2) College of Marine Science and Engineering, Nanjing Normal University, Nanjing 210023, China;
3) Marine Science Research Institute of Shandong Province (National Oceanographic Center, Qingdao), Qingdao 266104, China
Paralichthys olivaceus is a commercially important teleost fish that is widely farmed across the world, especially in China, Japan, and Korea (Xiu et al., 2019). Bacteria, parasites and viruses are well-known infectious pathogens (Kim et al., 2010), which cause mixed infectious illnesses, countless fatalities, and massive economic losses (Xu and Zhang, 2013). Edwardsiella tarda is a Gram-negative bacterium that can cause intestinal or extraintestinal infections in fish. In previous studies, Danio rerio (Liu et al., 2014), Sebastes schlegelii (Cao et al., 2020), tilapia (Wu et al., 2018), Scophthalmus maximus L. (Chen et al., 2020b) and P. olivaceus (Tang et al., 2019) with intestinal inflammation are more susceptible to E. tarda. Meanwhile, the antimicrobial response during E. tarda infection might inhibit the growth of intestinal microbiota. Particularly, P. olivaceus infected with E. tarda shows various intestinal and extraintestinal diseases, causing huge losses in the aquaculture industry (Xiu et al., 2019, 2021). Therefore, researches on the innate immune regulation mechanisms of flounder may contribute to the development of disease-control measures and enhance the development of flounder aquaculture.
MicroRNAs (miRNAs) are a class of non-coding short RNA molecules with 22 – 24 nucleotides which play the significant role in transcriptional regulation through blocking mRNA translating or encouraging mRNA breakdown (Hatfield et al., 2005; Wienholds and Plasterk, 2005). Previous studies have suggested that miRNAs were attractive tools and targets for novel therapeutic approaches in cancer. In human, miR-144 is involved in the pathological development and progress of many malignancy tumors (Shi et al., 2019; Kooshkaki et al., 2020). Moreover, miR-144 has the ability to promote the pathological progress of some cancers through suppressing or accelerating tumor cell proliferation, migration and invasion (Cui and Wang, 2015; Cheng et al., 2020). On the other hand, miR-144 also plays a vital role in regulating the maturity, differentiation and immune functions of immune cells (Li et al., 2018). Particularly, ectopic expression of miR-144 can effectively block cell barrier disruption and hyperpermeability to regulate activation of inflammatory pathway in response to proinflammatory agents (Siddiqui et al., 2019).
In fish, substantial progress was made in recognizing the role of miR-144 during the last decade. On the one hand, with the progress and development of high-throughput sequencing technologies, miR-144 was differentially expressed in response to external stimulus in several fish (Wu et al., 2021). Moreover, miR-144 regulated a variety of physiological activities in vivo by targeting different genes. For example, miR-144 regulats erythropoiesis via repressing Dicer (Kretov et al., 2020) as well as hematopoiesis and vascular development by targeting meis1 (Su et al., 2014) in zebra fish. Additionally, miR-144 also mediates high fatinduced changes of cholesterol metabolism via targeting C/EBPα in yellow catfish (Chen et al., 2020a). Moreover, in terms of antibacterial function, it has been observed that in response to the infection of different bacteria including S. iniae (Liu et al., 2020b), E. tarda (Li et al., 2019), and megalocytivirus (Zhang et al., 2014), pol-miR-144-5p expression varied dramatically, indicating miR-144-5p might be a regulator of antibacterial immune response. In innate immunity, miR-144 were proposed to influence innate immunity in fish by binding immunity-related genes. Xu' group have reported that miR-144 played regulatory roles in inflammatory responses by regulating the NF-κB signaling pathway via targeting NOD1 (Chu et al., 2021) and IL-1β (Yan et al., 2019) in miiuy croaker. Thus, miR-144 plays important roles with antimicrobial function as well as regulation of immune and inflammatory responses in innate immunity.
In this study, we aim to investigate the underlying regulation mechanism of miR-144-5p in intestinal mucosal immunity of flounder. Firstly, we effectively inhibited miR-144-5p expression by miR-144-5p interference in vivo. Then, we detected the differentially expressed genes (DEGs) by high-throughput sequencing technology. Next, in order to confirm the immunity-related signal pathways influenced by miR-144-5p, DEGs were analyzed by Gene Ontology (GO) functionality enrichment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. At last, we predicted protein-protein interaction networks. These findings will contribute to a better knowledge of the immunological regulation mechanisms of miR-144-5p in intestine of flounder and other fishes.
2 Materials and Methods 2.1 Experimental Fish and miRNA AntagomirAll healthy flounders were offered by the Huanghai Aquaculture Company (Haiyang, Shandong, China). The fish were approximately 24 weeks old, with a body length of 12 cm ± 4 cm (mean ± SD) and a weight of 10 g ± 3 g (mean ± SD). Before the trials, they have been habituated in a recirculating water system at 17℃ ± 1℃ for 7 days. miR-144-5p antagomir, and antagomir-NC (negative control of miR-144-5p antagomir) utilized in this study were synthesized by Genepharma, China.
2.2 In vivo Injection and qRT-PCRHealthy P. olivaceus were randomly separated into three groups (10 fish per group). Two experimental groups were interfered by 2 μg g−1 miR-144-5p antagomir or antagomir-NC, while the control group was without any treatment. Twenty four hours after injection, the intestine tissue was collected under aseptic conditions. To examine the expression level of miR-144-5p, the collected intestines of flounders was used for RNA isolation and qRT-PCR.
The PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Japan) has been used to create the cDNA. Then, based on the sequence of miR-144-5p, appropriate primers were constructed, with Po5s serving as an internal reference. The CFX96 real-time fluorescent quantitative PCR system (Bio-Rad, USA) with TB GreenTM Premix Ex TaqTM Ⅱ was used for the qRT-PCR (TaKaRa, Japan). The amplifying procedure included one cycle at 95℃ for 30 s, followed by 40 cycles of 5 s at 95℃ and 1 min at 60℃, then a melted curve from 60 to 95℃. The data were presented as means ± SE for three variables, and statistical analysis was conducted through SigmaPlot 11.0.
2.3 Transcriptome Sequencing Library ConstructionTRIzol Reagent (Invitrogen, USA) has been utilized to isolate total RNA according with the manufacturer guidelines. And 1% agarose gel electrophoresiswas employed to determine the quality and quantity of extracted RNA. Furthermore, the NanoPhotometer ® spectrophotometer (IMP-LEN, CA, USA), the RNA Assay Kit in the Qubit ® 2.0 Flurometer (Life Technologies, CA, USA) and the Bioanalyzer 2100 system's RNA Nano 6000 Assay Kit (Agilent Technologies, CA, USA) has been utilized to measure RNA purity, concentration and integrity, respectively. Approximately 22.335 μg of total RNA was subjected to isolate Poly(A) mRNA with poly-T oligo attached magnetic beads. Cracking has been performed in First Strand Synthesis Reaction Buffer (5X) utilizing divalent cations at high temperatures. RNaseH has been used to breakdown the RNA after the first strand cDNA was generated using a randomized hexamer primer and M-MuLV Reverse Transcriptase. Following that, second strand cDNA synthesis has been carried out utilizing DNA Polymerase Ⅰ and dNTP. The library fragments were reverse-transcribed to identify cDNA segments with 370 – 420 bp length (Beckman Coulter, Beverly, USA) and then confirmed the quality of the database. Finally, the libraries were analyzed on an Illumina NovaSeq 6000 platform, yielding 150 bp paired-end reads following cluster generation.
2.4 Transcriptome Assembly and Novel Transcripts PredictionRaw data were handled and then clean data were acquired via using in-house perl programs including remove adapter-containing readings, N base-containing reads, and low-quality reads in original data. Meanwhile, the Q20, Q30, and GC contents of clean data were determined. Afterwards, standard genome and genetic modeling annotation data were obtained directly from the genomic site (ftp: /ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/970/005/GCF 001970005.1 Flounder ref guided V1.0/). Hisat2 (v2.0.5) was used to create the reference genome index, and mapped clean reads to the reference genome. Furthermore, the mapped reads of each sample were assembled by String-Tie (v1.3.3b) in a reference-based approach.
2.5 Different Expression Genes Analysis and miRNA-mRNA PredictionThe featureCounts v1.5.0-p3 program was used to check the amount of reads that matched each transcript. The FPKM of every transcript was computed based on its length and the number of reads linked to it. FPKM stands for the assumed number of fragments per kilobase of transcript sequence per million base pairs sequenced, and it takes into account the impact of assembling depth and genomic length on read count simultaneously, which was presently used to estimate gene expression levels as the most commonly method. The DESeq2 R package (1.20.0) was used to analyze the different expressions of the genes in the three groups (3 biological replicas per condition). Padj ≤ 0.05 and |log2 (Fold change)| ≥ 1 were chosen as the cutoffs for significantly different expression. The potential target genes of miR-144-5p were predicted using the TargetScan 8.0 database. Total of 3003 differentially expressed transcripts were submitted and further predicted.
2.6 GO and KEGG Enrichment Analysis of Differentially Expressed GenesGO enrichment assessment and KEGG pathway studies have been conducted on all of the DEGs to better understand their functional relevance. GO enrichment analysis of differentially expressed genes was implemented by the ClusterProfiler R package (3.8.1). KEGG is a database for understanding key functions and utilities of organisms, including the cell, embryo, and ecosphere, through the molecular-level data, especially large-scale molecular datasets produced by genetic analysis and other high-throughput experimental techniques. The ClusterProfiler R package (3.8.1) was used to examine the statistical enrichment of pathways in KEGG networks.
2.7 Establishing Protein-Protein Interaction NetworksProtein-protein interaction networks were generated to evaluate the connection of genes in the immune regulatory pathway by using PPI (protein-protein interaction) and WGCNA (Weighted correlation network analysis). To further explore the relationship between the genes in the immunological regulatory system, protein-protein interaction networks were generated using Cytoscape 3.9.0 (https://github-releases.org/) with default settings.
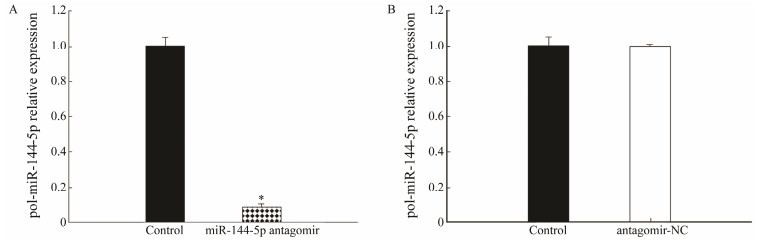
3 Results 3.1 Effect of miR-144-5p InterferenceTo examine the effect of miR-144-5p interference, the expression of miR-144-5p was determined by qRT-PCR. miR-144-5p in flounder can be knocked down by miR-144-5p antagomir. The qRT-PCR results showed that in the presence of miR-144-5p antagomir, the expression of miR-144-5p was significantly reduced (Fig. 1A). In contrast, the presence of antagomir-NC had no effect on miR-144-5p expression (Fig. 1B).
|
Fig. 1 The effect of miR-144-5p interference. (A) Compared with the control, miR-144-5p antagomir caused significant downregulation of miR-144-5p. (B) Compared with the control, antagomir-NC had no significant effect on miR-144-5p expression. |
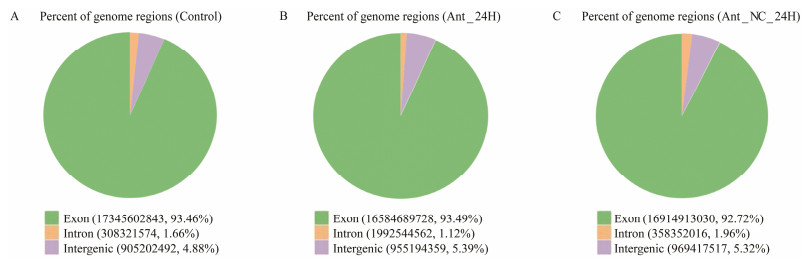
The Illumina-based RNA-Seq was performed on three groups each with three replicates in intestine of flounder 24 hours after the injection of antagomir (named as Ant_ 24H_1, Ant_24H_2, Ant_24H_3), antagomir-NC (named as Ant_NC_24H_1, Ant_NC_24H_2, Ant_NC_24H_3) or without any treatment (named as Control_1, Control_2, and Control_3). We acquired 137343982, 141233938 and 142285934 raw reads from the Ant_24H, Ant_NC_24H and control group, respectively. Then, these raw data were further filtered to obtain 134891344, 137509356, 139274542 clean reads. All the databases have high quality, with clean base values ≥ 6.5 G, error rates ≤ 0.03, Q20 ≥ 97.62% and Q30 ≥ 93.66%. As the result, all of the libraries were determined to be suitable for future research. The RNA-Seq results are summarized in Table 1. Then, from each group, a total of 118712851, 122177868 and 124245564 clean reads which accounted for approximately 88%, 88.85% and 89.21% of the total clean reads were matched to the reference sequence (Table 2). Moreover, on average 93.46%, 93.49%, 92.72% of the clean reads were found in the exon regions of the genome from three groups, as shown in Fig. 2. Finally, in three groups, a total of 24387 genes were identified for additional quantitative analysis and functional annotation.
|
|
Table 1 Summary of miR-144-5p sequencing data |
|
|
Table 2 Summary of sequencing and mapping |
|
Fig. 2 The regional distribution of sequenced reads in the genome. The green, yellow and purple areas in the pie chart represent ratios of sequenced reads to exon, intron and intergenic regions. |
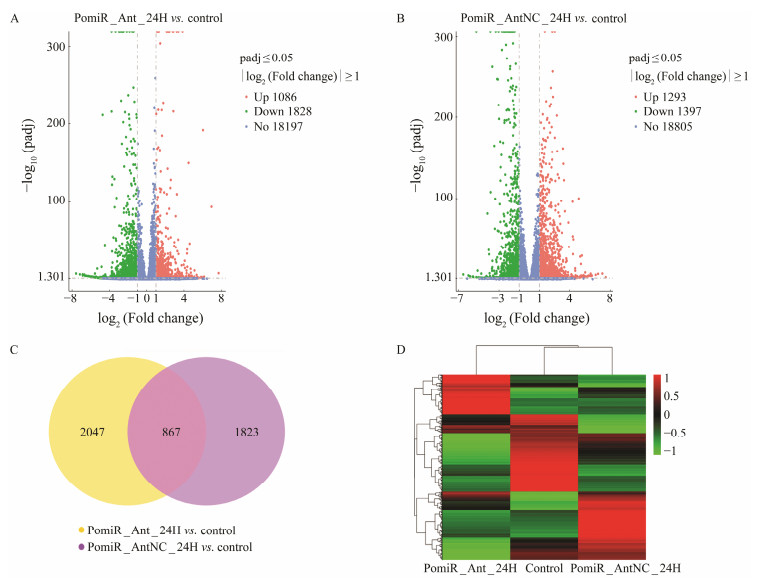
In contrast to the control group, there were 2914 DEGs as a result of miR-144-5p interference (P < 0.05), including 1828 significantly downregulated genes and 1086 significantly upregulated genes (Fig. 3A). Meanwhile, there were 2690 DEGs following antagomir-NC interference (P < 0.05), including 1397 significantly downregulated genes and 1293 significantly upregulated genes (Fig. 3B). According to the Venn diagram, 2047 transcripts were not affected in the Ant_NC_24H but changed in Ant_24H, accounting for about 39.3% of the identified genes (Fig. 3C). We utilized heatmap analyses of hierarchical clustering to identify the characteristics of the DEGs in this study. As shown in Fig. 3D, the heatmap analysis revealed that DEGs in the control and PomiR_Ant_NC_24H groups exhibited comparable clustering and expression patterns, whereas the Po-miR_Ant_24H group has distinct expression features. These findings suggest that miR-144-5p interference affected gene expression in flounder intestinal tissue, indicating that the impact of miR-144-5p on gene expression regulation should not be overlooked.
|
Fig. 3 Differentially expressed genes identification and analysis. (A) Volcano Plot of differently expressed genes distribution trends between PomiR_Ant_24H and control groups. (B) Volcano Plot of differently expressed genes distribution trends between PomiR_AntNC_24H and control groups. (C) Venn diagrams of DEGs shows the comparison of PomiR_ Ant_24H group vs. control group and PomiR_AntNC_24H group vs. control group. The yellow region of Venn diagrams shows the DEGs which were not affected in the Ant_NC_24H group but changed in Ant_24H group. (D) Heatmap analyses of hierarchical clustering of DEGs in the three groups (PomiR_Ant_24H, PomiR_AntNC_24H and control). |
In our study, a total of 2117 potential targeted genes were obtained. We selected 155 potential targeted genes which were stable in the Ant_NC_24H group but changed in Ant_ 24H group (Table 3), and further screened out 22 potential targeted genes in enriched immunity-related pathways. For example, apaf1 can participate in the regulation of various immune signaling pathways including p53 signaling pathway, Apoptosis and Herpes simplex virus 1 infection. LOC109641154 can be involved in the immune response of Focal adhesion, ErbB signaling pathway, C-type lectin receptor signaling pathway, MAPK signaling pathway and Salmonella infection, as shown in Table 3. These results suggested that miR-144-5p might regulate the immune response of flounder through targeting immunity-related genes.
|
|
Table 3 miR-144-5p target genes in the enrichment of immunity-related pathways |
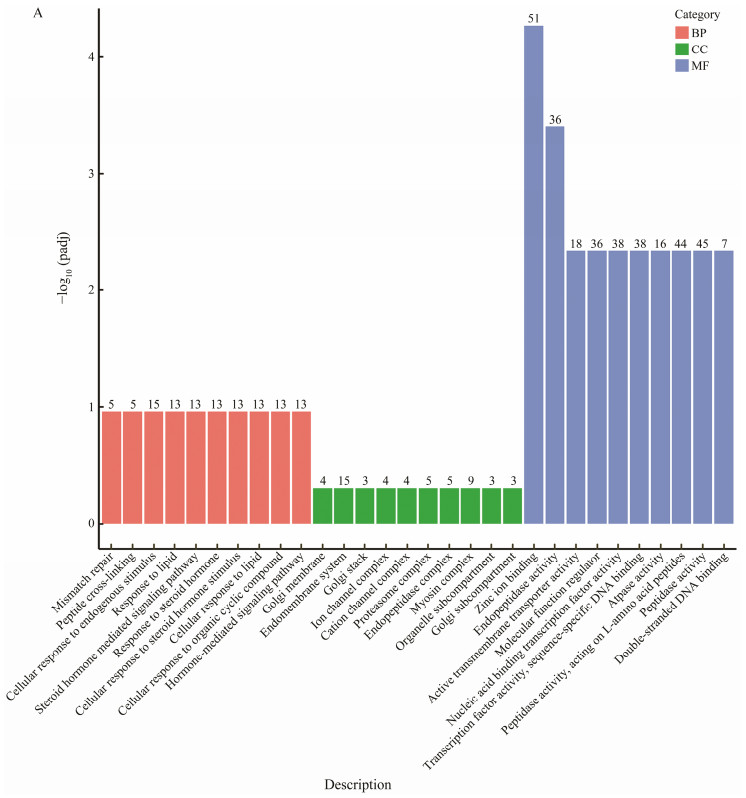
In order to investigate the potential role of DEGs, we performed GO enrichment and KEGG pathway analysis. DEGs, which were stable in the Ant_NC_24H group but changed in the Ant_24H group, were divided into three groups based on their GO functional enrichment analysis. A total of 2318 genes were matched to biological process (BP), 618 genes were matched to cellular component (CC), and 2084 genes were matched to molecular function (MF). The top 30 GO terms in these three categories are displayed in Fig. 4A. Most of the differential expression genes were enriched into cellular response to endogenous stimulus in biological process, endomembrane system in cellular component, and zinc ion binding in molecular function. Otherwise, several immunity-related GO terms, such as cell adhesion, regulation of cell death, regulation of apoptotic process, Wnt signaling pathway, cell-cell signaling by wnt, immune systemcomponents and immune response were significantly enriched in the biological process category (Table 4).
|
Fig. 4 (A) Functional annotation of DEGs which were stable in the Ant_NC_24H group but changed in Ant_24H group. Gene ontology (GO) enrichment analysis of DEGs. Distribution of GO annotation in three categories. The y-axis is the P-values of DEGs; the x-axis is the gene functional classification of GO. |
|
|
Table 4 immunity-related GO terms were significantly enriched in the biological process category |
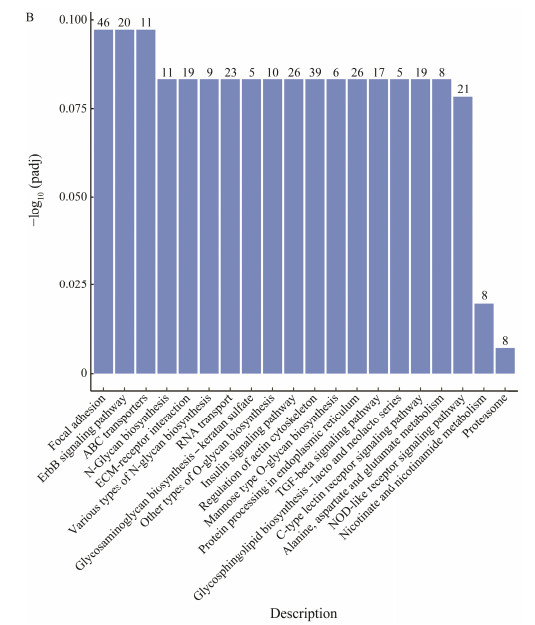
The KEGG enrichment analysis enabled us to get a greater comprehension of the biological functions of DEGs. In this study, 1355 DEGs among 2047 DEGs that were stable in the Ant_NC_24H group but changed in Ant_24H group were enriched in 147 signaling pathways. In order to depict the KEGG pathway enrichment data, the top 20 most significantly enriched pathways were shown in Fig. 4B. According to the KEGG pathway enrichment data, the DEGs have a substantial association with immunity-related signaling pathways, including NOD-like receptor, MAPK, Wnt, mTOR, Toll-like receptor signaling pathway. A summary of the 33 key immunity-related signaling pathways were showed in Table 5. In this study, function analysis of DEGs via GO and KEGG reveals that an abundance of immunity-related genes were regulated by miR-144-5p.
|
Fig. 4 (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs. The y-axis is the P-values of DEGs; the x-axis is KEGG classes. |
|
|
Table 5 immunity-related signaling pathways in KEGG enrichment analysis |
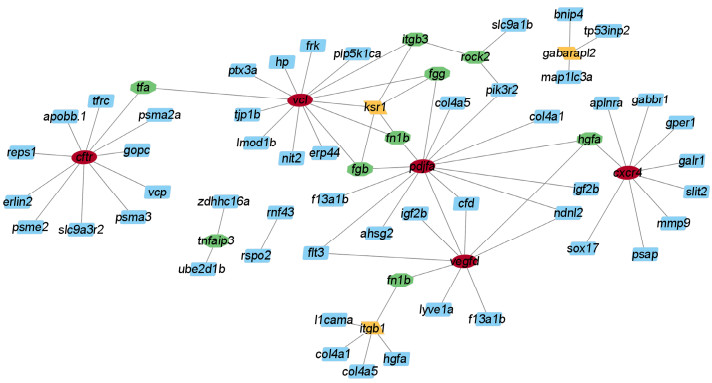
In vivo, proteins are the material underpinnings of physiological actions. The creation of immunity-related protein-protein interaction networks might aid in the discovery of key genes implicated in immunoreaction. Protein-protein function interactions were the core of regular physiological processes in P. olivaceus. The immunity-related protein-protein association networks have been constructed utilizing 67 DEGs from 33 immunity-related signaling pathways which were unaltered in the Ant_NC_24H but changed in Ant_24H (Table 5). As shown in Fig. 5, some nodes with several edges are thought to represent immune response hub genes and we opted to further investigate and validate these hub genes.
|
Fig. 5 Immunity-related protein-protein interaction networks. Network nodes represent proteins. Edges stand for protein-protein associations. |
As a result, 5 core genes with multiple connections (degree ≥ 8) were discovered, including platelet-derived growth factor alpha polypeptide b (pdjfa), vinculin (vcl), vascular endothelial growth factor D (vegfd), C-X-C motif chemokine receptor 4 (cxcr4) and CF transmembrane conductance regulator (cftr). The top 20 DEGs most associated with immunity identified by WGCNA are presented in Table 6.
|
|
Table 6 Summary of the 20 immunity-related DEGs identified by WGCNA |
In teleost fishes, miRNAs play a crucial role in immune cell differentiation, immune reaction, as well as the development of immunological disorders (Chu and Xu, 2020). The intestine of flounder represent an important organ for immunological response (Xiu et al., 2019). In order to research the immune regulation function of miR-144-5p in the intestine of flounder, we screened the differentially expressed genes by transcriptome sequencing after miR-144-5p interference. In this study, 2047 DEGs and 1823 DEGs were identified when comparing the fish in Ant_24H group with the control fish, and the fish in Ant_NC_24H group with the control fish, respectively. To further investigate the immunological response of miR-144-5p targeted DEGs, we selected 2047 genes that were unaffected in the Ant_ NC_24H group but changed in Ant_24H group. We discovered DEGs in the control and Ant_NC_24H groups had similar clustering and expression patterns, while the Ant_ 24H group has distinguishable expression features as a result of heatmap studies of hierarchical clustering. This result suggests that the immunological regulation of miR-144-5p should not be overlooked.
Based on the DEGs, GO and KEGG functional analyses display that there are many immunity-related genes in the intestinal transcriptome that respond to miR-144-5p interference, showing a complex immunological defensive response in P. olivaceus intestine. The thorough study of these GO enrichment and KEGG signaling pathways will help the research of the miR-144-5p immunoregulation in intestines of flounder, as well as give some significant hints for prevention and treatment of disease. We found 503 DEGs were highly enriched in 33 immunity-related pathways which were unaffected in the Ant_NC_24H group but changed in Ant_24H group.
Several signaling pathways were identified in our research, such as the NOD-like and Toll-like receptor signaling pathways, the phagosome, the intestinal immune network for IgT (IgA) production, as well as the p53 signaling pathway. In previous studies, miR-144 was discovered to be involved in the regulation of immune responses via regulating immunity-related signaling pathways. For example, in human, miR-144 promotes IL-1β-induced chondrocyte pyroptosis by regulating the NOD-like signaling pathway (Jiang et al., 2021). Additionally, miR-144-5p inhibits cancer cells proliferation by regulating the Toll-like signaling pathway via targeting Toll-like receptor 2 (TLR2) (Liu et al., 2020a). Moreover, miR-144 induced apoptosis and autophagy in lung cancer cells via regulating P53 signaling pathway by targeting TIGAR (Chen et al., 2015). In mouse, miR-144-5p agomirs mitigated M1 macrophage-associated inflammation by regulating the NF-κB signaling pathway via targeting TLR2 (Shi et al., 2020). In fish, miR-144 play regulatory roles in inflammatory responses after bacterial infection by regulating the NF-κB signaling via targeting NOD1 (Chu et al., 2021) and IL-1β (Yan et al., 2019) in miiuy croaker. Overall, we suggested that the miR-144 not only regulate cancer cells proliferation but also plays several roles including antimicrobial function and regulation of inflammatory responses in immune regulation.
In the miRNA-mediated immune response, functional connections between miRNA and genes are crucial. We discovered dynamic variations in the expression of a wide variety of immunity-related genes. On the basis of the 67 immunity-related DEGs, we constructed protein-protein interaction networks. As shown in Fig. 4, 67 immunity-related DEGs yielded 5 hypothesized hub genes, each of which was engaged in several protein-protein connections or KEGG pathways.
Depending on protein-protein association data, we determined that pdgfa, vegfd, and vcl are the three top hub genes (Fig. 4). Pdgfa was involved in cell survival, immunity and wound healing via regulating focal adhesion by interacting with vcl and vegfd (Wang et al., 2019). In our study, pdgfa, vegfd, and vcl were significantly down-regulated 24 h after miR-144-5p interference. Similar to previous observations, miR-93, miR-155 and miR-10b inhibitors suppress the expression of pdgfa, vegfd and vcl, respectively (Ibrahim et al., 2012; Chang et al., 2019; Feng et al., 2019). In a recent research, pdgfa connected with proinflammatory cytokines (TNF-α, IL-6, IL-1, IL-8) and chemokines (CXCL-1 and CCL2) was regulated by miR-29a (Wang et al., 2019). In zebrafish, vegfd is involved in angiogenesis and lymphangiogenesis through regulation by miR-194-5p (Song et al., 2007). Otherwise, deficiency of miR-663b-mediated vcl can promote epicardial hyperplasia and coronary vessel disruption (You et al., 2021). Our findings suggest that in fish, miR-144-5p can regulate the expressions of pdgfa, vegfd, and vcl which might play an important role to mitigate inflammatory responses against invading pathogens. Other hub genes, such as cftr and cxcr4, can regulate embryonic organizer formation in zebrafish (Liu et al., 2020c) and activate chemokine signalling pathway in immune responses to infection in channel catfish (Yeh and Klesius, 2010), respectively. Furthermore, miR-200b participates in the immunoreaction of hosts by regulating Wnt pathway via targeting cftr (Liu et al., 2020c) and TFA (Xu et al., 2018). Cxcr4 can regulate leukocyte chemotaxis, inflammatory response and numerous autoimmune diseases through the regulation by miR-23a (Pan et al., 2018) and participate in the intestinal immune network for IgT (IgA) production pathway (Mousavi, 2020). The findings in this research will be utilized to further research the miRNA-mediated immunological response to bacterial infections in flounder. The regulation function of miRNA need to be further studied in immune system of the flounder in the future.
5 ConclusionsIn our study, RNA-Seq was utilized to detect transcriptional expression in the intestine of flounder in response to miR-144-5p interference. Primarily, we detected 2047 DEGs which were unaffected in the Ant_NC_24H group but changed in Ant_24H group. Then, 503 DEGs were identified to be enriched in 33 immunity-related pathways, demonstrating a broad immune response mediated by miR-144-5p interference. Following that analysis, a protein association interaction network was constructed, and 5 immunity-related hub genes were discovered to be engaged in immune regulation. Our findings emphasize the regulation roles of miR-144-5p in the flounder immune reaction on inflammatory response, antibacterial function and autoimmune diseases.
AcknowledgementsThis work was supported by the National Natural Science Foundation of China (No. 32002421), the Advanced Talents Foundation of QAU (No. 6651118016), the Natural Science Foundation of Shandong Province (No. ZR20 19BC009), the Fish Innovation Team of Shandong Agriculture Research System (No. SDAIT-12-06), the Aquatic Animal Immunologic Agents Engineering Research Center of Shandong Province, Key Research and Development Program in Shandong (No. 2018YFJH0703), the Consulting Research Project of Shandong Research Institute, China Engineering Science and Technology Development Strategy (No. 202103SDYB08), the 'First Class Fishery Discipline' Programme in Shandong Province, and the special top talent plan 'One Thing One Decision (YishiYiyi)' Programme in Shandong Province, China.
Cao, M., Yan, X., Yang, N., Fu, Q., Xue, T., Zhao, S., et al., 2020. Genome-wide characterization of Toll-like receptors in black rockfish Sebastes schlegelii: Evolution and response mechanisms following Edwardsiella tarda infection. International Journal of Biological Macromolecules, 164: 949-962. DOI:10.1016/j.ijbiomac.2020.07.111 (  0) 0) |
Chang, Y., Cui, M., Fu, X., Zhang, L., Li, X., Li, L., et al., 2019. MiRNA-155 regulates lymphangiogenesis in natural killer/T-cell lymphoma by targeting BRG1. Cancer Biology & Therapy, 20(1): 31-41. DOI:10.1080/15384047.2018.1504721 (  0) 0) |
Chen, G., Wu, K., Zhao, T., Ling, S., Liu, W., and Luo, Z., 2020a. miR-144 mediates high fat-induced changes of cholesterol metabolism via direct regulation of C/EBPα in the liver and isolated hepatocytes of yellow catfish. Journal of Nutrition, 150(3): 464-474. DOI:10.1093/jn/nxz282 (  0) 0) |
Chen, J., Zhang, L., Yang, N., Cao, M., Tian, M., Fu, Q., et al., 2020b. Characterization of the immune roles of cathepsin L in turbot (Scophthalmus maximus L.) mucosal immunity. Fish & Shellfish Immunology, 97: 322-335. DOI:10.1016/j.fsi.2019.12.005 (  0) 0) |
Chen, S., Li, P., Li, J., Wang, Y., Du, Y., Chen, X., et al., 2015. MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cellular Physiology and Biochemistry, 35(3): 997-1007. DOI:10.1159/000369755 (  0) 0) |
Cheng, B., Zhang, Y., Wu, Z. W., Cui, Z. C., and Li, W. L., 2020. MiR-144 inhibits colorectal cancer cell migration and invasion by regulating PBX3. European Review for Medical and Pharmacological Sciences, 24(18): 9361-9369. DOI:10.26355/eur-rev_202009_23019 (  0) 0) |
Chu, Q., and Xu, T., 2020. MicroRNA regulation of Toll-like receptor, RIG-I-like receptor and Nod-like receptor pathways in teleost fish: Fish miRNAs in innate immune. Reviews in Aquaculture, 12(1): 2177-2193. (  0) 0) |
Chu, Q., Bi, D., Zheng, W., and Xu, T., 2021. MicroRNA negatively regulates NF-κB-mediated immune responses by targeting NOD1 in the teleost fish Miichthys miiuy. Science China Life Sciences, 64(5): 803-815. DOI:10.1007/s11427-020-1777-y (  0) 0) |
Cui, S. Q., and Wang, H., 2015. MicroRNA-144 inhibits the proliferation, apoptosis, invasion, and migration of osteosarcoma cell line F5M2. Tumour Biology, 36(9): 6949-6958. DOI:10.1007/s13277-015-3396-0 (  0) 0) |
Feng, S., Gao, L., Zhang, D., Tian, X., Kong, L., Shi, H., et al., 2019. MiR-93 regulates vascular smooth muscle cell proliferation, and neointimal formation through targeting Mfn2. International Journal of Biological Sciences, 15(12): 2615-2626. DOI:10.7150/ijbs.36995 (  0) 0) |
Hatfield, S. D., Shcherbata, H. R., Fischer, K. A., Nakahara, K., Carthew, R. W., and Ruohola-Baker, H., 2005. Stem cell division is regulated by the microRNA pathway. Nature, 435(7044): 974-978. DOI:10.1038/nature03816 (  0) 0) |
Ibrahim, S. A., Yip, G. W., Stock, C., Pan, J. W., Neubauer, C., Poeter, M., et al., 2012. Targeting of syndecan-1 by micro-RNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. International Journal of Cancer, 131(6): E884-896. DOI:10.1002/ijc.27629 (  0) 0) |
Jiang, J. M., Mo, M. L., Long, X. P., and Xie, L. H., 2021. MiR-144-3p induced by SP1 promotes IL-1β-induced pyroptosis in chondrocytes via PTEN/PINK1/Parkin axis. Autoimmunity, 1: 1-11. DOI:10.1080/08916934.2021.1983802 (  0) 0) |
Kim, J. S., Harikrishnan, R., Kim, M. C., Balasundaram, C., and Heo, M. S., 2010. Dietary administration of Zooshikella sp. enhance the innate immune response and disease resistance of Paralichthys olivaceus against Sreptococcus iniae. Fish & Shell-fish Immunology, 29(1): 104-110. DOI:10.1016/j.fsi.2010.02.022 (  0) 0) |
Kooshkaki, O., Rezaei, Z., Rahmati, M., and Vahedi, P., 2020. MiR-144: A new possible therapeutic target and diagnostic/ prognostic tool in cancers. International Journal of Molecular Sciences, 21(7): 1422-1467. DOI:10.3390/ijms21072578 (  0) 0) |
Kretov, D. A., Walawalkar, I. A., Mora-Martin, A., Shafik, A. M., Moxon, S., and Cifuentes, D., 2020. Ago2-dependent processing allows miR-451 to evade the global microRNA turnover elicited during erythropoiesis. Molecular Cell, 78(2): 317-328.e6. DOI:10.1016/j.molcel.2020.02.020 (  0) 0) |
Li, R. D., Shen, C. H., Tao, Y. F., Zhang, X. F., Zhang, Q. B., Ma, Z. Y., et al., 2018. MicroRNA-144 suppresses the expression of cytokines through targeting RANKL in the matured immune cells. Cytokine, 108: 197-204. DOI:10.1016/j.cyto.2018.03.043 (  0) 0) |
Li, W. R., Hu, Y. H., Jiang, S., and Sun, L., 2019. Global profiling and characterization of Japanese flounder (Paralichthys olivaceus) kidney microRNAs regulated by Edwardsiella tarda infection in a time-dependent fashion. Fish & Shellfish Immunology, 93: 766-780. DOI:10.1016/j.fsi.2019.07.078 (  0) 0) |
Liu, G. M., Lu, T. C., Sun, M. L., Jia, W. Y., Ji, X., and Luo, Y. G., 2020a. Ginsenoside Rd inhibits glioblastoma cell proliferation by up-regulating the expression of miR-144-5p. Biological & Pharmaceutical Bulletin, 43(10): 1534-1541. DOI:10.1248/bpb.b20-00338 (  0) 0) |
Liu, S., Ning, X. H., Guan, X. L., Li, X. P., and Sun, L., 2020b. Characterization of Streptococcus iniae-induced microRNA profiles in Paralichthys olivaceus and identification of pol-3p-10740_175 as a regulator of antibacterial immune response. Fish & Shellfish Immunology, 98: 860-867. DOI:10.1016/j.fsi.2019.11.045 (  0) 0) |
Liu, X., Chang, X., Wu, H., Xiao, J., Gao, Y., and Zhang, Y., 2014. Role of intestinal inflammation in predisposition of Edwardsiella tarda infection in zebrafish (Danio rerio). Fish & Shellfish Immunology, 41(2): 271-278. DOI:10.1016/j.fsi.2014.09.009 (  0) 0) |
Liu, Y., Lin, Z., and Sun, H., 2020c. Cystic fibrosis transmembrane conductance regulator (CFTR) regulates embryonic organizer formation during zebrafish early embryogenesis. International Journal of Developmental Biology, 64(7-8-9): 409-413. DOI:10.1387/ijdb.190373hs (  0) 0) |
Mousavi, A., 2020. CXCL12/CXCR4 signal transduction in diseases and its molecular approaches in targeted-therapy. Immunology Letters, 217: 91-115. DOI:10.1016/j.imlet.2019.11.007 (  0) 0) |
Pan, Z., Shan, Q., Gu, P., Wang, X. M., Tai, L. W., Sun, M., et al., 2018. miRNA-23a/CXCR4 regulates neuropathic pain via directly targeting TXNIP/NLRP3 inflammasome axis. Journal of Neuroinflammation, 15(1): 29-48. DOI:10.1186/s12974-018-1073-0 (  0) 0) |
Shi, F., Su, J., Liu, Z., Wang, J., and Wang, T., 2019. miR-144 reverses cisplatin resistance in cervical cancer via targeting LHX2. Journal of Cellular Biochemistry, 120(9): 15018-15026. DOI:10.1002/jcb.28763 (  0) 0) |
Shi, X., Ma, W., Li, Y., Wang, H., Pan, S., Tian, Y., et al., 2020. MiR-144-5p limits experimental abdominal aortic aneurysm formation by mitigating M1 macrophage-associated inflammation: Suppression of TLR2 and OLR1. Journal of Molecular and Cellular Cardiology, 143: 1-14. DOI:10.1016/j.yjmcc.2020.04.008 (  0) 0) |
Siddiqui, M. R., Akhtar, S., Shahid, M., Tauseef, M., McDonough, K., and Shanley, T. P., 2019. miR-144-mediated inhibition of ROCK1 protects against LPS-induced lung endothelial hyper-permeability. American Journal of Respiratory Cell and Molecular Biology, 61(2): 257-265. DOI:10.1165/rcmb.2018-0235OC (  0) 0) |
Song, M., Yang, H., Yao, S., Ma, F., Li, Z., Deng, Y., et al., 2007. A critical role of vascular endothelial growth factor D in zebrafish embryonic vasculogenesis and angiogenesis. Biochemical and Biophysical Research Communications, 357(4): 924-930. DOI:10.1016/j.bbrc.2007.04.033 (  0) 0) |
Su, Z., Si, W., Li, L., Zhou, B., Li, X., Xu, Y., et al., 2014. MiR-144 regulates hematopoiesis and vascular development by targeting meis1 during zebrafish development. International Journal of Biochemistry and Cell Biology, 49: 53-63. DOI:10.1016/j.biocel.2014.01.005 (  0) 0) |
Tang, X., Gong, J., Zeng, C., Sheng, X., Xing, J., and Zhan, W., 2019. Dynamic distribution of formalin-inactivated Edwardsiella tarda in flounder (Paralichthys olivaceus) post intraperitoneal vaccination. Fish & Shellfish Immunology, 89: 393-402. DOI:10.1016/j.fsi.2019.04.022 (  0) 0) |
Wang, M., Wei, J., Shang, F., Zang, K., and Ji, T., 2019. Platelet-derived growth factor B attenuates lethal sepsis through inhibition of inflammatory responses. International Immunopharmacology, 75: 105792. DOI:10.1016/j.intimp.2019.105792 (  0) 0) |
Wienholds, E., and Plasterk, R. H., 2005. MicroRNA function in animal development. FEBS Letters, 579(26): 5911-5922. DOI:10.1016/j.febslet.2005.07.070 (  0) 0) |
Wu, J., Liu, G., Sun, Y., Wang, X., Fang, H., Jiang, H., et al., 2018. The role of regulator FucP in Edwardsiella tarda pathogenesis and the inflammatory cytokine response in tilapia. Fish & Shellfish Immunology, 80: 624-630. DOI:10.1016/j.fsi.2018.06.011 (  0) 0) |
Wu, P., Chen, L., Cheng, J., Pan, Y., Zhu, X., Bao, L., et al., 2021. The miRNA expression profile directly reflects the energy metabolic differences between slow and fast muscle with nutritional regulation of the Chinese perch (Siniperca chuatsi). Com-parative Biochemistry and Physiology. Part A, Molecular and Integrative Physiology, 259: 111003, DOI: 10.1016/j.cbpa.2021.111003.
(  0) 0) |
Xiu, Y., Jiang, G., Zhou, S., Diao, J., Liu, H., Su, B., et al., 2019. Identification of potential immunity-related circRNA-miRNA-mRNA regulatory network in intestine of Paralichthys olivaceus during Edwardsiella tarda infection. Frontiers in Genetics, 10: 731-747. DOI:10.3389/fgene.2019.00731 (  0) 0) |
Xiu, Y., Li, Y., Liu, X., Su, L., Zhou, S., and Li, C., 2021. Identification and characterization of long non-coding RNAs in the intestine of olive flounder (Paralichthys olivaceus) during Edwardsiella tarda Infection. Frontiers in Immunology, 12: 623764. DOI:10.3389/fimmu.2021.623764 (  0) 0) |
Xu, T. T., and Zhang, X. H., 2013. Edwardsiella tarda: An intriguing problem in aquaculture. Aquaculture, 431: 129-135. DOI:10.1016/j.aquaculture.2013.12.001 (  0) 0) |
Xu, X., Liu, Y., Tang, M., Yan, Y., Gu, W., Wang, W., et al., 2018. The function of Eriocheir sinensis transferrin and iron in Spiroplasma eriocheiris infection. Fish & Shellfish Immunology, 79: 79-85. DOI:10.1016/j.fsi.2018.05.019 (  0) 0) |
Yan, X., Cui, J., Liu, X., and Xu, T., 2019. microRNA-144 regulates the NF-κB signaling in miiuy croaker via targeting IL1β. Developmental and Comparative Immunology, 96: 47-50. DOI:10.1016/j.dci.2019.02.018 (  0) 0) |
Yeh, H. Y., and Klesius, P. H., 2010. Sequence analysis, characterization and mRNA distribution of channel catfish (Ictalurus punctatus Rafinesque, 1818) chemokine (C-X-C motif) receptor 4 (CXCR4) cDNA. Veterinary Immunology and Immunopathology, 134(3-4): 289-295. DOI:10.1016/j.vetimm.2009.09.022 (  0) 0) |
You, X., Sun, W., Wang, Y., Liu, X., Wang, A., Liu, L., et al., 2021. Cervical cancer-derived exosomal miR-663b promotes angiogenesis by inhibiting vinculin expression in vascular endothelial cells. Cancer Cell International, 21(1): 684-697. DOI:10.1186/s12935-021-02379-9 (  0) 0) |
Zhang, B. C., Zhang, J., and Sun, L., 2014. In-depth profiling and analysis of host and viral microRNAs in Japanese flounder (Paralichthys olivaceus) infected with megalocytivirus reveal involvement of microRNAs in host-virus interaction in teleost fish. BMC Genomics, 15(1): 878-893. DOI:10.1186/1471-2164-15-878 (  0) 0) |
 2023, Vol. 22
2023, Vol. 22


