2) School of Modern Agriculture and Biotechnology, Ankang University, Ankang 725000, China;
3) Laboratory of Fish Immunology and Nutrigenomics, Applied Animal and Aquatic Sciences Research Unit, Division of Fisheries, Faculty of Technology, Mahasarakham University, Khamriang Sub-District, Kantarawichai, Mahasarakham 44150, Thailand
Plastic products are widely used in our daily life and industrial production due to their cheapness and durability (Sui et al., 2020). The demand for plastics has dramatically increased in recent years, especially used for the production of personal protective equipment, medical supplies and packaging since the 2019 New Coronavirus epidemic (Govender et al., 2022). It is estimated that over 40% manufactured plastics, 250000 t of plastic litter is floated into the oceans (Ragusa et al., 2022). The plastic litter will finally be decomposed into particles smaller than 5 mm, defined as microplastics (MPs) (Hidalgo-Ruz et al., 2012). Recently, the MPs have been detected in the human breastmilk (Ragusa et al., 2022), which attract global researchers' attention.
Estuaries are considered to be the main pathway for MPs to enter the oceans from freshwater environments (Zhang et al., 2019). It is not surprising to detect high abundance of MPs in estuaries. In 2017, the concentration of MPs in China's Pearl River Estuary reached 8902 items m−3 (Shabaka et al., 2022). In 2021, the abundance of MPs in the Nile Estuary ranged from 761 ± 319 to 1718 ± 1008 items m−3 (Yan et al., 2019). And, the abundance of MPs at the Tecolutla Estuary in the Gulf of Mexico reached 151000 items m−3 (Sánchez-Hernández, 2021). The MPs pollution in estuaries has become a worldwide problem. However, the environmental and health risks of MPs exposure need further investigation.
It had been confirmed that MPs could be ingested by fish and had malign influences on fish (Pedà et al., 2016; Rummel et al., 2016; Steer et al., 2017). 60 to 90 days MPs exposure lead to severe structural alterations of intestine of European sea bass (Dicentrarchus labrax), with finding that the serosa, muscularis mucose and submucosa/mucosa layers were edematous, with an evident ectasia of vessels and also leukocyte infiltration (Pedà et al., 2016). Acute exposure of MPs disrupted the swimming activity of zebrafish (Danio rerio) larvae, and chronic exposure of MPs reduced the growth and reproduction of zebrafish and mekada (Oryzias melastigma) (Cormier et al., 2022). In addition, MPs can also affect the immune system of fish. MPs could induce the histological changes and oxidative stress in liver of zebrafish (Lu et al., 2016) and induced the expressions of immune related genes (e.g., complements) of rare minnow (Gobiocypris rarus) (Wang et al., 2022). Totally 41 differentially expressed genes (17 upand 26 down-regulated) were attributed to immune response in adult zebrafish following MPs exposure (Limonta et al., 2019). The respiratory burst activity of headkideny leucocyte of gilthead seabream (Sparus aurata) was significantly increased by polyvinyl chloride (PVC) MPs and that of European sea bass was increased by polyethylene (PE) MPs (Espinosa et al., 2018). The levels of oxidants (superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPx), glutathione (GSH) and malondialdehyde (MDA)) and some immune-related (lysozyme, alkaline phosphatase (ALP)) were increased by MPs (Wen et al., 2018). However, rainbow trout (Oncorhynchus mykiss) maintained barrier functions following MPs exposure, without significant expressional changes observed for the immune-related genes (Ašmonaitė et al., 2018). Thus, different fish species might share varied immune response towards MPs exposure.
Soiny mullet (Liza haematocheila) is an economically important fish species that cultured in the estuaries (Ge et al., 2020; Qi et al., 2022). However, the molecular response of soiny mullet to microplastic stress remains unclear. Whole transcriptome sequencing provides an efficient method to understand the genetic response to environmental changes (Qi et al., 2016). In the present study, the RNA expression profiles of soiny mullet induce by 0.5 μm MPs were investigated, hoping to provide new insights into understand the consequence of MPs pollution in fish.
2 Materials and Methods 2.1 FishHealthy soiny mullet larvae with average body length of 2.67 ± 0.16 cm and average body weight of 0.14 ± 0.01 g were provided by a local fish farm in Sheyang Town, Yancheng City, Jiangsu Province, China. The fish were transported into our laboratory and reared at least two weeks prior to the formal experiments. The fish were cultured in artificial seawater simulating pristine water with conditions of temperature = 25℃, salinity = 15‰, pH = 8.1 and dissolved oxygen > 7.0 mg L−1, and fed with brine shrimp (Artemia salina) twice daily at the rate of 2% of the fish's body weight.
2.2 Polystyrene MPs Preparation and ExposureThe 0.5 μm (2.5 w/v, 10 mL, 0.5 ± 0.012 μm) monodisperse polystyrene (PS) microspheres were purchased from BaseLine ChromTech Research Centre (Tianjin, China) and used for challenge experiments. The Fourier transform infrared (FTIR) of PS MPs was shown in Fig. S1. To simulate extreme MPs pollution in the water, the MPs concentration in the MPs exposure group was set to be 0.1 mg L−1 (1.46 × 109 items L−1) (Yan et al., 2020). During the experiment, aerators and air stones were used to maintain the suspension of MPs.
The larvae were randomly divided into three groups: control group (CTRL), MPs-challenge group for 7 d (MPs-7d group), and MPs-challenge group for 14 d (MPs-14d group). Each group contained 3 replicated glass tanks (20 fish per tank). During the 14-d exposure period, completely replace artificial seawater daily to avoid the effects of MPs accumulation. At the same time, PS MPs suspended in water were quantified under a microscope (40 ×) using a hemocytometer daily. The quantitative calculation results of MPs concentration were shown in Table S1. Laboratory coats made of natural fibers, nitrile gloves, and face masks were worn to prevent plastic contamination during the counting process. All the glassware, containers, filtration units, and other necessary instruments were rinsed three times with ddH2O before use. At 7 d and 14 d post exposure, three larvae in each tank were randomly selected and transferred into cryovials under anesthesia with tricaine methane-sulfonate (MS-222) (Sigma, USA), and stored − 80℃ until use.
2.3 Total RNA Exaction and cDNA Library ConstructionTotal RNA of larvae was extracted using Trizol reagent (Invitrogen, USA) according to manufacturer's protocols. The quality and integrity of the purified RNA were assessed using the Fragment Analyzer with the Standard Sensitivity RNA Analysis Kit (DNF-471) (Agilent Technologies, USA). The total RNA was purified by Dynabeas mRNA Purification Kit (Invitrogen, USA) and DNA probe, and digested with RNase H to remove rRNA. The DNA probe was further removed by digested with DNase Ⅰ. After that, the mRNA was fragmented into 200 – 250 bp with fragment buffer (Ambion, USA) and transcribed into the first strand cDNA using SuperScript Ⅱ reverse transcriptase (Invitrogen, USA) and N6 primers, and followed by the second strand cDNA synthesis with the second strand master mix (Invitrogen, USA). The synthetic double-strand cDNA was end-repaired, A-tail added, adaptor ligated and further enriched by PCR amplification with adaptor-specific primers to get the final cDNA library. The sequencing was performed on the DNBSEQ platform (BGI, China) (Rao et al., 2020).
2.4 Transcriptome AssemblyThe clean reads were obtained by filtering the raw reads with SOAPnuke software (Version 1.5.2) to remove the reads that contained sequencing adapters, more than 5% unknown base ('N' base), and low-quality base (Chen et al., 2018). The clean reads were further de novo assembled with Trinity platform (Haas et al., 2013) and clustered with the TIGR Gene Indices Clustering Tools (TGICL) software (Pertea et al., 2003) to get unigenes. The completeness of transcriptome assembly was evaluated using the Benchmarking Universal Single-Copy Orthologue (BUSCO) tool (Manni et al., 2021). The coding sequences (CDSs) from the assembled transcriptome were detected using TransDecoder program (Version 3.0.1) (Kim et al., 2015).
2.5 Differently Expressed Genes, Gene Ontology and Pathway Enrichment AnalysisThe differently expressed genes (DEGs) between different groups were performed using DESeq2 (v 1.34.0) with Q-value (adjusted p-value) ≤ 0.05 (Love et al., 2014). The results produced by DEseq2 were expressed as the estimated log2 (fold change) of the expression level of unigenes between different groups. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of annotated DEGs was performed by hypergeometric test. The significant levels of terms and pathways were corrected by Q value with a rigorous threshold (Q value ≤ 0.05) by Bonferroni's correction (Curtin and Schulza, 1998).
3 Results and DiscussionMPs pollution has become a worldwide environmental problem (Akdogan and Guven, 2019). Fish live in the water and directly contact with microplastics in water. Evaluation the fish response to MPs is helpful to understand the toxicological effects of MPs. In the present study, we analyzed the effects of MPs exposure on the immune- and apoptosis-related pathways of soiny mullet larvae based on the transcriptomic analysis, hoping to providing basis for determining the toxic effects of microplastics (Chen et al., 2020).
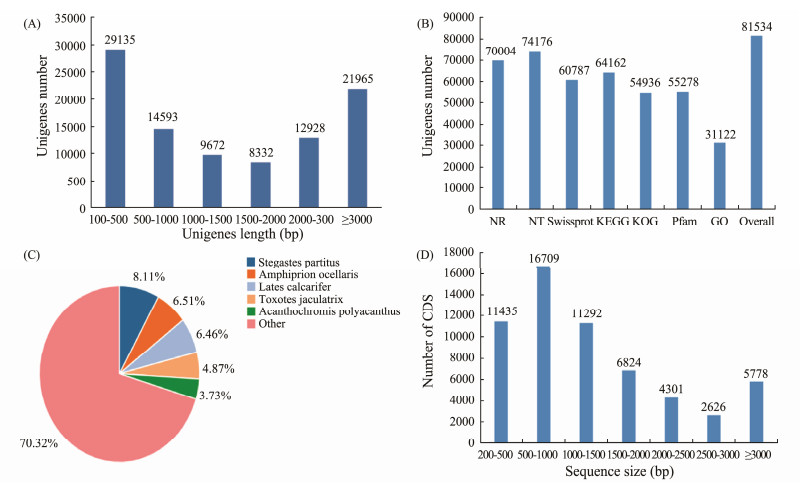
3.1 Transcriptome Sequencing and de novo AssemblyThe raw reads and clean reads of different groups were listed in Table 1. The Q20 and Q30 of clean reads were more than 80%, indicating the clean reads could be used. After assembled the clean reads using Trinity, a total number of 96585 unigenes were obtained, with mean length of 1925 bp, N50 of 3504 bp and GC content of 47.20% (Table 2). Further analysis revealed that 30.17% unigenes (19135) were 150 bp to 500 bp in length, followed by 22.74% (21965) unigenes were over 3000 bp, 15.11% (14593) unigenes were 500 bp to 1000 bp in length, 13.34% (12928) unigenes ranged from 2000 bp to 3000 bp, 10.01% (9672) unigenes ranged from 1000 bp to 1500 bp, and 8.63% (8332) unigenes ranged from 1500 bp to 2000 bp in length (Fig.1A). These results indicated that the transcriptomes were effectively constructed and could be used for further analysis.
|
|
Table 1 Statistics of transcriptomic sequences from different groups |
|
|
Table 2 Statistics of assembled unigenes |
|
Fig. 1 Basic analysis of the splenic transcriptome of soiny mullet larvae. (A), sequence length distribution of the assembled unigenes; (B), functional annotation of the assembled unigenes; (C), species distribution of the assembled unigenes; (D), sequence length distribution of CDS. |
96585 unigenes were compared against the NCBI non-redundant (NR), NCBI nucleotide (NT) and Swissprot database. Results showed that 72.48% (70004), 76.80% (74176), and 62.94% (60787) unigenes matched with the annotated sequences for NR, NT, and Swissprot database, respectively (Fig.1B). The species distribution showed that 70.32% (49229) unigenes hit to other vertebrates, followed by 8.11 to Stegastes partitus, 6.51% to Amphiprion ocellaris, 6.46% to Lates calcarifer (Fig.1C). The reason for the unigenes shared high similarity with other vertebrates might be the lack of genome or gene data for mugilidae, as soiny mullet belongs to the mugilid species (Qi et al., 2016). A total number of 58965 CDSs were obtained with average 53.03% GC content (Fig.1D).
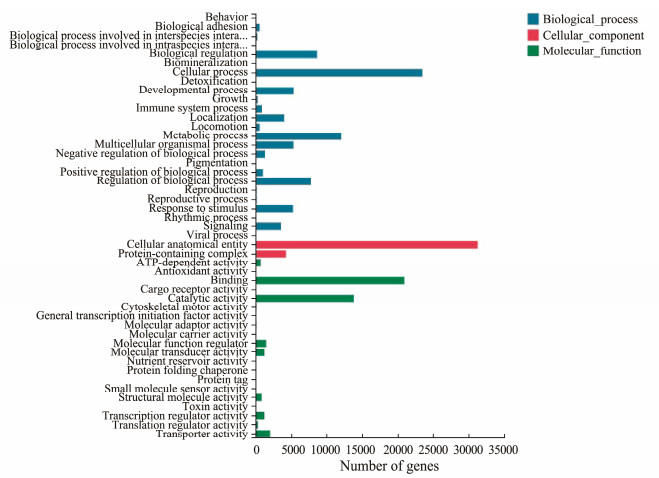
3.3 GO, KEGG and KOG Classification of Transcriptome SequencesThe GO knowledgebase is the world's largest source of information on the functions of genes (The Gene Ontology Consortium, 2019). Base on the NR annotation, 70004 unigenes were classified into 47 functional groups in three GO categories: biological process (81287 unigenes, 84.16%), cellular component (35602 unigenes, 36.86%) and molecular function (43179 unigenes, 44.71%) (Fig.2). In the biological process category, cellular process (23506 unigenes), metabolic process (12077 unigenes), biological regulation (8659 unigenes), regulation of biological process (7819 unigenes) and developmental process (5349 unigenes) was the five most abundant subcategories. In the cellular component category, the two most abundant subcategories were cellular anatomical entity (31309 unigenes) and protein-containing complex (4293 unigenes). The binding (20950 unigenes), catalytic activity (13828 unigenes), transporter activity (2048 unigenes), transporter activity (2048 unigenes), molecular function regulator (1485 unigenes), and molecular transducer activity (1238 unigenes) was the most five abundant subcategories in the molecular function category. A total number of 5284 and 889 unigenes were clustered into subcategories of response to stimulus and immune system process, respectively.
|
Fig. 2 GO classification of assembled unigenes. X-axis, the mumber of unigenes; Y-axis, three major functional GO categories including biological process (blue), cellular component (red) and molecular function (green). |
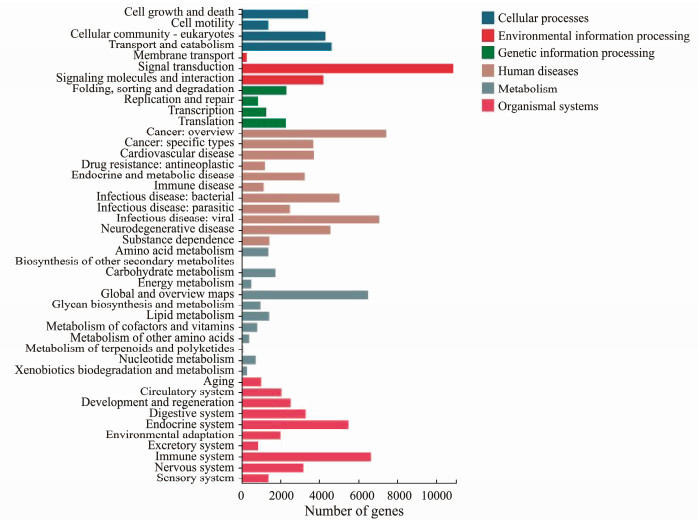
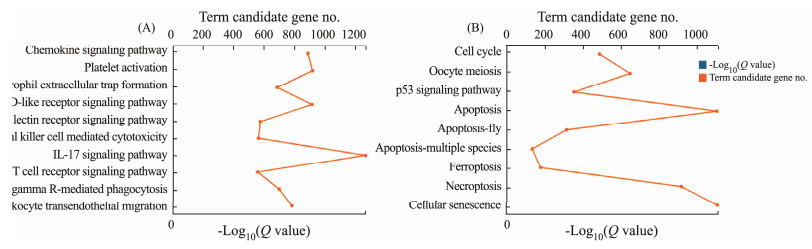
KEGG is a database resource for understanding highlevel functions and utilities of the biological system (Kanehisa et al., 2023). A total number of 64162 unigenes were grouped into 44 KEGG pathways in six main categories: cellular processes (13779 unigenes), environmental information processing (15361 unigenes), genetic information processing (6773 unigenes), human diseases (41229 unigenes), metabolism (15149 unigenes), and organismal system (28633 unigenes) (Fig.3). Among these unigenes, 12600 unigenes were assigned into the cluster of immune system and further classified into 22 immune-related pathways (Fig.4A), including interleukin (IL)-17 signaling pathway (1268 unigenes), platelet activation (920 unigenes), Nod-like receptor signaling pathway (916 unigenes), chemokine signaling pathway (890 unigenes), leukocyte transendothelial migration (787 unigenes), Fc gamma R-mediated phagocytosis (702 unigenes). In addition, 5255 unigenes were assigned into 9 cell growth and death related pathways, such as cellular senescence (1113 unigenes), apoptosis (1106 unigenes), necroptosis (919 unigenes) oocyte meiosis (649 unigenes) (Fig.4B).
|
Fig. 3 KEGG annotation of the assembled unigenes. |
|
Fig. 4 KEGG enrichment of unigenes that related to immune system (A), and cell growth and death (B). |
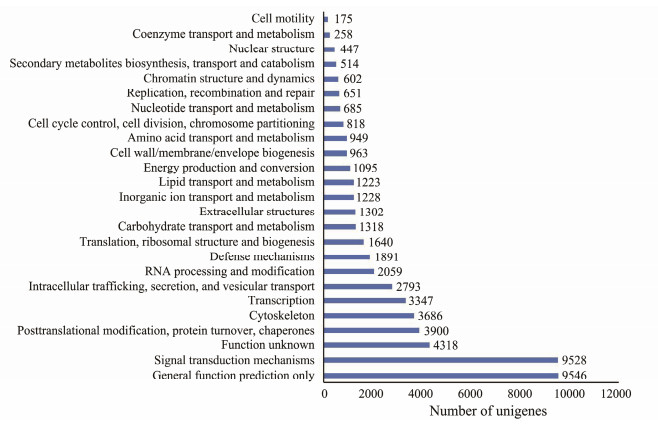
Eukaryotic Orthologous Groups (KOG) database is platform for functional annotation of newly sequenced genomes (Zheng et al., 2015). Based on sequence homology, a total number of 54936 unigenes (56.88%) were classified into 25 KOG functional categories (Fig.5), among which general function predication only (9546 unigenes), signal transduction mechanisms (9528 unigenes), function unknown (4318 unigenes), posttranslation modification, protein turnover, chaperons (3900 unigenes), cytoskeleton (3686 unigenes) are the five dominant categories. In addition, 1891 unigenes were assigned to the category of defense mechanisms.
|
Fig. 5 Clusters of KOG classification of assembled unigenes. X-axis, the number of unigenes annotated in the category; Y-axis, the name of 25 functional categories in KOG database. |
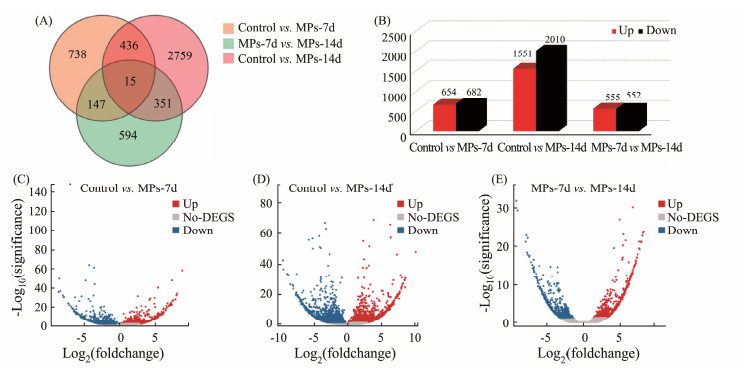
The expression profiles among the three groups were analyzed in pairwise comparisons with obtaining 6004 DEGs (Fig.6A). Of these DEGs, 654 up-regulated and 682 down-regulated in MPs-7d group, 1551 up-regulated and 2010 down-regulated in MPs-14d group were respectively identified when compared that of control group. Totally, 555 up-regulated and 552 down-regulated DEGs were identified when compared between MPs-7d group and MPs-14d group (Fig.6B). The overall difference level was visually displayed by volcano plots (Figs.6C – 6E). Our results were similar to previous study in medaka, with finding 4949 DEGs at 5 d post MPs exposure, and the number of DEGs increased to 5112 and 6490 at 9 and 19 d post MPs exposure (Chen et al., 2020). These results indicated that the effects of MPs on fish were time-dependent and the exposure time was also important factors when analyze the toxicity of MPs.
|
Fig. 6 Comparisons of DEGs among different groups. (A), the Venn diagram of number of shared DEGs between contrasts; (B), the up-regualtated and down-regulated DEGs between contrasts; X-axis, the comparison set between contrasts; Y-axis, the number of DEGs. (C – E), the volcano plots of DEGs between contrasts; X-axis, the fold change after Log2 transformation; Y-axis, the significance value after − Log10 transformation. |
A total number of 1960 DEGs were assigned into 37 terms of three main GO categories, with 991 DEGs into biological process, 358 DEGs into cellular component and 661 DEGs into molecular function, when compared between control group and MPs-7d group (Fig.S2A). In comparison between control group and MPs-14d group, 5709 DEGs were assigned into 39 terms of the three main GO categories, with 2940 DEGs into biological process, 1111 DEGs into cellular component, and 1658 DEGs into molecular function (Fig.S2B), indicating that the effects of MPs on the gene functions of soiny mullet larvae were aggravated along with the prolonging of MPs challenge time. Similarly, 1373 DEGs were assigned into 34 GO categories in the three GO categories (Fig.S2C). Importantly, 14, 43 and 9 DEGs involved in immune system, 83, 225 and 56 DEGs involved in response to stimulus were identified in three comparisons (control vs MPs-7d, control vs MPs-14d, MPs-7d vs MPs-14d) (Fig.S2). Our results were in line with previous studies in zebrafish and rare minnow (Lu et al., 2016; Veneman et al., 2017; Wang et al., 2022), indicating that the MPs challenge could trigger the fish immune response.
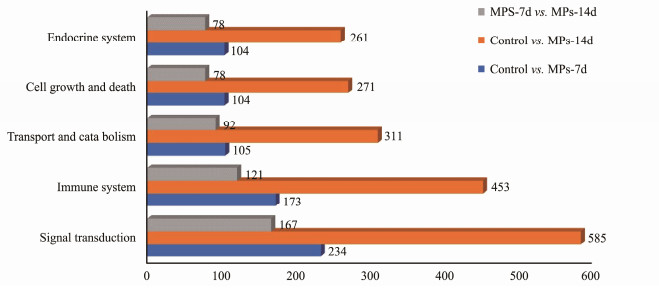
KEGG analysis revealed that a total number of 1589 DEGs were annotated to 31 metabolic pathways in five categories compared between control group and MPs-7d group, including cellular process (320 DEGs), environmental information processing (322 DEGs), genetic information processing (110 DEGs), metabolism (228 DEGs) and organismal systems (609 DEGs) (Fig.S3A). In the comparison between control group and MPs-14d group, 4132 DEGs were assigned into 33 metabolic pathways and the number of in the category of cellular process, environmental information processing, genetic information processing, metabolism and organismal systems were 765, 855, 265, 727 and 1520, respectively (Fig.S3B). Similarly, 1224 DEGs were assigned into 32 metabolic pathways compared between MPs-7d group and MPs-14d group (Fig.S3C). Notedly, the five dominant KEGG subcategories in the three comparisons were similar, including the signal transduction, immune system, transport and catabolism, cell growth and death, endocrine system (Fig.7).
|
Fig. 7 DEGs involved in five dominant KEGG sub-categories. X-axis, the number of DEGs annotated in the group; Y-axis, five dominant KEGG sub-categories. |
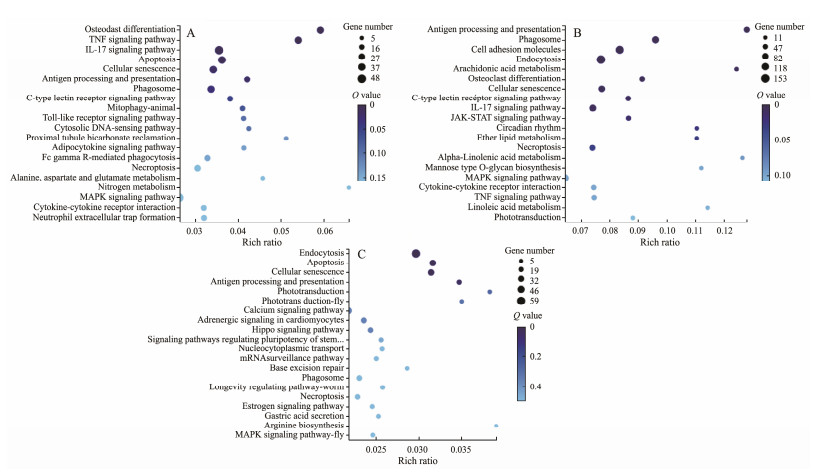
The pathway enrichment analysis based on the KEGG database were further performed to identify the biological pathways affected by MPs challenge. A total of 228, 249 and 219 pathways were enriched in the two comparison of control vs MPs-7d, control vs MPs-14d, and MPs-7d vs MPs-14d, respectively. The top 20 pathways with most abundant DEGs in the three comparisons were shown in Fig.8 In the comparison of control group and MPs-7d group, 8 pathways were significantly enriched (Q-value ≤ 0.05), including osteoclast differentiation, tumor necrosis factor (TNF) signaling pathway, IL-17 signaling pathway, apoptosis, cellular senescence, antigen processing and presentation, phagosome and C-type lectin receptor signaling pathway (Fig.8A). In the comparison between control group and MPs-14d group, 13 pathways were significantly enriched (Q-value ≤ 0.05), including antigen processing and presentation, phagosome, cell adhesion molecules, endocytosis, arachidonic acid metabolism, osteoclast differentiation, cellular senescence, C-type lectin receptor signaling pathway, IL-17 signaling pathway, Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway, circadian rhythm, ether lipid metabolism, necroptosis (Fig.8B). In the comparison between MPs-7d group and MPs-14d group, only four pathways were significantly enriched, including endocytosis, apoptosis, cellular senescence and antigen processing and presentation (Fig.8C). MPs could not enter into cells directly due to their large size, whilst the nano-MPs could directly enter into the cells (Azeem et al., 2021). It was not surprising to see the significant enrichment of endocytosis pathway, as previous studies found that endocytic or passive uptake processes was the mechanism for MPs enter the cells (Banerjee and Shelver, 2021; Kwon et al., 2022). These results revealed that the 0.5 μm MPs used in present study might also enter into the cells of soiny mullet larvae through endocytic processes.
|
Fig. 8 KEGG enrichment of DEGs. |
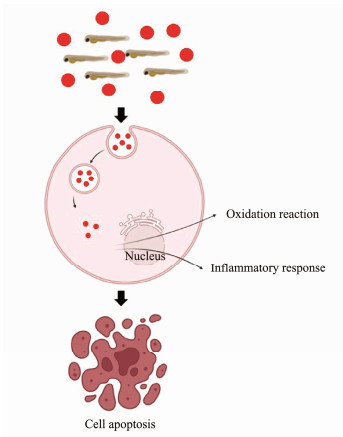
Among these significantly enriched signaling pathways, several immune-related pathways were significantly enriched, including TNF signaling pathway, IL-17 signaling pathway, antigen processing and presentation, C-type lectin receptor and JAK-STAT signaling pathway, which indicated that the MPs challenge exactly could trigger the immune response of soiny mullet larvae. Similar results were also observed in zebrafish and rare minnow (Lu et al., 2016; The Gene Ontology Consortium, 2019; Wang et al., 2022). The C-type lectin receptor pathway plays crucial roles in the immune response to self (endogenous) and non-self (exogenous) ligands (Brown et al., 2018). Significant enrichment of C-type lectin receptor pathway after MPs challenge (Fig.8) indicated that MPs could initiate the innate immunity of soiny mullet larvae. In addition, IL-17 and TNF which involve in mediating signaling synergistically to drive expression of inflammatory genes (Schmitt et al., 2021) were also activated. The JAK-STAT pathway which orchestrate the hematopoiesis, induce inflammation and control the immune response, were also enriched. All these results indicated that MPs challenge could drive the inflammatory response of soiny mullet larvae. Surprisingly, we did not observe the enrichment of adaptive immune related pathways in soiny mullet larvae after MPs challenge. More MPs enter into the cells would be handled by phagosome pathway to form membrane protrusions for covering the MPs and finally pinched out the cell (Kagan and Iwasaki, 2012; Uribe-Querol and Rosales, 2020). We also found that the phosphoinositide 3-kinase (PI3K) apoptosis pathway was enriched at 14 d post MPs challenge, which was similar to previous studies (An et al., 2021; Hou et al., 2021; Umamaheswari et al., 2021). The PI3K pathway has a central role in the cell survival and proliferation (Zhang and Zhang, 2019). We found that the expressions of several PI3K pathway related genes, including PI3K, mitogen-activated extracellular signal-regulated kinase (MEK) 1/2, IκB kinase (IKK) and IκBα were inhibited by MPs challenge (Fig.S4). These results indicated that MPs might induce the cell apoptosis through inhibit PI3K signaling pathway. Based on above analysis, we speculated that the 0.5 μm MPs mainly entered the cells through endocytic processes and could induce the inflammatory responses and oxidant reactions in fish. With more MPs enter the cells, the apoptosis processes would be triggered to eliminate the damaged cells (Fig.9).
|
Fig. 9 Schematic diagram of the effects of MPs on soiny mullet larvae. MPs were represented by red dots. |
In conclusion, the effects of MPs on the immune system of soiny mullet larvae were evaluated based on the transcriptomic analysis. Several immune related pathways, such as IL-17 pathway, TNF pathway, JAK-STAT pathway, were induced by MPs. And, the apoptosis pathway was also induced to eliminate the damaged cells. Our results provide solid basis for understanding the immune toxicity of MPs on fish.
AcknowledgementsThis study was supported by the Major Projects of Natural Science Research for University and Colleges in Jiangsu Province (No. 21KJA240001), the Key Research and Development Program of Jiangsu Province (No. BE2018348), and partially by the Jiangsu Agricultural Science and Technology Innovation Fund (No. CX (22) 3199) and the Projects for the High-Quality Development of Fishery Industry of Yancheng City (No. YCSCYJ2021 0014). Dr. Zhitao Qi was supported financially by the projects for 'Six Talents' of Jiangsu Province (No. NY115).
Author Contributions
Xuan Wei, Xiangyu Pi, Shengyuan Zhang, and Yanming Sui conducted the experiments; Qihuan Zhang and Zisheng Wang analyzed the data; Eakapol Wangkahart edited the figures; Zhitao Qi designed the experiments, wrote the paper, and supervised the research.
Data Availability
Supplementary material is available in the online version of this article at https://doi.org/10.1007/s11802-025-5793-3.
Declarations
Ethics Approval and Consent to Participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for Publication
Informed consent for publication was obtained from all participants.
Conflict of Interests
The authors declare that they have no conflict of interests.
Akdogan, Z., and Guven, B., 2019. Microplastics in the environment: A critical review of current understanding and identification of future research needs. Environmental Pollution, 254: 113011. DOI:10.1016/j.envpol.2019.113011 (  0) 0) |
An, R., Wang, X., Yang, L., Zhang, J., Wang, N., Xu, F., et al., 2021. Polystyrene microplastics cause granulosa cells apoptosis and fibrosis in ovary through oxidative stress in rats. Toxicology, 449: 152665. DOI:10.1016/j.tox.2020.152665 (  0) 0) |
Ašmonaitė, G., Sundh, H., Asker, N., and Almroth, B. C., 2018. Rainbow trout maintain intestinal transport and barrier functions following exposure to polystyrene microplastics. Environmental Science & Technology, 52: 14392-14401. (  0) 0) |
Azeem, I., Adeel, M., Ahmad, M. A., Shakoor, N., Jiangcuo, G. D., Azeem, K., et al., 2021. Uptake and accumulation of nano/ microplastics in plants: A critical review. Nanomaterial (Basel), 11: 2935. DOI:10.3390/nano11112935 (  0) 0) |
Banerjee, A., and Shelver, W. L., 2021. Micro- and nanoplastic induced cellular toxicity in mammals: A review. The Science of the Total Environment, 755: 142518. DOI:10.1016/j.scitotenv.2020.142518 (  0) 0) |
Brown, G. D., Willment, J. A., and Whitehead, L., 2018. C-type lec- tins in immunity and homeostasis. Nature Reviews Immunology, 18: 374-389. DOI:10.1038/s41577-018-0004-8 (  0) 0) |
Chen, J. C., Chen, M. Y., Fang, C., Zheng, R. H., Jiang, Y. L., Zhang, Y. S., et al., 2020. Microplastics negatively impact embryogenesis and modulate the immune response of the marine medaka Oryzias melastigma. Marine Pollution Bulletin, 158: 111349. DOI:10.1016/j.marpolbul.2020.111349 (  0) 0) |
Chen, Y., Chen, Y., Shi, C., Huang, Z., Zhang, Y., Li, S., et al., 2018. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience, 7: 1-6. (  0) 0) |
Cormier, B., Cachot, J., Blanc, M., Cabar, M., Clérandeau, C., Dubocq, F., et al., 2022. Environmental microplastics disrupt swimming activity in acute exposure in Danio rerio larvae and reduce growth and reproduction success in chronic exposure in D. rerio and Oryzias melastigma. Environmental Pollution, 308: 119721. DOI:10.1016/j.envpol.2022.119721 (  0) 0) |
Curtin, F., and Schulza, P., 1998. Multiple correlations and bonferroni's correction. Biological Psychiatry, 44: 775-777. DOI:10.1016/S0006-3223(98)00043-2 (  0) 0) |
Espinosa, C., Beltrán, J. M. G., Esteban, M. A., and Cuesta, A. G., 2018. In vitro effects of virgin microplastics on fish headkidney leucocyte activities. Environmental Pollution, 235: 30-38. DOI:10.1016/j.envpol.2017.12.054 (  0) 0) |
Ge, M., Liu, G., Liu, H., and Liu, Y., 2020. Levels of metals in fish tissues of Liza haematocheila and Lateolabrax japonicus from the Yellow River Delta of China and risk assessment for consumers. Marine Pollution Bulletin, 157: 111286. DOI:10.1016/j.marpolbul.2020.111286 (  0) 0) |
Govender, J., Naidoo, T., Rajkaran, A., Cebekhulu, S., Bhugeloo, A., and Sershen, Y., 2022. Towards characterising microplastic abundance, typology and retention in mangrove-dominated estuaries. Water, 12(10): 2802. (  0) 0) |
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., and Bowden, J., 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols, 8: 1494-1512. DOI:10.1038/nprot.2013.084 (  0) 0) |
Hidalgo-Ruz, V. L., Gutow, R. C., and Thompson, M. T., 2012. Microplastics in the marine environment: A review of the methods used for identification and quantification. Environmental Science & Technology, 46(6): 3060-3075. (  0) 0) |
Hou, J., Lie, Z., Cui, L., Hou, Y., Yang, L., An, R., et al., 2021. Polystyrene microplastics lead to pyroptosis and apoptosis of ovarian granulosa cells via NLRP3/Caspase-1 signaling pathway in rats. Ecotoxicology and Environmental Safety, 212: 112012. DOI:10.1016/j.ecoenv.2021.112012 (  0) 0) |
Kagan, J. C., and Iwasaki, A., 2012. Phagosome as the organelle linking innate and adaptive immunity. Traffic, 13: 1053-1061. DOI:10.1111/j.1600-0854.2012.01377.x (  0) 0) |
Kanehisa, M., Furumichi, M., Sato, Y. Y., Kawashima, M., and Ishiguro-Watanabe, M., 2023. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Research, 51: D587-D592. DOI:10.1093/nar/gkac963 (  0) 0) |
Kim, H. S., Lee, B. Y., Won, E. J., Han, J., Hwang, D. S., Park, H. G., et al., 2015. Identification of xenobiotic biodegradation and metabolism-related genes in the copepod Tigriopus japonicus whole transcriptome analysis. Marine Genomics, 24: 207-208. DOI:10.1016/j.margen.2015.05.011 (  0) 0) |
Kwon, W., Kim, D., Kim, H. Y., Jeong, S. W., Lee, S. G., Kim, H. C., et al., 2022. Microglial phagocytosis of polystyrene microplastics results in immune alteration and apoptosis in vitro and in vivo. Science of the Total Environment, 807(Pt 2): 150817. (  0) 0) |
Limonta, C., Mancia, A., Benkhalqui, A., Bertolucci, C., Abelli, L., Fossi, M. C., et al., 2019. Microplastics induce transcriptional changes, immune response and behavioral alterations in adult zebrafish. Scientific Reports, 9: 15775. (  0) 0) |
Love, M. I., Huber, W., and Anders, S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15: 550. (  0) 0) |
Lu, Y., Zhang, Y., Deng, Y., Jiang, W., Zhao, Y., Geng, J., et al., 2016. Uptake and accumulation of polystyrene microplastics in zebrafish (Danio rerio) and toxic effects in liver. Environmental Science & Technology, 50: 4054-4060. (  0) 0) |
Manni, M., Berkeley, M. R., Seppey, M., and Zdobnov, E. M., 2021. BUSCO: Assessing genomic data quality and beyond. Current Protocols, 1: e323. (  0) 0) |
Pedà, C., Caccamo, L., Fossi, M. C., Gai, F., Andaloro, F., Genovese, L., et al., 2016. Intestinal alterations in European sea bass Dicentrarchus labrax (Linnaeus, 1758) exposed to microplastics: Preliminary results. Environmental Pollution, 212: 251-256. (  0) 0) |
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al., 2003. TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics, 19: 651-652. (  0) 0) |
Qi, Z., Pi, X., Xu, Y., Zhang, Q., Wangkahart, E., Meng, F., et al., 2022. Molecular characterization of the evolutionary conserved signaling intermediate in Toll pathways (ECSIT) of soiny mullet (Liza haematocheila). Fish and Shellfish Immunology, 130: 79-85. (  0) 0) |
Qi, Z., Wu, P., Zhang, Q., Wei, Y., Wang, Z., Qiu, M., et al., 2016. Transcriptome analysis of soiny mullet (Liza haematocheila) spleen in response to Streptococcus dysgalactiae. Fish and Shellfish Immunology, 49: 194-204. (  0) 0) |
Ragusa, A., Notarstefano, V., Svelato, A., Belloni, A., Gioacchini, G., Blondeel, C., et al., 2022. Raman microspectroscopy detection and characterisation of microplastics in human breastmilk. Polymers (Basel), 14(13): 2700. (  0) 0) |
Rao, J., Peng, L., Liang, X., Jiang, H., Geng, C., Zhao, X., et al., 2020. Performance of copy number variants detection based on whole-genome sequencing by DNBSEQ platforms. BMC Bioinformatics, 21: 518. (  0) 0) |
Rummel, C. D., Löder, M. G. J., Fricke, N. F., Lang, T., Griebeler, E. M., Janke, M., et al., 2016. Plastic ingestion by pelagic and demersal fish from the North Sea and Baltic Sea. Marine Pollution Bulletin, 102: 134-141. (  0) 0) |
Sánchez-Hernández, L. J., Ramírez-Romero, P., Rodríguez- González, F., Ramos-Sánchez, V. H., Montes, R. A. M., Rubio, H. R. P., et al., 2021. Seasonal evidences of microplastics in environmental matrices of a tourist dominated urban estuary in Gulf of Mexico, Mexico. Chemosphere, 277: 130261. (  0) 0) |
Schmitt, H., Neurath, M. F., and Atreya, R., 2021. Role of the IL23/IL17 pathway in Crohn's disease. Frontier in Immunology, 12: 622934. (  0) 0) |
Shabaka, S., Moawad, M. N., Ibrahim, M. I. A., EI-Sayed, A. A. M., Ghobashy, M. M., Hamouda, A. Z., et al., 2022. Youssef. Prevalence and risk assessment of microplastics in the Nile Delta estuaries: 'The plastic nile' revisited. Science of the Total Environment, 852: 158446. (  0) 0) |
Steer, M., Cole, M., Thompson, R. C., and Lindeque, P. K., 2017. Microplastic ingestion in fish larvae in the western English Channel. Environmental Pollution, 226: 250-259. (  0) 0) |
Sui, Q., Zhang, L., Xia, B., Chen, B., Zhu, L., Wang, R. Y., et al., 2020. Spatiotemporal distribution, source identification and inventory of microplastics in surface sediments from Sanggou Bay, China. Science of the Total Environment, 723: 138064. (  0) 0) |
The Gene Ontology Consortium, 2019. The gene ontology resource: 20 years and still going strong. Nucleic Acids Research, 47: D330-D338. (  0) 0) |
Veneman, W. J., Spaink, H. P., Brun, N. R., Bosker, T., and Vijver, M. G., 2017. Pathway analysis of systemic transcriptome responses to injected polystyrene particles in zebrafish larvae. Aquatic Toxicology, 190: 112-120. (  0) 0) |
Umamaheswari, S., Priyadarshinee, S., Kadirvelu, K., and Ramesh, M., 2021. Polystyrene microplastics induce apoptosis via ROS- mediated p53 signaling pathway in zebrafish. Chemico-Biological Interactions, 345: 109550. (  0) 0) |
Uribe-Querol, E., and Rosales, C., 2020. Phagocytosis: Our current understanding of a universal biological process. Frontiers in Immunology, 11: 1066. (  0) 0) |
Wang, C., Hou, M., Shang, K., Wang, H., and Wang, J., 2022. Microplastics (polystyrene) exposure induces metabolic changes in the liver of rare minnow (Gobiocypris rarus). Molecules, 27: 584. (  0) 0) |
Wen, B., Jin, S. R., Chen, Z. Z., Gao, J. Z., Liu, Y. N., Liu, J. H., et al., 2018. Single and combined effects of microplastics and cadmium on the cadmium accumulation, antioxidant defence and innate immunity of the discus fish (Symphysodon aequifasciatus). Environmental Pollution, 243: 462-471. (  0) 0) |
Yan, M., Nie, H., Xu, K., He, Y., Hu, Y., Huang, Y., et al., 2019. Microplastic abundance, distribution and composition in the Pearl River along Guangzhou City and Pearl River Estuary, China. Chemosphere, 217: 879-886. (  0) 0) |
Yan, W., Hamid, N., Deng, S., Jia, P. P., and Pei, D. S., 2020. Individual and combined toxicogenetic effects of microplastics and heavy metals (Cd, Pb, and Zn) perturb gut microbiota homeostasis and gonadal development in marine medaka (Oryzias melastigma). Journal of Hazardous Materials, 397: 122795. (  0) 0) |
Zhang, J., Zhang, C., Deng, Y., Wang, R., Ma, E., Wang, J., et al., 2019. Microplastics in the surface water of small-scale estuaries in Shanghai. Marine Pollution Bulletin, 149: 110569. (  0) 0) |
Zhang, M., and Zhang, X., 2019. The role of PI3K/AKT/FOXO signaling in psoriasis. Archives of Dermatological Research, 311: 83-91. (  0) 0) |
Zheng, J., Hu, Z. H., Guan, X. L., Dou, D. Q., Bai, G., Wang, Y., et al., 2015. Transcriptome analysis of Syringa oblata Lindl. inflorescence identifies genes associated with pigment biosynthesis and scent metabolism. PLoS One, 10: e0142542. (  0) 0) |
 2025, Vol. 24
2025, Vol. 24


