set as -en -10, while those for RNAhybrid were set as -f 2, 8 -m 10000, -v 3, -e -20. Default parameters were used for targetscan.
2.6 GO and KEGG Enrichment AnalysisThe target gene candidates of DE-miRNAs were subjected to Gene Ontology (GO) enrichment analysis. GO-seq based Wallenius non-central hyper-geometric distribution (Young et al., 2010), which could adjust gene length bias, was implemented for GO enrichment analysis.
KEGG pathway enrichment analysis was conducted for miRNA target genes byKOBAS software (Mao et al., 2005) and P < 0.05 were considered significantly enriched.
2.7 Construction of miRNA-mRNA NetworkThe raw transcriptome data from the same experiments were analyzed to identify differentially expressed genes (DEGs) (Wan et al., 2022b). Subsequently, the DE-miRNA and DEGs interaction network was constructed using Cytoscape software.
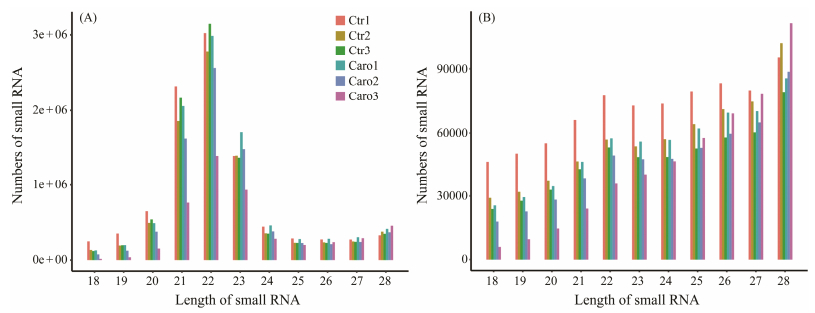
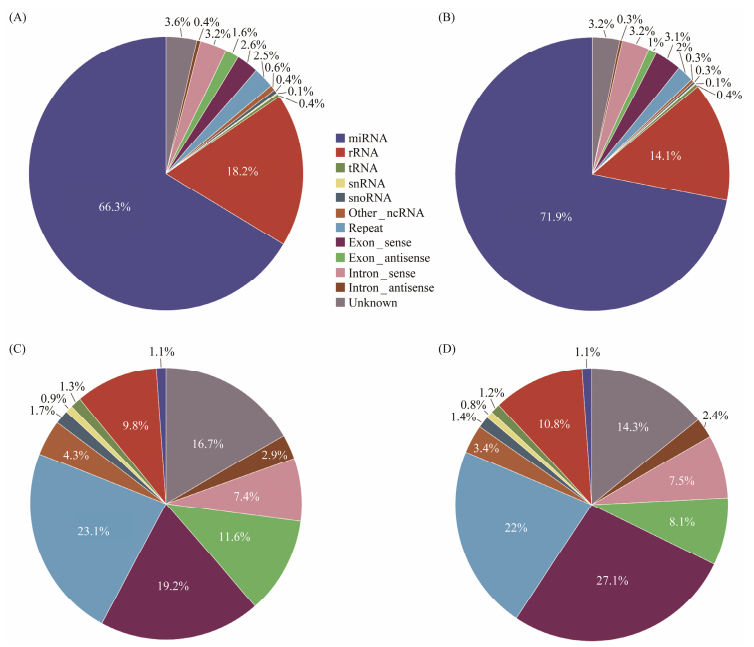
3 Results 3.1 Analysis and Identification of microRNAsA total of 49378012 and 49798903 raw reads were obtained from the small RNA (sRNA) libraries (caro and ctrl libraries) through high-throughput sequencing, respectively (Table 1). The raw data have been successfully uploaded to NCBI (PRJNA983684). After filtering the raw data and removing adapters, 47915498 (97.04%) and 48718271 (97.83%) clean reads were obtained. In the two small RNA libraries, the sRNA lengths ranged from 18 to 28 nt, with a predominance of 22-nt sequences followed by 23-nt and 21-nt sequences (Fig. 1A), while the most abundant unique read was 28 nt (Fig. 1B). After obtaining the filtered sRNAs, we mapped them to the genome and retrieved 26715883 and 21270716 miRNA reads from the two libraries respectively. The alignment rates were 65.77% and 64.65%, indicating a high degree of accuracy in the analysis. The analysis of different types of sRNA showed that miRNA accounted for the highest percentage of all reads, while the percentage of rRNA was below 20% in all cases, which indicated the high quality of the sequenced samples. The proportion of unique reads indicated that repeat and exon sequences account for the highest percentage, whereas miRNA only constituted 1.1% of the unique reads (Figs. 2A–D). A total of 849, 774, 813, 845, 772 and 609 miRNAs were identified as potential miRNAs for the Pacific oyster digestive gland by mapping sRNAs to known animal miRNA precursors.
|
|
Table 1 Statistics of small RNA sequences of the six libraries |
|
Fig. 1 Length distribution of small RNAs in Crassostrea gigas. (A), abundance of miRNAs in the two samples. (B), abundance of unique miRNAs in the two samples. The y-axis represents the numbers of small RNA identified in each library, the x-axis represents the length of small RNA. |
|
Fig. 2 Annotation distribution of sRNAs. (A) and (B) indicate the distribution of annotations of sRNAs in control and treatment groups, respectively. (C) and (D) indicate the distribution of annotations of unique sRNAs in control and treatment groups, respectively. |
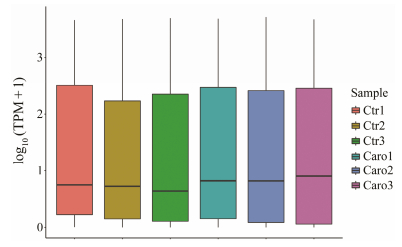
As a result of sequence alignment analysis, a total of 690 candidate miRNAs were identified from the Pacific oyster digestive gland, including 579 known and 111 novel miRNAs. The overall expression levels of miRNAs were higher in the caro group than in the control group (Fig. 3). In both libraries, miRNAs with TPM ≥ 1000 were classified as abundant, while those with TPM < 1 were categorized as rare. Among them, 65 miRNAs were abundantly expressed in both groups, while 271 miRNAs displayed low expression levels.
|
Fig. 3 Overall expression levels of miRNAs in two samples. |
A total of three DE-miRNAs were discovered through comparative analysis of treatment and control groups. In treatment group, rno-miR-200c-3p and novel0017 were upregulated and novel0025 was down-regulated.
3.3 Prediction of DE-miRNA Target Genes in the Pacific OysterThree software programs, miRanda, RNAhybrid and targetscan, were utilized to predict the target genes of the three DE-miRNAs. An intersecting of the three software predictions yielded 5544 miRNA-target gene pairs, of which 137 were identified as differentially expressed genes.
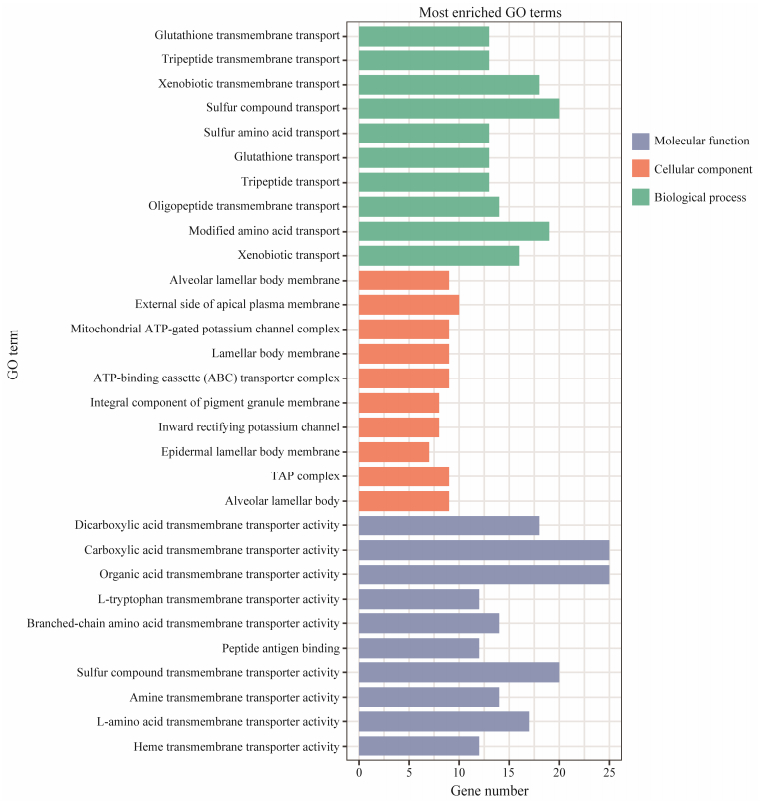
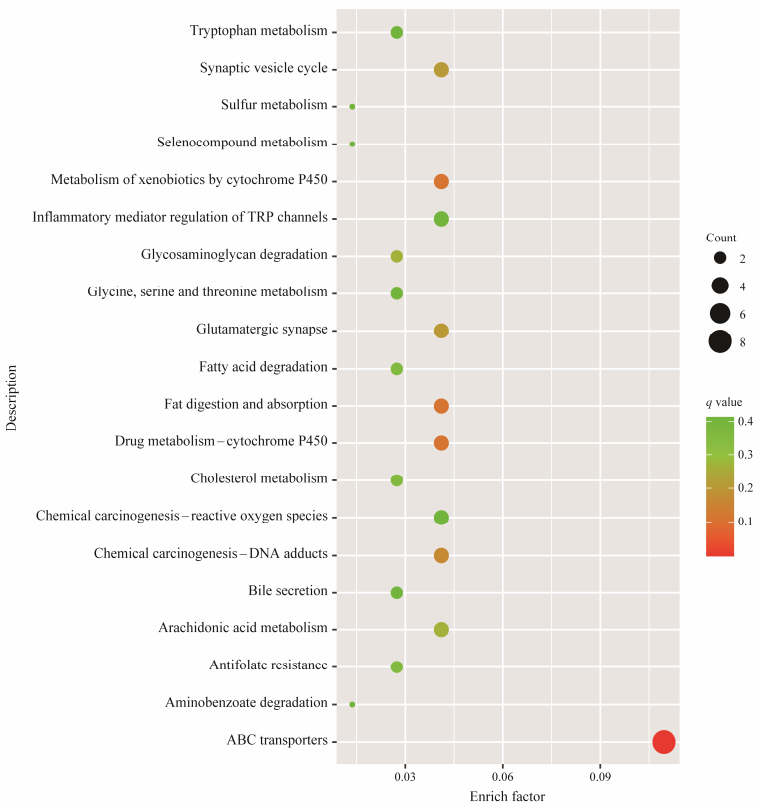
The enrichment analysis of miRNA target genes was performed in order to identify the pathways in which differentially expressed miRNAs may be involved and their impact on the treatment. GO enrichment analysis revealed that the target genes of DE-miRNA were mainly involved in transmembrane transport activity-related molecular function and biological processes associated with transport, as shown in Fig. 4. The target genes were significantly enriched to a KEGG pathway of ABC transporters, as shown in Fig. 5.
|
Fig. 4 Gene ontology distribution of the DE-miRNA target genes. |
|
Fig. 5 KEGG enrichment pathways of target genes of DE-miRNAs. The y-axis represents the KEGG enriched pathways, the x-axis represents the enrichment factor, which was calculated by ratio of the number of differentially expressed mRNAs divided by the number of annotated mRNAs in this pathway. |
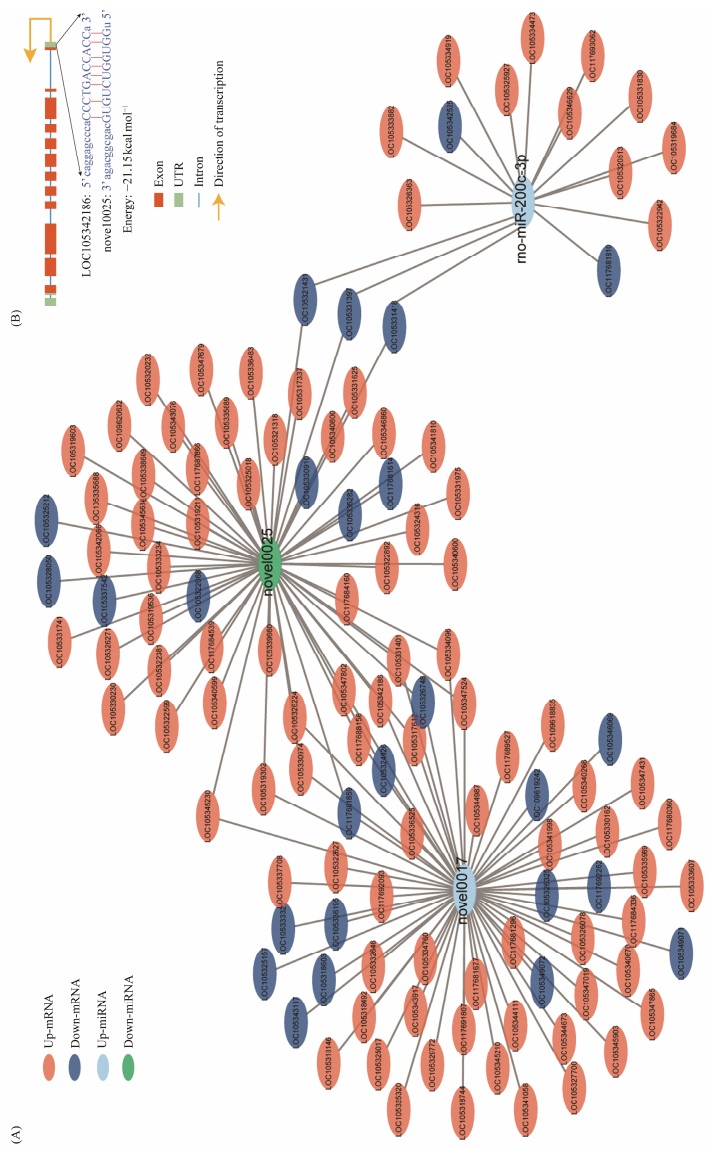
Among the DE-miRNAs, rno-miR-200c-3p and novel0017 were found to be up-regulated in treatment group, resulting in the down-regulation of 5 and 14 mRNAs, respectively. In the interaction network, novel0025 was found to have a central role and down-regulate the expression of 46 mRNAs in the treatment group (Fig. 6A). In addition, downregulation of novel0025 up-regulated the expression of the apolipoprotein gene LOC105342186. Therefore, a negative regulatory relationship exists between novel0025 and lipoprotein genes. Analysis with miRanda software showed that novel0025 was capable of binding to the 5'UTR region of the lipoprotein gene (Fig. 6B).
|
Fig. 6 Interaction network analysis of DE_miRNA and DEG. (A), interaction network of the differentially expressed miRNAs and their target genes. (B), binding sites of novel0025 and apolipoprotein gene predicted by miRanda software. |
To date, miRNAs were reported to function as key regulators in growth, development, immunity, shell color formation and glycogen synthesis in Pacific oysters (Feng et al., 2018; Rosani et al., 2020; Wang et al., 2021; Yue et al., 2022). Previously, we screened genes involved in carotenoid metabolism-related genes by transcriptome (Wan et al., 2022b). However, the regulatory mechanisms of miRNAs in carotenoid metabolism remain poorly understood. In this study, the miRNA transcriptomes of the microencapsulated group, which was fed with beta-carotene, and the normally fed group were compared, while the key miRNA and miRNA target genes in carotenoid metabolism were identified. These results laid the basis for further investigations associated with carotenoid anabolism.
In this study, the length of the sRNA library reads ranged mainly from 21 nt to 23 nt, in line with previous studies (Feng et al., 2020; Yue et al., 2022). The prevalence of 22 nt dicer cleavage products in Metazoa suggests the evolutionary conservation of miRNAs (Starega-Roslan et al., 2015). The small RNA statistics revealed a proportion of rRNAs below 40%, indicating the high quality of the sequencing data obtained (Qin et al., 2014). Interestingly, the most abundant reads in the category of small RNAs were 28 nt in length, which may indicate their classification as piRNAs given that piRNAs typically range from 25 to 31 nt (Girard et al., 2006).
To investigate the regulatory mechanism of miRNAs in carotenoid metabolism, we performed a comparative miRNA analysis of the digestive glands of the Pacific oysters in both the treatment and control groups. A total of 690 miRNAs were identified, which was more than the number of miRNAs previously identified in Pacific oysters (Feng et al., 2020). The previous studies focused on aligning small RNA sequences to known miRNA precursor sequences in invertebrates listed in miRbase (Feng et al., 2020; Li et al., 2021). In this study, small RNA sequences were aligned to miRNA precursor sequences across all animal species, resulting in a significantly expanded repertoire of identified miRNAs. However, we identified only 3 differentially expressed miRNAs from 690 candidates. These three miRNAs target over 5000 genes, yet only 137 of these genes exhibited differential expression between the treatment and control groups. A plethora of structurally diverse carotenoids that accumulate in shellfish are derived from the microalgae they ingest (Maoka, 2011). Moreover, their biosynthesis involves multiple genes for uptake, transport, metabolism and deposition (Parker, 1996). Many of the target genes identified were found to be associated with lipid transport and storage, carotenoid processing and retinoid metabolism, indicating a significant role of the digestive gland in carotenoid metabolism in Pacific oysters. The function of the ABC transport pathway in carotenoid transport has been found in several species (Verwaal et al., 2010; Roth et al., 2017). In this study, DEGs were only enriched in the ABC transport pathway, further illustrating the importance of the altered pathway for carotenoid transport. Collectively, miRNAs participate in the metabolic process of carotenoids by regulating genes in the transport process.
Carotenoid uptake, translocation, and metabolism are believed to primarily occur in the digestive glands due to the high levels of carotenoids found in the hepatopancreas of many shellfish species (Vershinin, 1996). A comparative transcriptome analysis between the digestive gland of the Pacific oyster and other tissues has revealed that genes enriched in the hepatopancreas match digestive functions, and these genes encode numerous proteins homologous to pancreatic digestive enzymes found in vertebrate. At the same time, numerous studies have demonstrated that the vertebrate liver and intestine are important sites for multiple genes to carry out their functions related to carotenoid metabolism (Raghuvanshi et al., 2015; Widjaja-Adhi et al., 2015). For instance, carotenoid lyase performs cleavage of carotenoids, while SRB and apolipoprotein perform the functions of carotenoid uptake, transport and storage of carotenoids in these organs (Widjaja-Adhi et al., 2015). In the present study, we found that miRNA novel0025 may be involved in the regulation of the apolipoprotein gene LOC105342186, which performs carotenoid transport and storage functions in Pacific oysters (Wan et al., 2022b). miRNAs typically match the 3'UTR of genes for functional inhibition of target genes. With further research, the miRNA recognition site has been investigated in the coding region or 5'UTR of the target gene (Pu et al., 2019). The results from miRanda software analysis showed that novel0025 mainly binds to the 5'UTR of the apolipoprotein gene, so it is hypothesized that novel0025 may function on the 5'UTR of this gene to inhibit its expression.
In summary, this study investigated carotenoid metabolism in Pacific oysters by supplementing their diet with betacarotene microcapsules. By conducting a comparative analysis of miRNAs in the digestive glands of Pacific oysters in the treatment and control groups, differentially expressed miRNAs related to carotenoid metabolism were identified and associated with a differentially expressed apolipoprotein gene.
AcknowledgementsThis study was supported by grants from the Shandong Science and Technology Small and Medium Enterprises Innovation Ability Improvement Project (No. 2021TSGC 1240), the Key R & D Program of Shandong Province, China (No. 2022TZXD002), and the China Agriculture Research System Project (No. CARS-49).
Chimsung, N., Lall, S. P., Tantikitti, C., Verlhac-Trichet, V., and Milley, J. E., 2013. Effects of dietary cholesterol on astaxanthin transport in plasma of Atlantic salmon (Salmo salar). Comparative Biochemistry And Physiology B: Biochemistry & Molecular Biology, 165(1): 73-81. (  0) 0) |
Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D. S., 2003. MicroRNA targets in Drosophila. Genome Biology, 5(1): R1. DOI:10.1186/gb-2003-5-1-r1 (  0) 0) |
Feng, D. D., Li, Q., Yu, H., Kong, L. F., and Du, S. J., 2018. Transcriptional profiling of long non-coding RNAs in mantle of Crassostrea gigas and their association with shell pigmentation. Scientific Reports, 8(1): 1436. DOI:10.1038/s41598-018-19950-6 (  0) 0) |
Feng, D. D., Li, Q., Yu, H., Liu, S. K., Kong, L. F., and Du, S. J., 2020. Integrated analysis of microRNA and mRNA expression profiles in Crassostrea gigas to reveal functional miRNA and miRNA-targets regulating shell pigmentation. Scientific Reports, 10(1): 20238. DOI:10.1038/s41598-020-77181-0 (  0) 0) |
Friedlander, M. R., Mackowiak, S. D., Li, N., Chen, W., and Rajewsky, N., 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Research, 40(1): 37-52. DOI:10.1093/nar/gkr688 (  0) 0) |
Girard, A., Sachidanandam, R., Hannon, G. J., and Carmell, M. A., 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature, 442(7099): 199-202. DOI:10.1038/nature04917 (  0) 0) |
Karadas, F., Pappas, A. C., Surai, P. F., and Speake, B. K., 2005. Embryonic development within carotenoid-enriched eggs influences the post-hatch carotenoid status of the chicken. Comparative Biochemistry and Physiology B: Biochemistry & Molecular Biology, 141(2): 244-251. (  0) 0) |
Kruger, J., and Rehmsmeier, M., 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Research, 34: W451-W454. DOI:10.1093/nar/gkl243 (  0) 0) |
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 10(3): R25. DOI:10.1186/gb-2009-10-3-r25 (  0) 0) |
Li, N., Hu, J. J., Wang, S., Cheng, J., Hu, X. L., Lu, Z. Y., et al., 2010. Isolation and identification of the main carotenoid pigment from the rare orange muscle of the Yesso scallop. Food Chemistry, 118(3): 616-619. DOI:10.1016/j.foodchem.2009.05.043 (  0) 0) |
Li, Z. Z., Li, Q., Liu, S. K., Han, Z. Q., Kong, L. F., and Yu, H., 2021. Integrated analysis of coding genes and non-coding rnas associated with shell color in the Pacific oyster (Crassostrea gigas). Marine Biotechnology (NY), 23(3): 417-429. DOI:10.1007/s10126-021-10034-7 (  0) 0) |
Liu, H. L., Zheng, H. P., Zhang, H. K., Deng, L. H., Liu, W. H., Wang, S. Q., et al., 2015. A de novo transcriptome of the noble scallop, Chlamys nobilis, focusing on mining transcripts for carotenoid-based coloration. BMC Genomics, 16(1): 44. DOI:10.1186/s12864-015-1241-x (  0) 0) |
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L., 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics, 21(19): 3787-3793. DOI:10.1093/bioinformatics/bti430 (  0) 0) |
Maoka, T., 2009. Recent progress in structural studies of carotenoids in animals and plants. Archives of Biochemistry and Biophysics, 483(2): 191-195. DOI:10.1016/j.abb.2008.10.019 (  0) 0) |
Maoka, T., 2011. Carotenoids in marine animals. Marine Drugs, 9(2): 278-293. DOI:10.3390/md9020278 (  0) 0) |
Maoka, T., 2020. Carotenoids as natural functional pigments. Journal of Natural Medicines, 74(1): 1-16. DOI:10.1007/s11418-019-01364-x (  0) 0) |
McGeary, S. E., Lin, K. S., Shi, C. Y., Pham, T. M., Bisaria, N., Kelley, G. M., et al., 2019. The biochemical basis of microRNA targeting efficacy. Science, 366(6472): eaav1741. DOI:10.1126/science.aav1741 (  0) 0) |
Moran, N. A., and Jarvik, T., 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science, 328(5978): 624-627. DOI:10.1126/science.1187113 (  0) 0) |
Olsen, R. E., Kiessling, A., Milley, J. E., Ross, N. W., and Lall, S. P., 2005. Effect of lipid source and bile salts in diet of Atlantic salmon, Salmo salar L., on astaxanthin blood levels. Aquaculture, 250(3-4): 804-812. DOI:10.1016/j.aquaculture.2005.03.013 (  0) 0) |
Parker, R. S., 1996. Absorption, metabolism, and transport of carotenoids. FASEB Journal, 10(5): 542-551. DOI:10.1096/fasebj.10.5.8621054 (  0) 0) |
Pu, M. F., Chen, J., Tao, Z. T., Miao, L. L., Qi, X. M., Wang, Y. Z., et al., 2019. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cellular and Molecular Life Sciences, 76(3): 441-451. DOI:10.1007/s00018-018-2940-7 (  0) 0) |
Qin, Q. H., Wang, Z. L., Tian, L. Q., Gan, H. Y., Zhang, S. W., and Zeng, Z. J., 2014. The integrative analysis of microRNA and mRNA expression in Apis mellifera following maze-based visual pattern learning. Insect Science, 21(5): 619-636. DOI:10.1111/1744-7917.12065 (  0) 0) |
Raghuvanshi, S., Reed, V., Blaner, W. S., and Harrison, E. H., 2015. Cellular localization of β-carotene 15, 15' oxygenase-1 (BCO1) and β-carotene 9', 10' oxygenase-2 (BCO2) in rat liver and intestine. Archives of Biochemistry and Biophysics, 572: 19-27. DOI:10.1016/j.abb.2014.12.024 (  0) 0) |
Rosani, U., Abbadi, M., Green, T., Bai, C. M., Turolla, E., Arcangeli, G., et al., 2020. Parallel analysis of miRNAs and mRNAs suggests distinct regulatory networks in Crassostrea gigas infected by Ostreid herpesvirus 1. BMC Genomics, 21(1): 620. DOI:10.1186/s12864-020-07026-7 (  0) 0) |
Roth, M. S., Cokus, S. J., Gallaher, S. D., Walter, A., Lopez, D., Erickson, E., et al., 2017. Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production. Proceedings of the National Academy of Sciences of the United States of America, 114(21): E4296-E4305. (  0) 0) |
Starega-Roslan, J., Galka-Marciniak, P., and Krzyzosiak, W. J., 2015. Nucleotide sequence of miRNA precursor contributes to cleavage site selection by Dicer. Nucleic Acids Research, 43(22): 10939-10951. DOI:10.1093/nar/gkv968 (  0) 0) |
Tan, K., Guo, Z., Zhang, H. C., Ma, H. Y., Li, S. K., and Zheng, H. P., 2021. Carotenoids regulation in polymorphic noble scallops Chlamys nobilis under different light cycle. Aquaculture, 531: 735937. DOI:10.1016/j.aquaculture.2020.735937 (  0) 0) |
Toews, D. P. L., Hofmeister, N. R., and Taylor, S. A., Trends in Genetics. 2017. The evolution and genetics of carotenoid processing in animals, 33(3): 171-182. (  0) 0) |
Vershinin, A., 1996. Carotenoids in mollusca: Approaching the functions. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 113(1): 63-71. DOI:10.1016/0305-0491(96)00104-6 (  0) 0) |
Verwaal, R., Jiang, Y., Wang, J., Daran, J. M., Sandmann, G., van den Berg, J. A., et al., 2010. Heterologous carotenoid production in Saccharomyces cerevisiae induces the pleiotropic drug resistance stress response. Yeast, 27(12): 983-998. DOI:10.1002/yea.1807 (  0) 0) |
Wan, S., Li, Q., Yu, H., Liu, S. K., and Kong, L. F., 2022a. A nuclear receptor heterodimer, CgPPAR2-CgRXR, acts as a regulator of carotenoid metabolism in Crassostrea gigas. Gene, 827: 146473. DOI:10.1016/j.gene.2022.146473 (  0) 0) |
Wan, S., Li, Q., Yu, H., Liu, S. K., and Kong, L. F., 2022b. Transcriptome analysis based on dietary beta-carotene supplement reveals genes potentially involved in carotenoid metabolism in Crassostrea gigas. Gene, 818: 146226. DOI:10.1016/j.gene.2022.146226 (  0) 0) |
Wang, X., Wang, W. J., Li, Z., Sun, G. H., Xu, X. H., Feng, Y. W., et al., 2021. Comprehensive analysis of differentially expressed ncRNA, mRNA, and their ceRNA networks in the regulation of glycogen content in the Pacific oyster, Crassostrea gigas. Aquaculture, 531: 735895. DOI:10.1016/j.aquaculture.2020.735895 (  0) 0) |
Widjaja-Adhi, M. A., Lobo, G. P., Golczak, M., and Von Lintig, J., 2015. A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Human Molecular Genetics, 24(11): 3206-3219. DOI:10.1093/hmg/ddv072 (  0) 0) |
Yabuzaki, J., 2017. Carotenoids database: Structures, chemical fingerprints and distribution among organisms. Database, 2017: bax004. (  0) 0) |
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A., 2010. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biology, 11(2): R14. DOI:10.1186/gb-2010-11-2-r14 (  0) 0) |
Yue, C. Y., Li, Q., and Yu, H., 2022. Integrated analysis of miRNA and mRNA expression profiles identifies potential regulatory interactions during sexual development of Pacific oyster Crassostrea gigas. Aquaculture, 546: 737294. DOI:10.1016/j.aquaculture.2021.737294 (  0) 0) |
Zhou, L., Chen, J., Li, Z. Z., Li, X., Hu, X. X., Huang, Y., et al., 2010. Integrated profiling of microRNAs and mRNAs: MicroRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One, 5(12): e15224. DOI:10.1371/journal.pone.0015224 (  0) 0) |
 2024, Vol. 23
2024, Vol. 23


