Novel peptaibiotics identified from Trichoderma clade Viride
Abstract
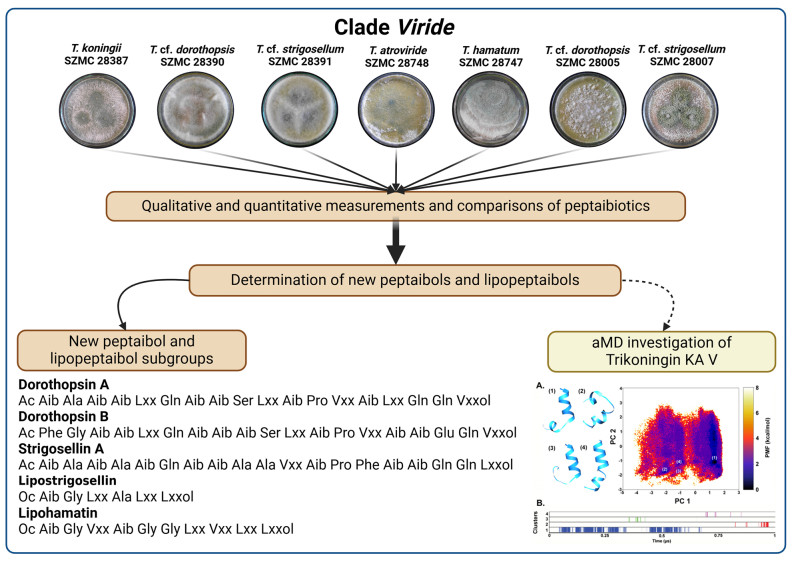
This study focuses on the peptaibiome produced by different species of Trichoderma belonging to clade Viride: T. koningii SZMC 28387 (CBS 979.70), T. cf. strigosellum SZMC 28007 (TUCIM 4886/IQ 191), T. cf. dorothopsis SZMC 28390 (TUCIM 416/TUB F-597), T. cf. strigosellum SZMC 28391 (TUCIM 423/DAOM 230018), T. atroviride SZMC 28748 (IMI 206040), T. hamatum SZMC 28747 (TUCIM 2730) and T. cf. dorothopsis SZMC 28005 (TUCIM 4882/IQ 11). We were able to identify new compounds with similarity to already known groups of peptaibiotics, as well as completely newly discovered compounds using high-performance liquid chromatography (HPLC) -mass spectrometry (MS). From the 367 peptaibiotics identified, 216 are peptaibols and 111 are lipopeptaibols. Out of all peptaibols, 55 are previously known, while 161 are newly discovered. The new peptaibol subgroups Strigosellin A, B and Dorothopsin A, B are introduced. Furthermore, besides 38 previously known lipopeptaibols, 73 new lipopeptaibol sequences, named Lipostrigosellins and Lipohamatins are also reported. In addition, 41 peptaibol-like compounds with unusual C-terminus were also found. Out of the 7 strains examined, 5 produced both peptaibols and lipopeptaibols, while 2 only peptaibols. The well-known compound, Trikoningin KA V (TRK-V) also produced by T. koningii SZMC 28387 (CBS 979.70), was studied for its folding dynamics using accelerated molecular dynamics simulations (aMD) for understanding the plausible three-dimensional structures adopted by these peptaibols of clade Viride. We observed a propensity to form kinked, right-handed helical structures when simulated in an aqueous environment.Graphical Abstract
Keywords
Trichoderma Clade Viride Peptaibol Lipopeptaibol Peptaibiome HPLC–MS Accelerated MD simulations1 1. Introduction
Clade Viride stands as one of the most extensive and varied group within the genus Trichoderma. Members of this clade exhibit a broad range of habitats and are found across diverse geographic regions [1]. They play significant roles in various sectors including industry, agriculture, and medicine [2]. One notable example is T. viride, which effectively combats decay in Obeche wood (Triplochiton sceleroxylon) caused by Gloeophyllum sp. and G. sepiarium through mycoparasitism and nutrient competition [3]. Certain species within this clade, such as T. asperellum and T. atroviride, were thought to be problematic for commercial mushroom cultivation, but eventually, they were not found to cause green mould infection of mushrooms even though they appeared in mushroom growing substrates [4]. Trichoderma green mould affects both Agaricus bisporus and Pleurotus ostreatus, two of the most important mushroom crops, leading to inquiries into the particular strains implicated and their preference for substrates. Green mould agents were described as T. aggressivum, T. pleuroti and T. pleuroticola belonging to clade Harzianum of the genus Trichoderma, while no green mould infections could be attributed to T. asperellum and T. atroviride [4–7]. Morphological characteristics of clade Viride representatives are consistent slow growth rate, globose to subglobose and strongly warted conidia, often solitary conidiophores and hooked phialides [8]. However, these morphological characteristics are not sufficient for exact species-level identification due to the overlap of morphology of Trichoderma species, and therefore, they need to be combined with molecular characterization. The mainly used one being integration of phenetic and phylogenetic characters, which is emphasized for species recognition, with a focus on concordance between both approaches, the translation elongation factor 1α (tef1α), internal transcribed spacer 1 and 2 (ITS1 and 2) regions, as well as, sequence analysis of fragments RNA polymerase Ⅱ gene (rpb2) genes with subsequent phylogenetic analysis. These altogether are essential for characterizing new species and understanding the genetic diversity within the clade [9].
Genus Trichoderma is recognized for generating secondary metabolites, including polyketides, alkaloids, terpenoids, non-ribosomally biosynthesized peptides, and mixed biogenesis metabolites [10]. Currently, extensive research on this genus is underway due to the production of bioactive peptaibiotics. Besides Trichoderma species, members of other closely related genera like Emericellopsis and Gliocladium are also significant producers of peptaibiotics [11]. The analytical exploration of peptaibiotics is referred to as peptaibiomics [12]. Peptaibiome is described as the entirety of fungal peptides which contain the amino acid residue α-aminoisobutyric acid (Aib) [12, 13]. These acyclic peptides are synthesized by non-ribosomal peptide synthetase (NRPS) enzymes [14].
In this study, peptaibols and lipopeptaibols were identified out of the 5 groups of peptaibiotics, which were categorized based on their length and chemical structures [12]. Peptaibols constitute a class of linear antibiotic peptides characterized by their length, usually ranging from five to twenty amino acids. Notable features of peptaibols include a high occurrence of non-proteinogenic C-alpha tetrasubstituted amino acids [15], such as the relatively uncommon isovaline (Iva) or the achiral Aib. Typically, the N-terminal amino acid of peptaibols is acylated, while the C-terminus consists of a β-amino alcohol. Among the most prevalent types of β-amino alcohols at the C-terminus are isoleucinol, phenylalaninol, valinol, and leucinol (Leuol). These compounds represent the most extensive category within peptaibiotics and several peptaibols are already known to be produced by members of clade Viride [16]. Lipopeptaibols represent naturally occurring short peptides possessing antimicrobial properties [17]. They are distinguished by a lipophilic acyl chain at the N-terminus, a significant presence of turn/helix forming Aib, and a 1,2-amino alcohol at the C-terminus. These peptides typically consist of 6 to 11 amino acid residues and feature fatty acyl moieties ranging from 8 to 15 carbon atoms, which is shorter compared to the nonlipidated peptaibols that typically contain 11 to 20 amino acid residues [17, 18]. Due to the presence of a lipophilic N-terminal group, these peptides are known as lipopeptaibols. They have been extracted from several fungal cultures including T. koningii (trikoningins) and T. viride (trichodecenins) from clade Viride, as well as T. longibrachiatum (trichogins), T. polysporum (trichopolyns), Tolypocladium geodes (antibiotics LP 237), and Mycogone rosea (helioferins) [17, 18]. It is then imperative that more unknown structures of peptaibiotics should be studied and analyzed to continue the growth of the scientific knowledge and industrial application of this unique and promising group of secondary metabolites.
2 Materials and methods
2.1 Fungal strains, media, culture conditions
Trichoderma strains were selected from the TU Collection of Industrially Important Microorganisms (TUCIM), Vienna, Austria and deposited at Szeged Microbiology Collection (SZMC) (Table 1). The selected strains were cultivated on yeast-glucose agar medium enriched with malt extract (3 g/L yeast extract, 10 g/L glucose, 20 g/L agar, and 50 mL of 20% liquid malt extract solution). To increase peptaibol production, the strains were cultivated on malt extract agar (MEA) medium containing 150 mL (20%) malt extract solution, 3 g/L soybean peptone, and 15 g/L agar. Incubation was carried out for 7 days at 25 ℃, and mycelial growth was subsequently observed.
Trichoderma strains selected for this study. The re-identification was based on the sequence analysis of the tef1α region
2.2 Extraction of peptaibols
Following the incubation period, peptaibols were extracted from the Trichoderma cultures using a solution composed of chloroform and methanol in a 2/1 (v/v) ratio. The resulting mixture was subjected to evaporation using an IKA RV 10 rotatory evaporator (IKA Works, USA) until complete dryness. The resultant dry mass was reconstituted in 1 mL of methanol and thoroughly mixed. Subsequently, the samples underwent centrifugation for 15 min (Heraeus Fresco 17, Thermo Scientific, CA, USA) to eliminate insoluble particles, and the clarified samples were stored at − 20 ℃ for further analysis.
2.3 Species reidentification
To cultivate mycelia of the Trichoderma strains, liquid medium was prepared in Eppendorf tubes on yeast-glucose agar medium enriched with malt extract mentioned above. The tubes were incubated for 2 days at 25 ℃. After incubation, approximately 100 mg of fungal tissue was introduced into sterile Eppendorf tubes. Glass beads with particle sizes of 0.4–0.6 mm and 0.9–0.15 mm were added to the tubes, and liquid nitrogen was employed for cell disruption. The tubes were placed in a cell disruptor for 4 min, and this process was repeated three times.
Following the initial procedures, DNA extraction was carried out using the E.Z.N.A.® Fungal DNA Mini Kit in accordance with the manufacturer's instructions. Subsequent to DNA purification, the tef1α region was amplified through polymerase chain reaction (PCR). The PCR master mixture for each sample (1 μL) consisted of 2 μL of Dream Taq buffer, 2 μL of dNTP mix, 4–4 μL of 1 μM tef1α primers (forward: 5’-CATCGAGAAGTTCGAGAAGG-3’ and reverse: 5’-AACTTGCAGGCAATGTGG-3’), 7 μL of water, and 0.1 μL of Dream Taq DNA polymerase. The PCR cycle was conducted using the MJ MiniTM Personal Thermal Cycler (BIO-RAD) following these steps: an initial step at 94 ℃ for 5 min; cycling steps at 94 ℃ for 30 s (denaturation), 57 ℃ for 30 s (annealing), and 72 ℃ for 30 s (elongation), repeated for 40 cycles; and a final elongation step at 72 ℃ for 4 min. Agarose gel electrophoresis was performed to check the success of the reaction on 1% agarose gel (35 mL TAE buffer, 0.35 g agarose, 2 μL ethidium bromide). Two μL of DNA samples were applied with 2 μL of bromophenol blue. The run time for gel electrophoresis was 30 min at 90 V. The DNA fragments were sent for sequence analysis to an external service (Eurofins Scientific). Sequences were analyzed by using Finch TV 1.4.0 (Geospiza Inc.) and NCBI Nucleotide BLAST softwares (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Multiple sequence alignment of partial tef1α sequences (provided in the supplementary file) was made by PAGAN v1.53 [24] with default settings. The final dataset consisted of two partitions for the intron and exon regions. For both partitions, the best fitting model was TrN + G4, determined by using ModelTest-NG v0.1.7 [25], based on the Bayesian Information Criterion (BIC). Maximum likelihood analysis with the selected models was carried out by RAxML-NG v1.2.2 [26], with 1000 bootstrap replicates.
2.4 Analytical procedures for peptaibols
The analysis of crude peptaibol extracts was conducted using high-performance liquid chromatography-mass spectrometry (HPLC–MS) based on a method previously described by Marik et al. [27]. For the analysis, an Orbitrap (QExactive Plus, Thermo Scientific, CA, USA) MS was used with a heated electrospray ion source (HESI) in positive mode coupled with an Dionex Ultimate 3000 (Thermo Scientific, CA, USA) HPLC containing a quaternary pump, a vacuum degasser, an autosampler, and a column heater. The instruments were controlled by the Xcalibur 4.2 Software (Thermo Scientific, CA, USA). The separation was achieved using a Purospher Star RP-15 HPLC column (100 × 2.1, 2 μm), while the solvent A consisted of H2O/MeOH/MeCN (8/1/1, v/v) with 5 mM ammonium acetate and 0.1% (v/v) acetic acid and solvent B comprised of acetonitrile/methanol (1/1, v/v) with 5 mM ammonium acetate and 0.1% (v/v) acetic acid. The flow rate was set to 0.2 mL/min, and the gradient program for Solvent B was as follows: 0 min—65%, 2 min—65%, 42 min—80%, 54 min—90%, 55 min—95%, 58 min—95%, 58.5—65% and 62.5—65%. The column temperature was maintained at 30 ℃, and the injection volume was 3 μL. The HESI parameters were as follows: spray voltage – 4.5 kV, sheath gas flow rate − 30 arbitrary units, aux gas flow rate − 12 arbitrary units, capillary temperature − 300 ℃, aux gas heater − 300 ℃. The acquisition mode was Full-MS-ddMS2. Full-MS parameters were: resolution − 70,000 at m/z 200, AGC target − 3e6, maximum injection time − 100 ms, scan range − 200–2000 m/z. The ddMS2 parameters: fixed first scan at m/z 80, resolution 17500 at m/z 200, AGC target − 1e6, maximum injection time − 50 ms, isolation window − 1 m/z, collision energy − 25 NCE. Loop count was set to 5.
2.5 Determination and nomenclature of peptaibol sequences
Peptaibols typically undergo fragmentation between Aib − Pro residues already in the ion source resulting in the generation of b- and y-ions. The identification of a specific peptaibol compound relies on its retention times (Rt) and on the mass of its b- and y-fragment ions as well as the mass of the hydrogen adduct of the primary mass of the compound [M + H]+ (b-ion + y-ion). During full scan measurements, the mass of the protonated molecular ion coexists with other adduct ions such as the primary mass and sodium adduct [M + Na]+, the primary mass and 2 hydrogen adducts [M + 2H]2+, the primary mass and 2 sodium adducts [M + 2Na]2+, and the primary mass and hydrogen and sodium adducts [M + H + Na]2+. In the longer peptaibols (16–19 residues), the fragmentation typically occurred between the centrally located Aib − Pro residues, leading to the formation of b11-12 and y5-7 ions. The analysis of MS2 results from b- and y-ion fragments provided insights into the complete sequences of the compounds (Supplementary Fig. 1).
Lipopeptaibols were identified based on the MS2 fragmentation of [M + H]+. The whole sequence could be observed on the MS2 spectra.
To assess the novelty of the obtained sequences, an initial comparison was made using the ‘Comprehensive Peptaibiotics Database’ [28]. This database is not available online since 2017, however, Prof. Rainer Schuhmacher kindly provided the offline version of the Peptaibiotics Database including the peptaibiotics records till 2017. Consequently, to gather more information and conduct a comprehensive comparison, an additional online literature search was conducted using PubMed (https://pubmed.ncbi.nlm.nih.gov/) with the keywords ‘peptaibol’ or ‘lipopeptaibol’.
The quantity of peptaibols were defined based on the integration of the peaks under the major y-ions originated from the decomposition of the Aib–Pro bond in the ion source on the extracted ion chromatograms (EIC) of the full scan MS measurement. Adding the separately integrated peaks resulted in the total peptaibol production of the strain, out of which, percentages were calculated for each compound. The quantity of lipopeptaibols was defined based on the integration of the peaks under the [M + H]+ on the EIC of the full scan MS measurement. Percentages of each compound were calculated as mentioned above.
The known peptaibols and lipopeptaibols were named using the closest identified compounds attached to a Roman numeral based on their elution order. The completely novel sequences were named after the producer species attached to a Roman numeral based on their elution order.
2.6 aMD simulations to reveal folding dynamics of Trikoningin KA V (TRK-V)
Trikoningin KA V was selected because the isotype of Vxx and Lxx residues of this compound was determined by Goulard et al. [29]. For non-standard amino acid residues, Aib and Leuol, the partial charges and force fields were calculated using a previously developed method reported by Tyagi et al. [30]. The unfolded peptide topology was prepared using the ‘tleap’ module of AmberTools20 using the Amberff19SB force field and solvated in TIP3P water solvent model [31]. The peptide solvation added 2324 water residues with the periodic box size of 45.96 × 44.35 × 46.88 Å and volume of 95587 Å3. The system preparation, minimization and equilibration steps were carried out as described in Tyagi et al. [32]. To calculate boost parameters, total number of atoms in the system (Natoms = 19467), number of residues (Nres = 20) and coefficients a1, a2 (4.5) and b1, b2 (0.20) were utilized. The aMD simulation was run for 1000 ns or 1 µs in total. The resulting trajectory files were prepared by the removal of water molecules. Dihedral angles were calculated for every residue from each frame of the simulation and the distribution was reweighted using Maclaurin series expansion.
3 Results and discussion
3.1 Reidentification of the investigated Trichoderma strains
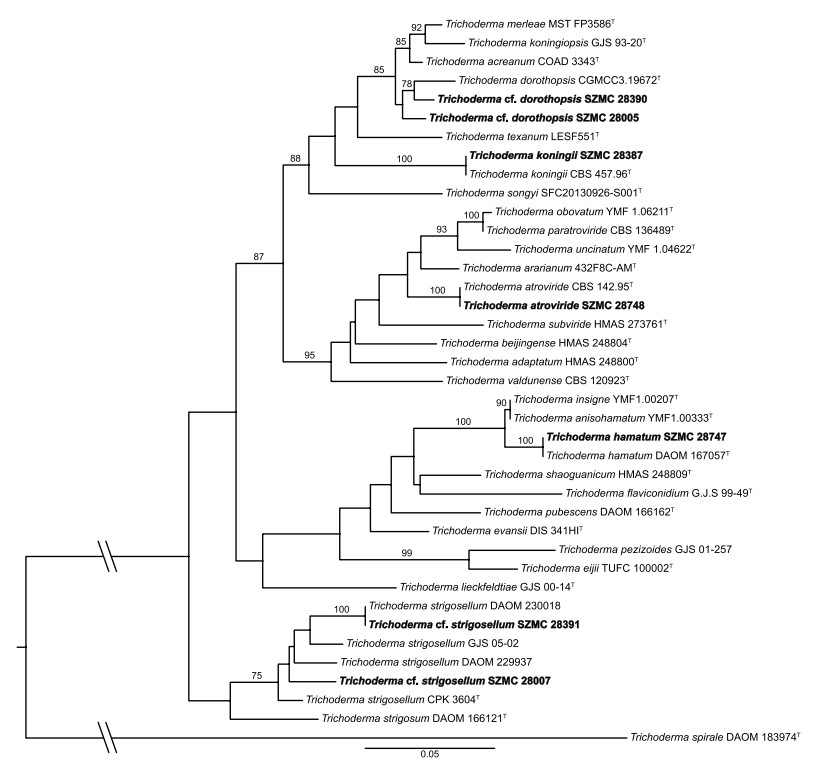
The examined isolates were assigned to species based on sequence analysis of partial tef1α sequences (provided in the supplementary file). Figure 1 shows the phylogenetic relation between the strains investigated in this article and the type strains of closely related species. T. koningii SZMC 28387 and T. atroviride SZMC 28748 showed 100% identity with their type strains. Strain SZMC 28747 originally was identified as T. fertile, however, it showed 100% identity with the type strains of T. hamatum. T. cf. strigosellum SZMC 28391 and SZMC 28007 could not be identified to species level, therefore, cf. strigosellum was introduced. T. taiwanense SZMC 28390 and T. cf. dorothopsis SZMC 28005 did not match with their type strains either, but were similar to T. dorothopsis type strain and were named as T. cf. dorothopsis SZMC 28390 and T. cf. dorothopsis SZMC 28005.
Phylogenetic tree of the examined strains from clade Viride based on their partial tef1α sequences
3.2 Peptaibiome of the investigated Trichoderma strains
Out of the 7 strains investigated, (Table 1), 5 produced both peptaibols and lipopeptaibols (T. cf. strigosellum SZMC 28391 and SZMC 28007, T. koningii SZMC 28387, T. hamatum SZMC 28747, and T. cf. dorothopsis SZMC 28005), while T. cf. dorothopsis SZMC 28390 and T. atroviride SZMC 28748 did not produce any lipopeptaibols, their peptaibiome consisted of exclusively SF1 peptaibols (Table 2, Supplementary Fig. 1). The two T. cf. strigosellum strains produced ~40/60% ratio of peptaibols/lipopeptaibols T. koningii SZMC 28387 and T. cf. dorothopsis SZMC 28005 produced one third ratio of peptaibols of the peptaibiome, while T. hamatum SZMC 28747 produced only one fourth of peptaibols of the peptaibiome (Table 2). Based on their length, lipopeptaibols could be categorised into the short (6–7-residue long), medium (10–11- residue long) or long (15-residue long) groups. The most amount of short lipopeptaibols was produced by T. cf. strigosellum SZMC 28007 with 61.35% of the peptaibiome, while the greatest number of long lipopeptaibols were produced by T. hamatum SZMC 28747 with 40.36% of the peptaibiome. The amount of medium lipopeptaibols was ranging from 30 to 65% of the total production (Table 2).
Ratio of peptaibiotics (peptaibiome) produced by the members of clade Viride investigated in this study
3.3 Peptaibol profiles of the investigated Trichoderma strains
Tables 3 and 4 were obtained from the chromatographic and MS analysis of the investigated Trichoderma strains and show the subfamily 1 (SF-1) peptaibols of the investigated Trichoderma strains. The diagnostic fragment ions resulted by MS2 fragmentation are collected in Supplementary Table 1.
SF1 peptaibols produced by Trichoderma strains from clade Viride
Similarities of SF1 peptaibols produced by Trichoderma strains from clade Viride to known peptaibols
3.3.1 Identification of new Strigaibol sequences and the new groups of Strigosellins from T. cf. strigosellum
Investigation of the peptaibol profile of T. cf. strigosellum SZMC 28391 revealed twenty-one 19-residue peptaibols belonging to the SF-1 peptaibol family (Tables 3 and 4). Appart from the known Strigaibol sequences, 14 proved to be new. Their closest positional isomers are Strigaibol sequences described by Park et al. [33]. MS and chromatographical analysis of T. cf. strigosellum SZMC 28007 revealed the production of 14 peptaibols novel to science and were named as Strigosellin A as well as further 6 sequences named as Strigosellin B peptaibols. These peptaibols showed a great similarity to the sequences of Trichokonin TKO-V, Trichokonin V, Tricholongin B and Tricholongin LBⅠ sequences [34–37].
Strain T. cf. strigosellum SZMC 28391 produced two peptaibols in higher quantities, which were Strigaibol Ⅻ and ⅩⅣ (36.15% and 27.59% of the total peptaibol production, respectively). Four more sequences were produced above 4%, Strigaibol Ⅴ, Ⅵ, Ⅹ and ⅩⅩ (5.14%, 6.44%, 4.34% and 7.2%, respectively). Strain T. cf. strigosellum SZMC 28007 produced Strigosellin A Ⅸ covering 14.55% of the total peptaibol production, while Strigosellins A Ⅻ, ⅩⅢ, and ⅩⅣ were also produced in relatively higher quantities (10.73%, 13.01% and 10.95%, respectively). Strigosellins A Ⅱ, Ⅴ, Ⅵ, and Ⅷ, were also produced in significant quantities between 2.78 and 7.41%, while Strigosellins A Ⅰ, Ⅲ, Ⅳ, Ⅶ, Ⅹ, and XI were produced at 0.48–1.78% of the total peptaibol production. Strigosellins B Ⅰ, Ⅱ and Ⅴ were produced also in significant quantities (5.38%, 8.2% and 6.51%, respectively), while Strigosellin B Ⅰ, B Ⅲ, B Ⅳ and B Ⅵ were below 2.44%.
3.3.2 New Trikoningin KA-like peptaibol compounds from T. koningii
T. koningii SZMC 28387 produced thirty-eight new sequences, while nineteen were positionally isomeric with the previously described Trikoningin KA Ⅴ from T. koningii described by Goulard et al. [29], and Trikoningin KA-like compounds Ⅰa, Ⅳa, Ⅳb, Ⅴa, Ⅵa and Ⅺ from T. gamsii SZMC 1656 described by Marik et al. [38] (Tables 3 and 4).
T. koningii SZMC 28387 also produced two peptaibols, Trikoningin KA-like ⅩLⅢ and ⅩLⅤ (25.83% and 28.48%, respectively) in high quantities, while apart from Trikoningin KA-like ⅩL, ⅩLⅥ and ⅩLⅧ (4.65%, 6.25% and 6.74%, respectively), the rest of the peptaibols were produced for less than 3% and mostly below 1%.
3.3.3 Identification of the new group of Dorothopsins from T. cf. dorothopsis
Strain T. cf. dorothopsis SZMC 28390 produced thirteen 19-residue peptaibols also belonging to the SF-1 peptaibol family. All the thirteen compounds were completely new (Tables 3, 4) and their closest positional isomers are Trichostrigocins TSG-A and TSG-B described by Degenkolb et al. [37]. These new compounds were named as Dorothopsins A-a Ⅰ-XIII (Table 3). However, 18-residue peptaibols, Dorothopsins A-b Ⅰ-Ⅷ and Dorothopsins A-d Ⅰ-Ⅲ, as well as, the 17-residue Dorothopsin A-c Ⅰ and Dorothopsin A-f Ⅰ-Ⅱ sequences, furthermore, a 16-residue Dorothopsin A-e Ⅰ sequence were also produced by this strain (Table 3). All Dorothopsin A sequences are also new additions to literature (Table 4). Strain T. cf. dorothopsis SZMC 28390 produced Dorothopsin A-a Ⅶ covering more than half of the total peptaibol production (54.05%). After that, Dorothopsins A-a Ⅷ and A-d Ⅰ, were produced the second highest with 10.48% and 8.35%, respectively. Among the Dorothopsin A-a sequences, a few peptaibols were 1.5%–4.5%, but the rest and all Dorothopsin A-b, -c, -d, -e and -f sequences were below 1.5% apart from Dorothopsin A-d Ⅰ with 8.35%.
T. cf. dorothopsis SZMC 28005 produced thirty-four new sequences, which were the most unique among the already known SF1 peptaibols and named as Dorothopsin B. They had the most varied sequences, sometimes with differences in 6 positions from the already known Tricholongin LBⅠ sequence described by Degenkolb et al. [37]. The most interesting characteristic of these sequences is the substitution of the acetylated Aib1 (Ac-Aib1) and Phe3 known from Tricholongin sequences to the acetylated Phe1 (Ac-Phe1) and Aib3. Furthermore, Phe1 was also substituted with Trp1 in several Dorothopsin B sequences. The most produced sequence by T. cf. dorothopsis SZMC 28005 was Dorothopsin B ⅩⅩⅥ with a relatively low quantity of 10.15%, which was followed by Dorothopsins B Ⅶ, ⅩⅢ, ⅩⅤ, ⅩⅩⅤ, ⅩⅩⅩⅠ, and ⅩⅩⅩⅣ (5.05%, 9.71%, 7.95%, 6.16%, 4.01% and 4.53%, respectively). All the other sequences were produced below 4%.
3.3.4 Identification of new Trichorzianin TA sequences and peptaibol-like compounds from T. atroviride
T. atroviride SZMC 28748 produced thirty-three peptaibol compounds, as well as fourty-one incomplete peptaibol sequences named as Peptaibol-like compounds in Tables 3 and 4. The thirty-three peptaibol sequences were positionally isomeric with the Trichorzianin TA sequences [41], out of which, thirteen proved to be new. The Peptaibol-like sequences were also identical with the thirty-three trichorzianin TA sequences [41], but unusual at their C-terminus, where the last part of the sequences is shown with the remaining mass difference, which were Δm = 145, 146, 160, or 222 D, furthermore, all MS2 spectra contain the water loss [y6–H2O]+ fragment ions (Supplementary Fig. 2), which usually occurs when the C-terminus is an aminoalcohol. The sequences contain only two variable positions, R5 contains Aib/Vxx, while R14 Vxx/Lxx. All other residues are the same in the sequences, compared to each other. These sequences were produced in a relatively low concentration (all below 0.25%), therefore, they were not investigated further.
The two most produced peptaibols of T. atroviride SZMC 28748 were Trichorzianin TA-like ⅩⅩⅤ and ⅩⅩⅨ with 15.24% and 17.59% of the total peptaibol production. They were followed by Trichorzianin TA-like Ⅶ, ⅩⅤ, ⅩⅩ, ⅩⅩⅡ, ⅩⅩⅦ and ⅩⅩⅧ peptaibols with 7.68%, 8.99%, 5.26%, 5.44%, 6.19% and 5.34%, respectively. The rest of the peptaibols produced by this strain were around or below 1–2%. All the 41 Peptaibol-like compounds produced by T. atroviride SZMC 28748 were below 0.25% of the total peptaibiome.
3.3.5 Identification of new Tricholongin sequences from T. hamatum
The chromatographic and MS analysis of the T. hamatum SZMC 28747 extract revealed the production of twenty-one 19-residue-peptaibols belonging to the SF-1 peptaibol family (Tables 3 and 4). Among them, 6 peptaibols were new to science and the other sequences were identical to already determined sequences of Tricholongins LB Ⅰ, Ⅱ, Ⅲ, and Ⅳ [37]. The newly identified sequences also shared similarities with the sequences of Tricholongins LB Ⅰ, Ⅱ, Ⅲ, and Ⅳ. The known peptaibols are produced by T. cf. strigosum BBA 69577 and T. cf. pubescens BBA 66989 strains [37].
Tricholongin LB-like ⅩⅢ and ⅩⅥ sequences produced by T. hamatum SZMC 28747 covered 25.68% and 29.09% of the total peptaibol production, which were followed by Tricholongin LB-like ⅩⅦ and ⅩⅨ sequences (7.13% and 10.03%, respectively). All other sequences were produced mostly around 1%.
3.3.6 Module skipping hypothesis
The microheterogeneity of non-ribosomal peptides like peptaibols arises from the ability of NRPSs to incorporate various amino acids [42]. Usually, Trichoderma species produce 1 to 4 main peptaibol compounds in high quantities, while the rest of the peptaibol variants are in much lower quantities, which can be the result of the modular synthesis and the misassembly of the peptaibol compounds by NRPS and the lack of repair mechanisms compared to ribosomal synthesis. This may be the reason for the wide range of new peptaibols that can be identified even from already known peptaibol subgroups. This hypothesis needs a thorough genetic data support based on the number of NRPS genes that can be found in the available Trichoderma full genomes.
Based on the results of this study, we hypothesize that the 16, 17 and 18-residue peptaibols produced by T. cf. dorothopsis SZMC 28390 also occured through a ‘module skipping’ event, which means that the NRPS enzyme skipped a module during the synthesis [27, 39]. Two skipping mechanisms were proposed in the literature: direct intermediate transfer between nonadjacent modules caused by a “loss of function mutation” [40], or nonfunctional thiolation or condensation domains of the NRPSs during biosynthesis [39]. Module skipping is usually seen in the case of 10-, 13-, 18-, and 19-residue peptaibols. Similar results were found in the case of SF-1 peptaibols produced by T. flagellatum, T. sinense and T. parareesei, which produced 19-residue peptaibols among the 20-residue peptaibols, but the investigation of the genome of the producer strains revealed no extra 19-module NRPS synthetases, only the gene of a 20-module enzyme was present [27]. Based on this hypothesis, the positions at which module skipping most probably occurred are shown in Table 4 with a minus sign. Our prediction is based on comparing the sequences of 16-residue (Dorothopsin A-e), 17-residue (Dorothopsin A-c) and 18-residue (Dorothopsins A-b and A-d) peptaibols to the sequences of 19-residue peptaibols (Dorothopsin A-a) (Tables 3 and 5).
The 1most produced sequences, 2consensus sequences and 3amino acid varieties of the peptaibols produced by each strain
3.4 Comparative analysis of peptaibols produced by species of clade Viride
The microheterogeneity within the sequences occurs in the positions where one amino acid is substituted by another one. The strictest positions were found to be in the middle of the sequences usually appearing as the Gln6-Aib7-Aib8-Aib9-Ser10-Lxx11-Aib12-Pro13-Vxx14 motif. This motif was different only in Strigosellin A sequences, where it was substituted with Gln6-Aib7-Aib8-Ala9-Ala10-Vxx11-Aib12-Pro-13-Phe14/Lxx14 produced by T. cf. strigosellum SZMC 28007. R1 usually contained an acetylated Aib except for a few T. cf. strigosellum SZMC 28391 and T. cf. dorothopsis SZMC 28390 sequences, where it was often substituted with Vxx. Acetylated Vxx is not present in SF-1 peptaibols, only in 11–16-residue peptaibols [10, 34, 37, 39]. Mostly, AcAib can be found in SF-1 peptaibols, however, several compounds produced by these strains contained another amino acid residue at the N-terminus (Table 3). Dorothopsin B sequences produced by T. cf. dorothopsis SZMC 28005 always contained Phe1 or Trp1. Boletusin 1, Chrysospermins and Peptaivirins, a group of another 19 residue-peptaibols identified from Boletus spp., Apiocrea chrysosperma and Trichoderma spp. (KGT142), respectively, also contained AcPhe at their N-termini [43–45]. Phe occurred strictly in R1, R3, R14 and R19 positions in T. cf. dorothopsis SZMC 28005, T. hamatum SZMC 28747, T. cf. strigosellum SZMC 28007 and T. atroviride SZMC 28748, respectively, while the other strains investigated did not contain Phe at all. The difference in Strigosellin A and B sequences relies in Phe14 and Lxx14 from T. cf. strigosellum SZMC 28007. Interestingly, both types are produced almost equally by this strain. Regularly, Phe is close to one of the termini of the sequences in all peptaibols, however, Phe is in the middle of the sequences in Strigosellin A peptaibols. This strain shows production of both Strigosellin A (Phe14) and B (Lxx14) sequences, as if the production of the regular Lxx14 containing peptaibols were replaced to the rare Phe14 sequences. AcPhe1 was often substituted with AcTrp1 in Dorothopsin sequences, also observed in the literature for 15–16-residue peptaibols (Ampullosporins and Zervamicins) produced by Sepedonium ampullosporum (HKI-0053) and Emericellopsis salmosynnemata, respectively [46, 47]. Sequences of T. hamatum SZMC 28747 contained Ala1 instead of Aib1 in case of only 2 sequences. Only three 20-residue sequences (Trichoaureocin 1a, Trichokonin IIb and Atroviridin D) with AcAla were identified from T. aureoviride, T. koningii Oudemans and an unidentified Trichoderma sp., respectively [35, 48, 49], out of which, the first two species belong to clade Viride, while the latter one was later reidentified as T. arundinaceum belonging to clade Brevicompactum [50]. Gly2 was substituted to Ala or Ser in several sequences of all strains investigated, while R3 contained mostly Ala or Aib. However, in the sequences of T. hamatum SZMC 28747, Phe or Tyr was always identified at the R3 position. Tyr has never been identified from peptaibols at this position, only from lipopeptaibols and other peptaibiotics, such as LP237 F5 and Trichoatrokontins, respectively [18, 51, 52], as well as from all-Aib-replaced peptides by Amycolatopsis azurea, used for bioconversion of AS 1387392 [53]. R4 was strictly Aib in all sequences except for a few sequences produced by T. hamatum SZMC 28747. At R5, mostly Vxx and Lxx were identified, except for T. hamatum SZMC 28747 and T. cf. strigosellum SZMC 28007 sequences, which mostly contained Aib5. This position was followed by the R6-R7-R8-R9-R10-R11 and R12-R13-R14 motifs discussed above. The module skipping occurred at R9 position in all Dorothopsin A-b, -c and -e sequences. R15 contained mostly Aib like in R4, except for a few sequences produced by T. cf. strigosellum SZMC 28007, where it was substituted to Vxx in several sequences. R16 contained either Ala, Aib, Vxx or Lxx which determined the mass of the y7 ion fragments. Gln16 was often substituted with Glu which, apart from the mass of the y7 ion fragments, defines the acidity of the peptaibols as Gln is neutral and Glu shows acidic characteristics. Dorothopsin A sequences also showed the 2nd module skipping at this position. One Dorothopsin A sequence (Dorothopsin A-e Ⅰ from Table 3) showed in total 3 module skippings at R9, R16 and R17. R18 mostly contained Gln except for Dorothopsin A compounds, where it was often substituted with Gly or Ala. The aminoalcohol was mostly Vxxol or Lxxol, except for Dorothopsin A and Tricholongin LB-like sequences, which often contained Alaninol (Alaol) or phenylalaninol (Pheol)/tryptophanol (Trpol) at the C-terminus, respectively. Pheol is often identified as the C-terminus of peptaibols. The first naming of peptaibols by Pandey et al. [54] as ‘peptaibophols’ was also based on the inclusion of Pheol, which was defined as peptide antibiotics containing the marker amino acid Aib and a C-terminal Pheol residue [54, 55]. After this, additional 2-alcohols such as valaninol (Valol) in TXT-A40 and Leuol in Hypelcin from Hypocrea peltata (current name: T. peltatum) were detected as C-terminal constituents, the above definition of ‘peptaibophols’ needed to be revised and updated. Nevertheless, Trpol is rare at the C-terminus of peptaibols. It was only identified from Boletusin 1, Chrysospermins, Peptaivirin A, as well as Trichorzianin TA and PA sequences [41, 43–45, 56, 57].
In a few cases, the consensus sequence (which is a created sequence based on the most abundant amino acid present at each position) was different from the most produced sequence of the strain. These residues are underlined in Table 5. Trikoningin KA-like peptaibols mostly contained Gln at R17 while the most produced Trikoningin KA-like ⅩLⅤ had Glu at this position. Exactly this could be observed among Dorothopsin B sequences where Dorothopsin B ⅩⅩⅥ contained Glu instead of Gln. Similarly, Trichorzianin TA-like peptaibols at R14 contained Lxx instead of Vxx in the most produced Trichorzianin TA-like ⅩⅩⅨ. Tricholongin LB-like sequences equally produced Aib and Vxx containing sequences at R16, however, the most produced Tricholongin LB-like ⅩⅥ had Vxx at this position.
3.5 Lipopeptaibol production of the investigated Trichoderma strains
Strain T. cf. dorothopsis SZMC 28390 and T. atroviride SZMC 28748 did not produce any lipopeptaibols (Table 1), their peptaibiomes consist of exclusively SF1 peptaibols. In Tables 6 and 7 the sequences are ordered based on their similarities to each other, the producer strains are shown in the 2nd column. The diagnostic fragment ions resulted by MS2 fragmentation are collected in Supplementary Table 2.
Lipopeptaibols produced by Trichoderma strains from clade Viride The area percentages are calculated based on the total lipopeptaibol production of each strain
Similarities to known compounds of lipopeptaibols produced by Trichoderma strains from clade Viride
3.5.1 Identification of the new group of Lipostrigosellins from T. cf. strigosellum
T. cf. strigosellum SZMC 28391 produced a total of 12 lipopeptaibols, out of which, four were 6-residue long, while eight were 7-residue long (Table 6). Seven sequences were new, while five were positionally isomeric with already identified Lipostrigocin LSG sequences [37] (Table 7). All the 6-residue lipopeptaibols were new to science and named as Lipostrigosellin Ⅰ-Ⅳ. They are different from 7-residue lipopeptaibols in skipping of Aib/Vxx4. Other Trichoderma species were not found to produce 6-residue lipopeptaibols. Only Halovir A-E sequences from Scytalidium sp. CNL240 [58] and Lipohexin from Moeszia lindtneri HKI-0054 [59] were identified as 6-residue lipopeptaibols. Strain T. cf. strigosellum SZMC 28391 also produced a unique compound in a relatively high amount, the amino acid composition of which could not be revealed. The mass differences (Δm) on the MS2 spectra are shown in Table 6, which is a repetition of 72 and 127 mass differences. Strain T. cf. strigosellum SZMC 28007 produced nine already known and seven new 7-residue sequences which were named as Lipostrigaibols Ⅰ-ⅩⅥ. The sequences always started with an OcGly only known from Lipostrigaibols [33]. It is followed by an Ala2, which is again unique, because this position is usually taken by Gly2. R3 and 4 were highly variable, Ala/Aib/Vxx/Lxx and Ala/Aib/Vxx were at these positions. R5 was strictly Ala and R6-7 contained either Vxx-Lxxol, or Lxx-Lxxol, which are usual in the lipopeptaibols (Table 7).
Strain T. cf. strigosellum SZMC 28391 produced two lipopeptaibols in higher quantities, which were Lipostrigocin LSG-like Ⅷb and Ⅵ, with 37.08% and 31.96% of the total lipopeptaibol production, respectively (Table 6). The “unidentified compound” mentioned above was also produced in a relatively higher amount, 17.45%, compared to the rest of the lipopeptaibols, which were all below 4% (Table 6). The 7-residue lipopetaibols were more produced than the 6-residue lipopeptaibols, the latter all being below 2.17%. Three lipopeptaibols were produced in high quantities by T. cf. strigosellum SZMC 28007, Lipostrigaibols ⅩⅤ, ⅩⅢ and ⅩⅥ (23.7%, 22.15% and 20.8%, respectively) followed by Lipostrigaibols ⅩⅣ and Ⅻ with 9.97% and 8.77%, respectively. The rest of the sequences were all produced below 5.22%.
3.5.2 New Lipostrigocin LSG-like sequences peptaibol compounds from T. koningii
T. koningii SZMC 28387 produced a total of 25 lipopeptaibols, out of which seven were 7-residue long while eighteen were 11-residue long (Table 6). Twelve sequences were new, while thirteen were positionally isomeric with already identified Lipostrigocin LSG sequences [37].
T. koningii SZMC 28387 produced more 11-residue than 7-residue lipopeptaibols, out of which Lipostrigocin LSG-like ⅩⅣ was the most produced (37.46%). Lipostrigocin LSG-like ⅩⅤ, ⅩⅥ and ⅩⅩ were also produced in a relatively high amount, at 9.6%, 11.01% and 11.37%, respectively. The 7-residue lipopeptaibols were produced below 1.28%.
3.5.3 Identification of the new group of Lipohamatins from T. hamatum
T. hamatum SZMC 28747 produced sixty-two 7-, 10-, 11- and 15-residue lipopeptaibols, out of which 16 compounds were known, while 47 were newly identified, out of which all the 10-residue lipopeptaibols were completely new to science and were named as Lipohamatin I-IV. They were similar to Linopubescin LPB A, B and C [37]. The 7-, and 11- residue sequences showed similarity to the Lipostrigocin LSG sequences, while the 15-residue compounds to Trichogin GB IX [37, 60], which was the only 15-residue lipopeptaibol reported.
T. hamatum SZMC 28747 produced the most diverse lipopeptaibol profile. The lipopeptaibol most produced by this strain was Lipostrigocin LSG-like ⅩⅩⅩⅨ with 11.1%. Trichogin GB IX-like ⅩⅩⅥ was the most produced 15-residue lipopeptaibol by this strain (6.67%). All 7- and 10-residue lipopeptaibols were produced below 1.71%.
3.5.4 New Lipostrigocin LSG-like peptaibol sequences from T. cf. dorothopsis
Strain T. cf. dorothopsis SZMC 28005 produced only nine lipopeptaibols, out of which five were 11-residue, four were 15-residue sequences. Similarly to the lipopeptaibols of T. hamatum SZMC 28747, the 11-residue sequences were positionally isomeric with Lipostrigocin LSG B3 and B11 sequences, while all the 15-residue sequences were new but similar to Trichogin GB IX [37, 60]. Two sequences, Lipostrigocin LSG-like ⅩⅩⅩⅡ and ⅩⅩⅩⅤ were also produced by T. hamatum SZMC 28747 (Table 6).
Strain SZMC 28005 produced four 11-residue lipopeptaibols in high quantities, which were Lipostrigocin LSG-like ⅩⅩⅩⅡ, ⅩLⅥ, ⅩⅩⅩⅤ and ⅩLⅦ (24.4%, 21.86%, 15.84% and 15.58%, respectively), as well as a further 11-residue compound, Lipostrigocin LSG-like ⅩLⅤ at 1.94%. All the other 15-residue sequences were produced between 3.57% and 6.67% of the total lipopeptaibol production.
3.6 Comparative analysis of lipopeptaibols produced by species of clade Viride
The microheterogeneity within the lipopeptaibols were different from the SF1 peptaibols (Table 8). The repetitive motif Vxx/Lxx-Aib-Gly appeared in all sequences between R3-R5 in 7- and 10-residue lipopeptaibols, except for T. cf. strigosellum SZMC 28007 lipopeptaibols, where Gly5 was substituted with Ala5. In 11-residue lipopeptaibols, the R3-R5 Vxx/Lxx-Aib-Gly motif continued with a Gly with the repetition of the Vxx/Lxx-Aib-Gly motif between R7-9. In 15-residue lipopeptaibols, the Vxx/Lxx-Aib-Gly motif appeared from R3-R5, R7-9, and R11-13. Between these motifs, Gly was at positions R6 and R10. This phenomenon can also be the result of module skipping based on the sequence similarities, that the “original” lipopeptaibols are the longer ones [27, 39], however, it needs further research and proof.
The 1most produced sequences, 2consensus sequences and 3amino acid varieties of lipopeptaibols produced by each strain (Lipostrigocin LSG like lipopeptaibols from T. cf. strigosellum were produced by SZMC 28391, while Lipostrigaibols were from T. cf. strigosellum SZMC 28007)
The 6-residue lipopeptaibols contained only one variable position at R4 which could be Gly/Ala (Table 8). Gly and Ala were present at this position equally. The same Gly/Ala variability is also present in 7-residue lipopeptaibols at R5, with Gly in Lipostrigocin LSG-like sequences and Ala in the Lipostrigaibol sequences. R1 could be Oc/Aib/OcVxx, while R6 was Vxx/Lxx. Lipostrigocin LSG-like Ⅳb produced by T. cf. strigosellum SZMC 28391 is unique, because it has Phe at R3 instead of Lxx (Tables 6, 7 and 8). Only LP237 F7 and LP237 F8 lipopeptaibols from Tolypocladium geodes contain Phe3 in their sequences, however, they are 11-residue long [18, 51]. The microheterogeneity of 7-residue Lipostrigaibols appeared at R4, R5 and R6, which is discussed above. In the 10-residue Lipohamatins, Vxx/Lxx variability could be detected at R3, R7 and R8. The 11-residue Lipostrigocin LSG-like sequences showed microheterogenity at R1, R3, R4, R7 and R10 (Table 8). R1 was OcAla/OcAib/OcVxx, R4 Ala/Aib, while R3, R7 and R10 were Vxx/Lxx. The microheterogenity of the 15-residue Trichogin GA IX like lipopeptaibols was similar to the 11-residue lipopeptaibols, however, R4 was only Aib and R11 and R14 were also Vxx/Lxx (Table 8). Trichogin GB IX-like Ⅰ was unique among these sequences, it contained Vxxol instead of Lxxol at its C terminus (Tables 6 and 7).
The consensus sequence was different from the most produced sequence of the two T. cf. strigosellum strains at R4, which are underlined in Table 8. At R3, Lipostrigocin LSG-like peptaibols containing mostly Lxx differed from the most produced Vxx containing Lipostrigocin LSG-like Ⅶa. This could also be observed among Lipohamatins where Lipohamatin II contained Vxx instead of the most abundant Lxx at R3. Lipohamatins also differed from the most produced Lipohamatin II at their R7, they contained equally Vxx and Lxx (Table 8). Lipostrigocin LSG-like peptaibols produced by T. cf. strigosellum SZMC 28391 contained Aib4, while the most produced Lipostrigocin LSG-like Ⅻb contained Vxx4 (Table 8). Lipostrigaibols produced by T. cf. strigosellum SZMC 28007 contained Aib4, while the most produced Lipostrigaibol-like ⅫbV contained Vxx4 (Table 8). Similarly, Lipostrigocin LSG-like lipopeptaibols produced by T. koningii SZMC 28387 contained Lxx3, while the most produced Lipostrigocin LSG-like Ⅻa Vxx3 (Table 8). The most produced Lipohamatin II contained Vxx3 instead of the most abundant Lxx3 and Lxx7, while R7 was equally Vxx or Aib (Table 8). The most produced Trichogin GA IX like ⅩⅩⅥ contained Lxx3 and Lxx11 instead of the most abundant Vxx3 and Vxx11 in lipopeptaibols produced by T. hamatum SZMC 28747. OcAib1was detected in Trichogin GB IX-like ⅩⅩ, while OcAib and OcVxx was equally present in Trichogin GB IX-like sequences produced by T. cf. dorothopsis SZMC 28005.
3.7 Quantitative analysis of the whole peptaibiome produced by the members of clade Viride
Tables 3 and 6 also contain the integrated area of each peak belonging to each compound produced by the strains investigated. The values are based on the integrated area under each peak. In the previous sections, the peptaibols and lipopeptaibols were quantified based on the percentages of the total peptaibol and lipopeptaibol production separately, however, it is also important to quantify the entire peptaibiom altogether. T. cf. dorothopsis SZMC 28390 produced exculively peptaibols, which quantity was discussed above, while T. atroviride SZMC 28748 also produced peptaibol-like sequences apart from peptaibols, but no lipopeptaibols. Within the strains investigated in clade Viride, one peptaibol subgroup was produced by each strain, on the other hand, a few lipopeptaibols were the same produced by different strains, which are shown in Table 6. Strains T. koningii SZMC 28387, T. cf. strigosellum SZMC 28007 and T. hamatum SZMC 28747 all produced the 7-residue Lipostrigocins LSG Ⅵ and Ⅶ, while only T. koningii SZMC 28387 and T. hamatum SZMC 28747 produced Lipostrigocins LSG Ⅲa, Ⅳa, Ⅴ, Ⅷa and Ⅸa. T. cf. strigosellum SZMC 28007 produced similar lipopeptaibols only differing in 1 amino acid exchange from the ‘a’ type variants, which are Lipostrigocins LSG Ⅲb, Ⅳb, Ⅷb and Ⅸb (Table 6). Strains T. hamatum SZMC 28747 and T. cf. dorothopsis SZMC 28005 produced the same 11-residue Lipostrigocin LSG-like ⅩⅩⅩⅡ and ⅩⅩⅩⅤ, and the 15-residue Trichogin GB IX-like ⅩⅢ and ⅩⅩ lipopeptaibols.
T. koningii SZMC 28387 produced only two peptaibol compounds in higher abundances, Trikoningins KA-like ⅩLⅤ and ⅩLⅢ (9.14% and 8.28%, respectively). More lipopeptaibols were produced in higher quantities, Lipostrigocin LSG-like ⅩⅣ (25.44%) is the most produced, followed by Lipostrigocin LSG-like ⅩⅩ (7.72%), Lipostrigocin LSG-like ⅩⅥ (7.48%) and Lipostrigocin LSG-like ⅩⅤ (6.52%). The rest of the peptaibiome for T. koningii ranges from 0.002% to 4.17%. Strigaibol-like Ⅻ from T. cf. strigosellum SZMC 28391 appeared as the most produced peptaibol (14.19%), followed by Strigaibol-like ⅩⅣ (10.83%). The most produced lipopeptaibol by this strain is Lipostrigocin LSG-like Ⅷb (22.53%), followed by Lipostrigocin LSG-like Ⅵ (19.42%). The “unidentified compound” was also produced at higher quantity (10.6%). The rest of the peptaibiome ranges from 0,03 to 2.53%. Among T. atroviride SZMC 28748 peptaibols, the most produced compound is Trichorzianin TA-like ⅩⅩⅨ (8.56%) followed by Trichorzianin TA-like ⅩⅩⅤ (7.42%). The rest of the peptaibiome ranged from 0.03 to 4.37%. The peptaibol-like compounds were produced in much less quantities ranging from 0.003 to 0.22%. T. hamatum SZMC 28747 produced Tricholongin LB-like ⅩⅥ (6.87%) as the most produced peptaibol, which was followed by Tricholongin LB-like ⅩⅢ (6.07%). This strain also produced several lipopeptaibols in high amounts, the most produced being Lipostrigocin LSG-like ⅩⅩⅩⅨ (8.48%) followed by Lipostrigocin LSG-like ⅩⅩⅩⅣ (4.8%) and Lipostrigocin LSG-like ⅩLⅡ (4.62%). Other lipopeptaibols, Lipostrigocin LSG-like ⅩⅩⅩⅦ, Trichogin GB IX-like Ⅺ, Trichogin GB IX-like ⅩⅩⅢ, Trichogin GB IX-like ⅩⅩⅢ, Trichogin GB IX-like ⅩⅩⅤ and Trichogin GB IX-like ⅩⅩⅥ were also produced in high quantities rarnging from 2.87% to 4.08%. This was the only investigated strain, which produced these many compounds in high amounts. The most produced peptaibol of T. cf. dorothopsis SZMC 28005 is Dorothopsin B XXVI (3.38%) and Dorothopsin B ⅩⅢ (3.23%), which is much less than the most produced lipopeptaibols, Lipostrigocin LSG-like ⅩⅩⅩⅡ (16.29%), Lipostrigocin LSG-like ⅩLⅥ (14.59%), Lipostrigocin LSG-like ⅩⅩⅩⅤ (10.57%) and Lipostrigocin LSG-like ⅩLⅦ (10.4%). T. cf. strigosellum SZMC 28007 produced less peptaibols than lipopeptaibols similarly to T. cf. dorothopsis SZMC 28005. Strigosellin A Ⅸ (5.62%) was the most produced peptaibol followed by Strigosellin A ⅩⅢ (5.03%), Strigosellin A ⅩⅣ (4.23%) and Strigosellin A Ⅻ (4.15%). The most produced lipopeptaibols are Lipostrigaibol-like ⅩⅤ (14.54%), Lipostrigaibol-like ⅩⅢ (13.59%) and Lipostrigaibol-like ⅩⅥ (12.76%). The rest of the peptaibiome ranges from 0.05% to 6.12%.
3.8 Structural investigation of a representative peptaibol of clade Viride, Trikoningin KA V produced by T. koningii
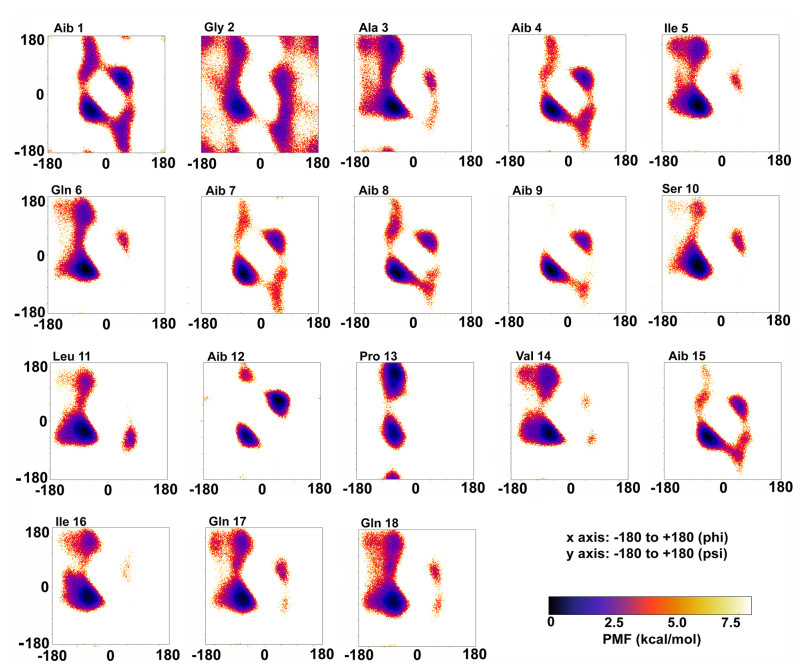
Strain T. koningii SZMC 28387 produced Trikoningin KA-like peptaibols out of which Trikoningin KA-like ⅩⅨ and its positional isomers (Trikoningin KA-like ⅩⅩⅩⅤ, ⅩL, ⅩLⅢ and L) from Tables 3 and 4 show the same sequences as Trikoningin KA (TRK-V) identified by Goulard et al. [29]. They also determined the isomers of Lxx and Vxx, and since TRK-V is a good representation of peptaibols produced by members of clade Viride, this sequence was selected for aMD simulations. Auvin-Guette et al. [61] firstly reported the structure of TRK-V to be “right-handed helical form” using circular dichroism spectroscopy and was found to membrane active and displayed antibiotic activity against Staphylococcus aureus. We previously reported the classical MD simulations-based results of TRK-V [38]. Using the newly optimized aMD parameters, the previously reported TRK-V molecule was elucidated again to obtain the complete canonical ensemble and compare with short classical MD conducted earlier. TRK-V was previously identified as a peptaibol produced by T. koningiopsis along with two other 11-residue sequences, Trikoningins KB Ⅰ and KB Ⅱ [62]. Another study by our group identified novel peptaibols which were named as “Koningiopsins” with TRK-V as the closest sequence [38]. TRK-V, positionally isomeric with sequences Pept-Ⅴb, -Ⅵb, and -Ⅶ of T. gamsii, is a 19-residue peptaibol with seven Aib residues in its sequence. Aib is an achiral residue, which has been shown to promote helix formation and can exist in both right- and left-handed helix regions on the Ramachandran plot. To determine the propensities of each residue for a given secondary structure type, their relative free energies were calculated which clearly describe an energetically favourable conformation (Fig. 2). The spread of dihedral angle scatter during the simulation indicates that the system underwent through all the conformations. The darkest regions indicate energetically preferable conformations.
The reweighted phi-psi torsional distribution for each residue of TRK-V. Most residues show an energy minimum in the right-handed α-helical region
Our previous results of Ramachandran plots of TRK-V [38] showed overall secondary structural propensity towards a left-handed helical conformation. However, upon comparing the reweighted phi-psi populations in Fig. 2 with the same plots from Fig. 1 from Marik et al. [38] (100 ns long classical MD), it becomes clear that most residues like Aib1-Ile5, Ser10, Leu11, Pro13, and Gln17-Gln18 lie in the same free energy minima. However, many others flanking the central region like Gln6-Aib9, Aib12, and Val14-Ile16 show shifts from the predominant left-handed helical regions to the right-handed helical region. It is understandable for all Aib residues as the probability of occurring in both left-handed and right-handed helical quadrants is the same due to its achiral nature. A significant shift was observed in the free energy minima of Gln6 and Val14 residues from the left-handed to the right-handed helical region.
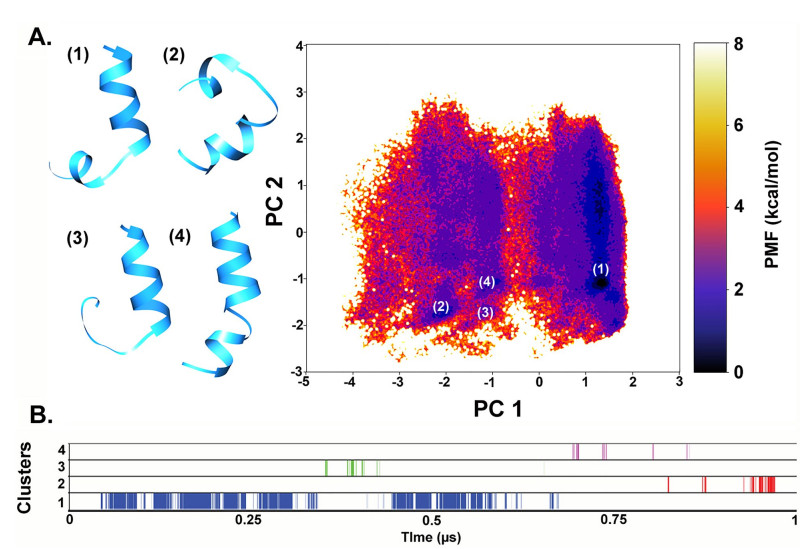
Figure 3A describes the reweighted free-energy landscape of TRK-V as obtained from the first two principal components of dihedral PCA of 1 μs long aMD simulation. It is clear that the largest cluster lies at the energy minimum and the corresponding representative structure presumes a helical shape with the C-terminus showing a hinge-like bend. The loss of helical fold before the C-terminus is responsible for this hinge-like motion and is a characteristic of the Aib-Pro bond found in all long peptaibols. The next two largest clusters 2 and 3 correspond to highly bent and C-terminus loss-of-helix conformations, respectively, which probably indicates intermediate states. The 4th cluster with the smallest population lies at a separate region on the free-energy landscape and corresponds to the highly helical, slightly bent conformation. This structure is most likely to be observed using experimental methods like X-ray crystallography along with the curved backbone conformations. The 1st cluster appears to populate a separate region on the FEL map which can be accessed under 2 kcal mol−1. Its representative structure can be explained by the presence of a helix-destabilizing residue Ile16 at the C-terminus [63]. Their occurrence along the course of the simulation can be visualized in Fig. 3B. Interestingly, Ile residues at the 5th and 16th positions also show a strong right-handed α-helical propensity (Fig. 2). Li and Deber [64] measured the helical propensity of amino acid residues in membrane and reported that ß-branched Ile and Val can act as ‘helix-promoters’ in membrane environment but as ‘helix-destabilizers’ in aqueous environment. The effect of seemingly similar residues Leu/Ile/Val substitution on peptaibol sequences must be studied in detail. There is evidence in case of other antimicrobial peptides where a single substitution could affect interaction with lipid membranes and resulting bioactivity. For example, Aurein 2.2 and 2.3 vary only at a single position between Leu/Ile and show different bioactivities against bacterial species [65]. Studies on derivatives of another peptide, δ-lysin, showed that peptides rich in Ile may bind bilayer membranes more efficiently than Leu-rich peptides [66]. Deber and Stone [67] published a systematic study of the effect of Leu/Ile substitution on membrane-active peptides and found that peptides with all Leu/Ile placed on one side (lipophilic side) in contrast to the less hydrophobic face on the other side, showed that the Ile-rich peptide had a higher capability to insert in the membrane than the Leu-rich peptide. Similarly, Ile-rich peptides were found to be more protected from the action of proteinases than Leu peptides in the presence of lipid bilayers. Recently, Nakatani et al. [68] also reported the construction of derivatives of Trichorovin-XII with varying Ile content where all of them folded into α-helix like β-bend spiral. However, the compound found to be the most bioactive contained the highest number of Ile in its sequence. Ile analogs also showed greater ion channel activity compared to Leu analogs.
A Reweighted FEL of the first two principal components calculated from dihedral angles, phi-psi, for better clustering based on internal motions. The representative structures of TRK-V corresponding to various energy minima have also been provided, B Diagrammatic representation of cluster distribution along the simulation trajectory
3.9 Predicted bioactivity of peptaibiotics produced by the members of Trichoderma clade Viride
A detailed investigation of the bioactivity and function of the reported peptaibols is required to evaluate their application potential in agriculture or as therapeutics. However, their potential bioactivity can be predicted based on their sequences and the already available knowledge. First of all, the longer peptaibols (19-residues) are supposed to show higher bioactivity against bilayer membranes than their shorter counterparts. It is known that the 20-residue Alamethicin folds into a strict α-helix at the N-terminus and a 310-helix at the C-terminus, and the resulting length of the folded peptide exactly spans the transmembrane width to form ion-channels. On the other hand, the 15-residue Ampullosporin A rather shows a detergent-like activity in membranes and is less active [69]. Adam et al. [70] reported on the conclusive effect of Aib-foldamers (peptaibol) length and C-terminal lipophilicity on ionophoric and antibacterial activity. The longer foldamers with a hydrophobic C-terminus were found to be more active, suggesting that the length of the peptides is an important criterion that may affect their activity. The 20-residue Paracelsins investigated in our previous work [27] folded into strict α-helices very similarly to Alamethicin, and thus, can be expected to have a similar mechanism of action.
Another major marker is the amount of Aib residues in the sequence which imparts a helical structure but not in a highly strict conformation. It can fluctuate between α-helical and 310-helical conformations, thereby, expanding the length of the folded peptide to span the transmembrane region and also provide space to fit in bulky neighbouring side chains [71]. In the newly reported subgroups of Strigosellins and Dorothopsins, a total of 8 Aib residues could be found in the sequences. All peptaibol subgroups, except for Strigosellins, have 3 consecutive Aib residues from R7-R9. The presence of three consecutive helix-shape promoting Aib residues [72, 73] provides a strong helical character to the structure which seems to be crucial for most membrane-active peptides. Therefore, the presence of helix-promoting residues is also one of the main crucial features.
Lastly, the presence and position of helix-breaking residues like Gly and Pro can give us crucial clues towards helical stability. Peptide curvature or “kink” may also be a crucial feature in determining the biological activity. Cheng and Chang [74] discussed the implication of the more stable kinked form of Alamethicin than the energetically less stable linear form in voltage-gating. They proposed that linearization of the Alamethicin helices from the kinked structure upon increasing electric potential beyond a threshold may be the first event in voltage-gating mechanism. The kinks are introduced by helix-breaker residues like Pro and Gly in peptaibols. Kaduk et al. [75] showed that the substitution of Gly11 and Pro14 in Alamethicin did not affect channel formation but reduced conductance levels and significantly reduced lifetimes. Duclohier [76] in his review noted that without any applied voltage to the membrane system, Alamethicin with a higher kink angle than Trichotoxin will have a greater embedment of the N-terminus in the hydrocarbon core while the C-terminus lies flat at the bilayer interface. This greater embedment of the N-terminus, in turn, explains the higher voltage dependence of Alamethicin. Similarly, Trikoningin KA-V with a helical N-terminus, 19-residue length and a slight kink at the Aib-Pro bond also seems to be ideal for transmembrane orientation and ion channel formation. In this study, all sequences have an Aib-Pro bond which is expected to introduce this influential ‘kink’ to the folded structure. Moreover, a high bioactivity is expected from lipopeptaibols due to the presence of the fatty acid chain which is prone to strongly interact with the lipid bilayer [77]. Due to their shorter length they may not form transmembrane ion channels, but may have a different mechanism of action than peptaibols.
4 Conclusions
The peptaibiotic production of Trichoderma clade Viride revealed known and new subgroups of peptaibiotics. The new subgroups of peptaibols are Strigosellins A and B with 14 and 6, Dorothopsins A-a, -b, -c, -d, -e and -f with 13, 8, 1, 3, 1 and 2, as well as, Dorothopsin B compounds with 36 completely new sequences, respectively. The already known and certain new compounds belong to the Trikoningin KA, Trichorzianin TA and Tricholongin LB subgroups with 51, 33 and 21 identified sequences, respectively. New lipopeptaibol subgroups are Lipostrigosellins and Lipohamatins with 4, and 4 completely new sequences. The other identified new and already known lipopeptaibols belong to the Lipostrigocin LSG and Trichogin GB subgroups with a total of 103 sequences. The peptaibol subgroups produced by the examined species were completely different from each other, while a few lipopeptaibols were produced by more than one species within the clade. We selected the TRK-V sequence for structure elucidation as a good representation of peptaibols produced by T. koningii SZMC 28387 which also has been elucidated before. Enhanced sampling of MD simulations allowed to observe that TRK-V takes a right-handed helical shape with a strong helix-disrupting effect of Ile residues when simulated in an aqueous environment. Previous research indicates towards a better membrane interactive potential of Ile-containing peptides in a hydrophobic environment. The results provide an idea about the three-dimensional structural folding of all Trikoningin-like peptaibols mentioned in this work.
Notes
Acknowledgements
We acknowledge the use of supercomputing facility by the Governmental agency for IT development (Kormányzati Informatikai Fejlesztési Ügynökség, KIFÜ), Hungary, for aMD simulations. B.G. and A.V. were supported by the Stipendium Hungaricum Scholarship by the Tempus Public Foundation during this research.
Author contributions
Conceptualization: [Tamás Marik], [Chetna Tyagi], [László Kredics]; Methodology: [Tamás Marik], [Sándor Kocsubé], [Mónika Varga], [Chetna Tyagi]; Formal analysis and investigation: [Tamás Marik], [Bonaya Gufu], [Anusha Vishwanathula], [Dóra Balázs], [Ákos Rozsnyói], [Gergő Terna], [Fanni Kovács]; Writing—original draft preparation: [Tamás Marik], [Bonaya Gufu]; Writing—review and editing: [Tamás Marik], [Bonaya Gufu], [Chetna Tyagi], [László Kredics], [András Szekeres], [Irina. S. Druzhinina]; Funding acquisition: [László Kredics], [András Szekeres], [Tamás Papp], [Csaba Vágvölgyi]; Resources: [László Kredics], [András Szekeres], [Tamás Papp], [Csaba Vágvölgyi]; Supervision: [Tamás Marik], [László Kredics], [András Szekeres].
Funding
T.M. and D. B. was supported by the Sholarship Program of the Ministry of Culture and Innovation, Financed from the National Research, Development and Innovation Fund (EKÖP-24-4SZTE-629, EKÖP-24-4-SZTE-605, respectively). C.T. is supported by the short-term KGYNK-2024 grant from the Hungarian Academy of Sciences (HAS). S.K., C.V. and T.P. were supported by the grants HUN-REN 2001007 and TKP2021-EGA-28.
Data availability
All data generated or analysed during this study are included in this published article.
Declarations
Competing interests
The authors declare that there are no conflicts of interest associated with this work.
References
-
1.Kredics L, Hatvani L, Naeimi S, Körmöczi P, Manczinger L, Vágvölgyi C, Druzhinina I. Biodiversity of the genus Hypocrea/Trichoderma in different habitats. In: Gupta VK, Schmol M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohi MG, editors. Biotechnology and biology of Trichoderma. Elsevier: Amsterdam; 2014. p. 3–24. PubMed Google Scholar
-
2.Schuster A, Schmoll M. Biology and biotechnology of Trichoderma. Appl Microbiol Biotechnol. 2010;87: 787-99. CrossRef PubMed Google Scholar
-
3.Ejechi BO. Biological control of wood decay in an open tropical environment with Penicillium sp. and Trichoderma viride. Int Biodeterior Biodegrad. 1997;39(4): 295-9. CrossRef PubMed Google Scholar
-
4.Hatvani L, Antal Z, Manczinger L, Szekeres A, Druzhinina IS, Kubicek CP, Nagy A, Nagy E, Vágvölgyi C, Kredics L. Green mold diseases of Agaricus and Pleurotus spp. are caused by related but phylogenetically different Trichoderma species. Phytopathology. 2007;97(4): 532-7. CrossRef PubMed Google Scholar
-
5.Kredics L, Antal Z, Dóczi I, Manczinger L, Kevei F, Nagy E. Clinical importance of the genus Trichoderma. Acta Microbiol Immunol Hung. 2003;50(2–3): 105-17. CrossRef PubMed Google Scholar
-
6.Komon-Zelazowska M, Neuhof T, Dieckmann R, von Döhren H, Herrera-Estrella A, Kubicek CP, Druzhinina IS. Formation of atroviridin by Hypocrea atroviridis is conidiation associated and positively regulated by blue light and the G protein GNA3. Eukaryot Cell. 2007;6(12): 2332-42. CrossRef PubMed Google Scholar
-
7.Druzhinina IS, Komoń-Zelazowska M, Kredics L, Hatvani L, Antal Z, Belayneh T, Kubicek CP. Alternative reproductive strategies of Hypocrea orientalis and genetically close but clonal Trichoderma longibrachiatum, both capable of causing invasive mycoses of humans. Microbiology. 2008;154(11): 3447-59. CrossRef PubMed Google Scholar
-
8.Jaklitsch WM, Samuels GJ, Dodd SL, Lu BS, Druzhinina IS. Hypocrea rufa/Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Stud Myol. 2006;56(1): 135-77. CrossRef PubMed Google Scholar
-
9.Cai F, Druzhinina IS. In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 2021;107(1): 1-69. CrossRef PubMed Google Scholar
-
10.Mukherjee PK, Wiest A, Ruiz N, Keightley A, Moran-Diez ME, McCluskey K, Pouchus YF, Kenerley CM. Two classes of new peptaibols are synthesized by a single non-ribosomal peptide synthetase of Trichoderma virens. J Biol Chem. 2011;286(6): 4544-54. CrossRef PubMed Google Scholar
-
11.Hou X, Sun R, Feng Y, Zhang R, Zhu T, Che Q, Zhang G, Li D. Peptaibols: diversity, bioactivity, and biosynthesis. Eng Microbiol. 2022;2(3): 100026. CrossRef PubMed Google Scholar
-
12.Degenkolb T, Brückner H. Peptaibiomics: towards a myriad of bioactive peptides containing Cα-dialkylamino acids. Chem Biodivers. 2008;5(9): 1817-43. CrossRef PubMed Google Scholar
-
13.Brückner H, Maisch J, Reinecke C, Kimonyo A. Use of α-aminoisobutyric acid and isovaline as marker amino acids for the detection of fungal polypeptide antibiotics. Screening of Hypocrea Amino Acids. 1991;1(2): 251-7. CrossRef PubMed Google Scholar
-
14.Mukherjee PK, Horwitz BA, Kenerley CM. Secondary metabolism in Trichoderma–a genomic perspective. Microbiology. 2012;158(1): 35-45. CrossRef PubMed Google Scholar
-
15.Degenkolb T, Fog Nielsen K, Dieckmann R, Branco-Rocha F, Chaverri P, Samuels GJ, Thrane U, von Döhren H, Vilcinska A, Brückner H. Peptaibol, secondary-metabolite, and hydrophobin pattern of commercial biocontrol agents formulated with species of the Trichoderma harzianum complex. Chem Biodivers. 2015;12(4): 662-84. CrossRef PubMed Google Scholar
-
16.Neumann NK, Stoppacher N, Zeilinger S, Degenkolb T, Brückner H, Schuhmacher R. The peptaibiotics database–a comprehensive online resource. Chem Biodivers. 2015;12(5): 743-51. CrossRef PubMed Google Scholar
-
17.Degenkolb T, Berg A, Gams W, Schlegel B, Gräfe U. The occurrence of peptaibols and structurally related peptaibiotics in fungi and their mass spectrometric identification via diagnostic fragment ions. J Pept Sci. 2003;9(11–12): 666-78. CrossRef PubMed Google Scholar
-
18.Toniolo C, Crisma M, Formaggio F, Peggion C, Epand RF, Epand RM. Lipopeptaibols, a novel family of membrane active, antimicrobial peptides. Cell Mol Life Sci. 2001;2001(58): 1179-88. CrossRef PubMed Google Scholar
-
19.Samuels GJ, Dodd SL, Lu BS, Petrini O, Schroers HJ, Druzhinina IS. The Trichoderma koningii aggregate species. Stud Mycol. 2006;56(1): 67-133. CrossRef PubMed Google Scholar
-
20.Kubicek CP, Bissett J, Druzhinina I, Kullnig-Gradinger C, Szakacs G. Genetic and metabolic diversity of Trichoderma: a case study on South-East Asian isolates. Fungal Genet Biol. 2003;38(3): 310-9. CrossRef PubMed Google Scholar
-
21.López-Quintero CA, Atanasova L, Franco-Molano AE, Gams W, Komon-Zelazowska M, Theelen B, Müller WH, Boekhout T, Druzhinina I. DNA barcoding survey of Trichoderma diversity in soil and litter of the Colombian lowland Amazonian rainforest reveals Trichoderma strigosellum sp. nov. and other species. Antonie Van Leeuwenhoek. 2013;104: 657-74. CrossRef PubMed Google Scholar
-
22.Atanasova L, Crom SL, Gruber S, Coulpier F, Seidl-Seiboth V, Kubicek CP, Druzhinina IS. Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genomics. 2013;14: 1-15. CrossRef PubMed Google Scholar
-
23.Gazis R, Chaverri P. Wild trees in the Amazon basin harbor a great diversity of beneficial endosymbiotic fungi: is this evidence of protective mutualism. Fungal Ecol. 2015;17: 18-29. CrossRef PubMed Google Scholar
-
24.Löytynoja A, Vilella AJ, Goldman N. Accurate extension of multiple sequence alignments using a phylogeny-aware graph algorithm. Bioinformatics. 2012;28(13): 1684-91. CrossRef PubMed Google Scholar
-
25.Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 2020;37(1): 291-4. CrossRef PubMed Google Scholar
-
26.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21): 4453-5. CrossRef PubMed Google Scholar
-
27.Marik T, Tyagi C, Balázs D, Urbán P, Szepesi Á, Bakacsy L, Endre G, Rakk D, Szekeres A, Andersson MA, Salonen H, Druzhinina IS, Vágvölgyi C, Kredics L. Structural diversity and bioactivities of peptaibol compounds from the Longibrachiatum clade of the filamentous fungal genus Trichoderma. Front Microbiol. 2019;10: 1434. CrossRef PubMed Google Scholar
-
28.Stoppacher N, Neumann NK, Burgstaller L, Zeilinger S, Degenkolb T, Brückner H, Schuhmacher R. The comprehensive peptaibiotics database. Chem Biodivers. 2013;10(5): 734-43. CrossRef PubMed Google Scholar
-
29.Goulard C, Hlimi S, Rebuffat S, Bodo B. Trichorzins HA and MA, antibiotic peptides from Trichoderma harzianum I. Fermentation, isolation and biological properties. J Antibiot. 1995;48(11): 1248-53. CrossRef PubMed Google Scholar
-
30.Tyagi C, Marik T, Vágvölgyi C, Kredics L, Ötvös F. Accelerated molecular dynamics applied to the peptaibol folding problem. Int J Mol Sci. 2019;20(17): 4268. CrossRef PubMed Google Scholar
-
31.Case DA, Aktulga HM, Belfon K, Ben-Shalom I, Brozell SR, Cerutti DS, et al. Amber 2021. University of California, San Francisco. PubMed Google Scholar
-
32.Tyagi C, Marik T, Szekeres A, Vágvölgyi C, Kredics L, Ötvös F. Tripleurin XIIc: Peptide folding dynamics in aqueous and hydrophobic environment mimic using accelerated molecular dynamics. Molecules. 2019;24(2): 358. CrossRef PubMed Google Scholar
-
33.Park YS, Kim ES, Deyrup ST, Lee JW, Shim SH. Cytotoxic peptaibols from Trichoderma strigosum. J Nat Prod. 2024;87: 2081-94. CrossRef PubMed Google Scholar
-
34.Degenkolb T, Dieckmann R, Nielsen KF, Gräfenhan T, Theis C, Zafari D, Chaverri P, Ismaiel A, Brückner H, von Döhren H, Thrane U, Petrini O, Samuels GJ. The Trichoderma brevicompactum clade: a separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol Progress. 2008;7: 177-219. CrossRef PubMed Google Scholar
-
35.Huang Q, Tezuka Y, Hatanaka Y, Kikuchi T, Nishi A, Tubaki K. Studies on metabolites of mycoparasitic fungi. V. ion-spary ionization mass spectrometric analysis of Trichokonin-Ⅱ, a peptaibol mixture obtained from the culture broth of Trichoderma koningii. Chem Pharm Bull. 1996;44(3): 590-3. CrossRef PubMed Google Scholar
-
36.Rebuffat S, Prigent Y, Auvin-Guette C, Bodo B. Tricholongins BI and BII, 19-residue peptaibols from Trichoderma longibrachiatum: solution structure from two-dimensional NMR spectroscopy. Eur J Biochem. 1991;201(3): 661-74. CrossRef PubMed Google Scholar
-
37.Degenkolb T, Gräfenhan T, Berg A, Nirenberg HI, Gams W, Brückner H. Peptaibiomics: screening for polypeptide antibiotics (peptaibiotics) from plant-protective Trichoderma species. Chem Biodivers. 2006;3(6): 593-610. CrossRef PubMed Google Scholar
-
38.Marik T, Tyagi C, Racić G, Rakk D, Szekeres A, Vágvölgyi C, Kredics L. New 19-residue peptaibols from Trichoderma clade Viride. Microorganisms. 2018;6(3): 85. CrossRef PubMed Google Scholar
-
39.Degenkolb T, Karimi Aghcheh R, Dieckmann R, Neuhof T, Baker SE, Druzhinina IS, Kubicek CP, Brückner H, von Döhren H. The production of multiple small peptaibol families by single 14-module peptide synthetases in Trichoderma/Hypocrea. Chem Biodivers. 2012;9: 499-535. CrossRef PubMed Google Scholar
-
40.Wenzel SC, Meiser P, Binz TM, Mahmud T, Müller R. Nonribosomal peptide biosynthesis: point mutations and module skipping lead to chemical diversity. Angew Chem Int Edit. 2006;45(14): 2296-301. CrossRef PubMed Google Scholar
-
41.El Hajji M, Rebuffat S, Lecommandeur D, Bodo B. Isolation and sequence determination of trichozianines A antifungal peptides from Trichoderma harzianum. Int J Pept Protein Res. 1987;29: 207-15. CrossRef PubMed Google Scholar
-
42.Süssmuth RD, Mainz A. Nonribosomal peptide synthesis—principles and prospects. Angew Chem Int Edit. 2017;56(14): 3770-821. CrossRef PubMed Google Scholar
-
43.Lee SJ, Yeo WH, Yun BS, Yoo ID. Isolation and sequence analysis of new peptaibol, boletusin, from Boletus spp. J Pept Sci. 1999;5(8): 374-8. CrossRef PubMed Google Scholar
-
44.Dornberger K, Ihn W, Ritzau M, Gräfe U, Schlegel B, Fleck WF, Metzger JW. Chrysospermins, new peptaibol antibiotics from Apiocrea chrysosperma Ap101. J Antibiot. 1995;48(9): 977-89. CrossRef PubMed Google Scholar
-
45.Yun BS, Yoo ID, Kim YH, Kim YS, Lee SJ, Kim KS, Yeo WH. Peptaivirins A and B, two new antiviral peptaibols against TMV infection. Tetrahedron Lett. 2000;41(9): 1429-31. CrossRef PubMed Google Scholar
-
46.Kronen M, Kleinwaechter P, Schlegel B, Haertl A, Graefe U, Ampullosporins B. C, D, E1, E2, E3 and E4 from Sepedonium ampullosporum HKI-0053: structures and biological activities. J Antibiot. 2001;54(2): 175-8. CrossRef PubMed Google Scholar
-
47.Rinehart KL Jr., Gaudioso LA, Moore ML, Pandey RC, Cook JC Jr., Barber M, Sedgwick RD, Bordoli RS, Tyler AN, Green BN. Structures of eleven zervamicin and two emerimicin peptide antibiotics studied by fast atom bombardment mass spectrometry. J Am Chem Soc. 1981;103(21): 6517-20. CrossRef PubMed Google Scholar
-
48.Brückner H, Kirschbaum J, Jaworski A. Sequences of peptaibol antibiotics trichoaureocins from Trichoderma aureoviride. Proceedings of the 27th European Peptide Symposium, Sorrento, 2002. p. 362–363. PubMed Google Scholar
-
49.Ayers S, Ehrmann BM, Adcock A, Kroll DJ, Carcache de Blanco EJ, Shen Q, Swanson SM, Falkinham JO 3rd, Wani MC, Mitchell SM, Pearce CJ, Oberlies NH. Peptaibols from two unidentified fungi of the order Hypocreales with cytotoxic, antibiotic, and anthelmintic activities. J Pept. 2012;18(8): 500-10. CrossRef PubMed Google Scholar
-
50.Kyle KE, Puckett SP, Caraballo-Rodríguez AM, Rivera-Chávez J, Samples RM, Earp CE, Raja HA, Pearce CJ, Ernst M, van der Hooft JJJ, Adams ME, Oberlies NH, Dorrestein PC, Klassen JL, Balunas MJ. Trachymyrmex septentrionalis ants promote fungus garden hygiene using Trichoderma-derived metabolite cues. Proc Natl Acad Sci U S A. 2023;120(25): e2219373120. CrossRef PubMed Google Scholar
-
51.Tsantrizos YS, Pischos S, Sauriol F, Widden P. Peptaibol metabolites of Tolypocladium geodes. Can J Chem. 1996;74(2): 165-72. CrossRef PubMed Google Scholar
-
52.Stoppacher N, Zeilinger S, Omann M, Lassahn PG, Roitinger A, Krska R, Schuhmacher R. Characterisation of the peptaibiome of the biocontrol fungus Trichoderma atroviride by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(12): 1889-98. CrossRef PubMed Google Scholar
-
53.Sasamura S, Muramatsu H, Takase S, Fujie A, Fujii T, Hino M, Sakamoto K, Hashimoto M. Bioconversion of AS1387392: screening and characterization of actinomycetes that convert AS1387392 to AS1429716. J Antibiot. 2010;63(11): 637-42. CrossRef PubMed Google Scholar
-
54.Pandey RC, Cook JC, Rinehart KL. High resolution and field desorption mass spectrometry studies and revised structures of alamethicins Ⅰ and Ⅱ. J Am Chem Soc. 1977;99: 8469-83. CrossRef PubMed Google Scholar
-
55.Rinehart KL Jr, Cook JC Jr, Meng H, Olson KL, Pandey RC. Mass spectrometric determination of molecular formulas for membrane-modifying antibiotics. Nature. 1977;269(5631): 832-3. CrossRef PubMed Google Scholar
-
56.Rebuffat S, El Hajji M, Hennig P, Davoust D, Bodo B. Isolation, sequence, and conformation of seven trichorzianines B from Trichoderma harzianum. Int J Pept Prot Res. 1989;34: 200-10. CrossRef PubMed Google Scholar
-
57.Duval D, Rebuffat S, Goulard C, Prigent Y, Becchi M, Bodo B. Isolation and sequence analysis of the peptide antibiotics trichorzins PA from Trichoderma harzianum. J Chem Soc Perkin Trans. 1997;1: 21. CrossRef PubMed Google Scholar
-
58.Rowley DC, Kelly S, Kauffman CA, Jensen PR, Fenical W. Halovirs A-E, new antiviral agents from a marine-derived fungus of the genus Scytalidium. Bioorg Med Chem. 2003;11: 4263-74. CrossRef PubMed Google Scholar
-
59.Christner C, Zerlin M, Gräfe U, Heinze S, Küllertz G, Fischer G. Lipohexin, a new inhibitor of prolyl endopeptidase from Moeszia lindtneri (HKI-0054) and Paedlomyces sp. (HKI-0055; HKI-0096) Ⅱ. inhibitory activities and specificity. J Antibiot. 1997;50(5): 384-9. CrossRef PubMed Google Scholar
-
60.China N, Blond A, Goulard C, Bodo B, Rebuffat S. Structure and membrane properties of trichogin GB IX from Trichoderma longibrachiatum, the longest sequence among lipopeptaibols. In: Peptides 2000, 26th European Peptide Symposium, Montpellier, France, 5 pages. PubMed Google Scholar
-
61.Auvin-Guette C, Rebuffat S, Vuidepot I, Massias M, Bodo B. Structural elucidation of trikoningins KA and KB, peptaibols from Trichoderma koningii. J Chem Soc Perk T. 1993;1(2): 249-55. CrossRef PubMed Google Scholar
-
62.McMullin DR, Renaud JB, Barasubiye T, Sumarah MW, Miller JD. Metabolites of Trichoderma species isolated from damp building materials. Can J Microbiol. 2017;63(7): 621-32. CrossRef PubMed Google Scholar
-
63.Lyu PC, Sherman JC, Chen A, Kallenbach NR. Alpha-helix stabilization by natural and unnatural amino acids with alkyl side chains. Proc Natl Acad Sci U S A. 1991;88(12): 5317-20. CrossRef PubMed Google Scholar
-
64.Li SC, Deber CM. A measure of helical propensity for amino acids in membrane environments. Nat Struct Biol. 1994;1(6): 368-73. CrossRef PubMed Google Scholar
-
65.Cheng JT, Hale JD, Elliot M, Hancock RE, Straus SK. Effect of membrane composition on antimicrobial peptides aurein 2.2 and 2.3 from Australian southern bell frogs. Biophys J. 2009;96(2): 552-65. CrossRef PubMed Google Scholar
-
66.Cherry MA, Higgins SK, Melroy H, Lee HS, Pokorny A. Peptides with the same composition, hydrophobicity, and hydrophobic moment bind to phospholipid bilayers with different affinities. J Phys Chem B. 2014;118(43): 12462-70. CrossRef PubMed Google Scholar
-
67.Deber CM, Stone TA. Relative role(s) of leucine versus isoleucine in the folding of membrane proteins. Peptide Sci. 2019;111(1): e24075. CrossRef PubMed Google Scholar
-
68.Nakatani T, Koga A, Goto S, Inoue M, Shigedomi K, Seki K, Araki K, Taira J, Kodama H, Osada S. Importance of isoleucine residue in ion channel formation ability of 11-residue peptaibols. Bioorg Med Chem. 2024;110: 117839. CrossRef PubMed Google Scholar
-
69.Ritzau M, Heinze S, Dornberger K, Berg A, Fleck W, Schlegel B, Hartl A, Grafe U. Ampullosporin, a new peptaibol-type antibiotic from Sepedonium ampullospomm HKI-0053 with neuroleptic activity in mice. J Antibiot. 1997;50: 722-8. CrossRef PubMed Google Scholar
-
70.Adam C, Peters AD, Lizio MG, Whitehead GF, Diemer V, Cooper JA, Cockroft SL, Clayden J, Webb SJ. The role of terminal functionality in the membrane and antibacterial activity of peptaibolmimetic aib foldamers. Chemistry. 2018;24: 2249-56. CrossRef PubMed Google Scholar
-
71.Crisma M, Formaggio F, Moretto A, Toniolo C. Peptide helices based on α-amino acids. Pept Sci. 2006;84: 3-12. CrossRef PubMed Google Scholar
-
72.Banerjee R, Basu G. A short Aib/Ala-based peptide helix is as stable as an Ala-based peptide helix double its length. Chem Bio Chem. 2002;3: 1263-6. CrossRef PubMed Google Scholar
-
73.Tsuji G, Misawa T, Doi M, Demizu Y. Extent of helical induction caused by introducing α-aminoisobutyric acid into an oligovaline sequence. ACS Omega. 2018;3: 6395-9. CrossRef PubMed Google Scholar
-
74.Cheng SF, Chang DK. Proline-induced kink in a helix arises primarily from dihedral angle energy: a molecular dynamics simulation on alamethicin. Chem Phys Lett. 1999;301: 453-7. CrossRef PubMed Google Scholar
-
75.Kaduk C, Dathe M, Bienert M. Functional modifications of alamethicin ion channels by substitution of glutamine 7, glycine 11 and proline 14. Biochim Biophys Acta. 1998;1373: 137-46. CrossRef PubMed Google Scholar
-
76.Duclohier H. Helical kink and channel behaviour: a comparative study with the peptaibols alamethicin, trichotoxin and antiamoebin. Eur Biophys J. 2004;33: 169-74. CrossRef PubMed Google Scholar
-
77.De Zotti M, Biondi B, Park Y, Hahm KS, Crisma M, Toniolo C, Formaggio F. Antimicrobial lipopeptaibol trichogin GA Ⅳ: role of the three Aib residues on conformation and bioactivity. Amino Acids. 2012;43: 1761-77. CrossRef PubMed Google Scholar
Copyright information
© The Author(s) 2025
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.