The need for smart microalgal bioprospecting
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sector.
Abstract
Microalgae's adaptability and resilience to Earth's diverse environments have evolved these photosynthetic microorganisms into a biotechnological source of industrially relevant physiological functions and biometabolites. Despite this, microalgae-based industries only exploit a handful of species. This lack of biodiversity hinders the expansion of the microalgal industry. Microalgal bioprospecting, searching for novel biological algal resources with new properties, remains a low throughput and time-consuming endeavour due to inefficient workflows that rely on non-selective sampling, monoalgal culture status and outdated, non-standardized characterization techniques. This review will highlight the importance of microalgal bioprospecting and critically explore commonly employed methodologies. We will also explore current advances driving the next generation of smart algal bioprospecting focusing on novel workflows and transdisciplinary methodologies with the potential to enable high-throughput microalgal biodiscoveries. Images adapted from (Addicted04 in Wikipedia File: Australia on the globe (Australia centered).svg. 2014.; Jin et al. in ACS Appl Bio Mater 4:5080–5089, 2021; Kim et al. in Microchim Acta 189:88, 2022; Tony et al. in Lab on a Chip 15, 19:3810–3810; Thermo Fisher Scientific INC. in CTS Rotea Brochure).Graphical Abstract
Keywords
Microalgae Bioprospecting Fluorescent probing1 Introduction
In recent years, the CO2 emission and climate change-induced necessity of developing new Green Circular Economies and CO2 fixation technologies has put microalgae in the spotlight [6, 7]. Microalgae, a term used to describe a biodiverse polyphyletic group comprised of both eukaryotic (Glaucophyta, Rhodophyta, Chlorophyta, Haptophyta, Cryptophyta, Ochrophyta, Miozoa, Cercozoa and Euglenozoa) and prokaryotic (Cyanobacteria) taxa [8, 9], are predominantly photosynthetic microorganisms [7, 10–12] which can be ubiquitously found across all of Earth's habitats [7].
Microalgae's valuable biochemical composition, photosynthetic and CO2 fixation efficiency, fast growth rates and their lack of competition for arable land have spawned the creation of novel microalgal industries such as biomanufacturing, atmospheric carbon capture and its conversion into high-value bioproducts and/or biofuels [6, 7, 13]. Despite microalgae's biodiversity [8, 9], these nascent industries only employ a limited number of species, restricting their abilities to exploit the diverse biological properties present across the wide range of microalgal taxa [14–16].
Bioprospecting, the systematic search for novel microalgae with biotechnological and/or commercial potential, is an essential requirement for the expansion of the microalgal industry [17]. However, current microalgal bioprospecting suffers from low-throughput, inefficient, time-consuming and non-scalable methodologies. This review aims to provide an updated overview of the current state of microalgal bioprospecting, addressing its limitations and pitfalls. By providing a comprehensive perspective on currently used methodologies, identifying and critically analysing innovative and transdisciplinary approaches, this review aims to serve as a source of inspiration for advancing the implementation of smart microalgal bioprospecting.
2 Climate change: a substrate for a microalgal revolution
2.1 Global carbon engineers
In the late Archean Eon, Cyanobacteria pioneered oxygenic photosynthesis, culminating in the Great Oxidation Event that transformed Earth's atmosphere and facilitated the evolution of aerobic life [18, 19]. Microalgae have successfully colonized Earth's photic habitats [20–26]. Adapting to this broad habitat range has exposed microalgae to diverse selective pressures, fostering their remarkable phenotypic and ecological diversity [12]. Regardless of their versatility, all microalgae play a pivotal role in global nutrient cycling [12, 27]. Microalgae annually contribute 50% of the total globally assimilated carbon [28] and drive the biological carbon pump [29] responsible for the long-term storage of carbon in the deep ocean [30]. As primary producers, microalgae form the basis of aquatic food webs [30] and are responsible for the biosynthesis of essential biomolecules such as Docosahexaenoic Acid (DHA), the bioaccumulation of which across higher trophic levels led to early human brain evolution [31, 32].
Like the rest of Earth's species, microalgae are threatened by climate change [33] driven by anthropogenic CO2 emissions [34]. The eventual translation of global warming consequences into ocean ecosystems will reduce marine CO2 availability due to ocean acidification and salinification [30]. These environmental shifts will pose complex challenges to microalgae, potentially altering their physiology, ecological interactions, and community compositions, which may cascade to higher trophic levels and devastatingly disrupt ecological and geochemical processes [28, 30, 33, 35].
2.2 Microalgal cell factories for every industry
Chlorella vulgaris, isolated and studied in 1890, marked the beginning of scientific microalgal exploration [36–38]. However, human-microalgae interactions date back centuries, for instance, the consumption of Arthospira (Spirulina) can be traced back to the fourteenth century [39, 40]. Microalgae have shaped human evolution [31, 32], but they might also allow us to evade the CO2 emission-driven path of climate change-led collapse that the industrially developed world is currently following [34, 41, 42]. International efforts now strive for carbon neutrality [42, 43], an endeavour necessitating innovative biotechnological technologies [44–46] and, as in the Archean Eon [18], CO2 has again put microalgae in a central position. Microalgae are considered a CO2 mitigation strategy [7, 47] due to their rapid growth and CO2 fixation rates, not requiring arable land for cultivation and being a cellular factory of biotechnologically valuable carbon molecules [47–49] (Fig. 1).
3 Microalgal bioprospecting
3.1 Bioprospecting microalgae's biodiversity dark matter
Microalgae support a diverse array of industries [16]. Regardless of microalgae's high biodiversity [9], only a limited number of microalgal species are currently exploited in industrial processes [15, 16, 51–54] (Table 1).
Described microalgal biodiversity encompasses 50, 000 species [55], with conservative estimates placing total diversity at around 200, 000 species [56]. Despite this high biodiversity, the number of commercially cultivated species remains in the dozens, with highly economically developed regions like the European Union only cultivating 46 microalgal species [51]. This under-utilization of microalgal biodiversity constrains the nascent industry, impedes diversification into novel applications and reduces industrial resilience.
Bioprospecting, systematically sampling and selecting nature's biological resources—taxonomic species, biomolecules, biosynthetic pathways, genes or genomes—offers the possibility of harnessing microalgal biodiversity by exploring novel microalgae. Leveraging microalgae's sustainable nature, bioprospecting not only supports industrial, commercial or research microalgal applications [57], but also aligns with the accomplishment of several United Nations Sustainable Development Goals (UN SDGs) [58] (Fig. 2). This presents a new opportunity for discovering novel species, biometabolites and genes using novel biotechnological methodologies to exploit microalgae's untapped potential.
3.2 Current state of microalgal bioprospecting
Bioprospecting, like microalgal consumption as a food source [39, 40], has been omnipresent throughout human history [60]. The expansion of the commercial use of microalgae beyond the food industry, sparked by the US oil crises in the latter half of twentieth century [61], propelled large-scale algal cultivation and intensified microalgal bioprospecting. This, in conjunction with academic advances, has led to the discovery and characterization of diverse taxonomical alternatives to develop the ever-widening microalgae-supported industrial applications.
Microalgal bioprospecting methodologies involve two distinct phases: [1] microalgae sampling and isolation and [2] characterization to select microalgae with desired traits. Workflows can be classified further into univariate approaches, limited to widespread sampling and characterization for a specific trait, and multivariate methodologies that integrate multiple selection strategies in the search of strains combining several traits to narrow down the pool of possible candidate taxa [62].
3.2.1 Sampling and isolation
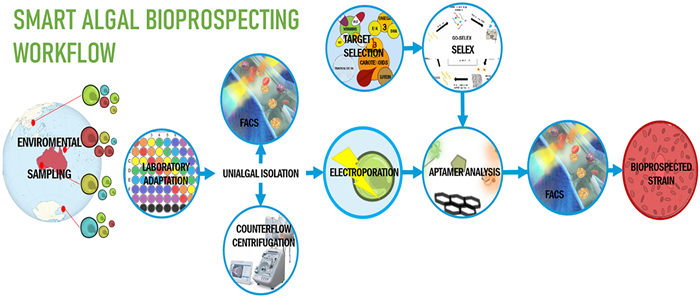
The first step in bioprospecting is sampling, which is typically achieved by directly collecting water samples in aquatic environments. Despite this, many other available methodologies have been adapted to non-aquatic sampling [63–65]. Upon arrival at the laboratory, samples generally undergo nutrient enrichment with different media; typically, ES, F/2 or BG11SW, to promote specific taxa [63]. Subsequently, single-cell or agar isolation methodologies, often combined with dilution techniques, isolate specific microalgae to establish mono-algal cultures [63]. Depending on factors like sampling location, identified microalgal taxa, and intended use of the culture, axenic cultures may be obtained through selective treatments with antimicrobials such as antibiotics, antifungals, antivirals, and/or enzymatic treatments [63].
3.2.2 Traditional characterization methodologies
Varied characterization methodologies can be applied depending on the methodology and the desired phenotypic trait/s, an observable and quantifiable characteristic/s, used for selection. Contrarily, growth rates, commonly estimated from correlation between Optical Density (OD) measurements with a spectrophotometer; typically at 680–750 nm [66–68], and cell counts; performed with a haemocytometer chamber [69, 70], are a point of consensus across microalgal bioprospecting studies (Table 2). Growth kinetics can also be employed in a multivariate manner [66, 69, 71–73]. For instance, Rezaei, et al. (2023) simultaneously isolated, grew and selected high growing strains bioprospected from high-mountain lakes and grown in cold conditions [66] and Condori, et al. (2024) bioprospected microalgae from contaminated water environments to characterize their growth and nutrient removal efficiency when grown in explosive industry wastewater [69]. These manifold combinations of screening criteria enable the selection of algal strains both successfully and rapidly growing under a selected set of stringent culture conditions [66, 69, 71, 73]. Similarly prevalent is the weighting of dry biomass; performed by sun, hot air, oven, spray or freeze drying [68, 70, 74, 75].
Commonly employed characterization methodologies in microalgal bioprospecting
Microalgae's taxonomic biodiversity has turned these microorganisms into cellular powerhouses to produce an even more diverse range of biometabolites [47–49]. As such, biometabolite characterization; the identification, study and quantification of the industrially relevant molecules present in newly bioprospected isolates, is a common step in microalgal bioprospecting workflows.
Bioprospecting efforts in microalgae, driven by the pursuit of economically viable biofuels, have consistently focused on lipid and fatty acid characterization [7, 66, 68–70, 74, 76–78]. Traditionally, multi-step gravimetric methods have been employed to quantify lipid fractions, known for their precision but limited in providing a comprehensive lipidomic profile [66, 69, 74, 78–80]. Alternatively, chromatographic techniques such as Thin-Layer (TLC), High-Performance Liquid (HPLC), or Gas Chromatography (GC) [66, 68, 70, 76] [79] coupled with Mass Spectroscopy (MS), Flame Ionization Detector (FID) or Time-Of-Flight (TOF) [79, 81] offer detailed analyses of transesterified or transmethylated fatty acids [81], enhancing the effectiveness of the lipidomic assessment for biofuel potential [85, 87]. This is the case of Ammar, et al. (2024), which simultaneously gravimetrically quantified and GC–MS profiled the lipid and Fatty Acid Methyl Ester (FAME) fractions of bioprospected Tunisian algal isolates [68].
Photosynthetic microalgal pigments—chlorophylls, carotenoids, and phycobilins—are pivotal in microalgal biometabolite production, attracting significant bioprospecting interest [82]. Pigments are analysed after having been extracted using organic solvents [83] through spectrophotometric readings at pigment-specific autofluorescence wavelengths, often using equations to extrapolate these reads for total pigment quantification [67–69, 84]. Variations include HPLC-based quantification as seen in Grubišic, et al. (2022) or Patel, et al. (2022), who also extrapolated the total pigment content of their bioprospected microalgae by using established equations [70, 78].
Due to their diverse applications [47, 49, 85] microalgal carbohydrates are also repeatedly bioprospected [67, 69, 74, 86]. Commonly, carbohydrate quantification is performed through the phenol–sulfuric acid method by Dubois, et al. (1956) [87] or similar derivatives [86, 88]. However, just as with gravimetric lipid quantification, Dubois and derivative methods do not explore the carbohydrate profile; a knowledge gap also solved by GC–MS or GC-FID [88, 89].
Microalgal proteins; yet another highly sought after algal biometabolite [90, 91], are quantified using spectrophotometric methodologies [67, 70, 74]. Both Assobhi, et al. (2024) and Grubišic, et al. (2022) employed the Lowry method [92] for protein quantification [70, 74], a technique based on colorimetric change [93]. Comparably, Araj-Shirvani, et al. (2024) read the OD at 595 nm of their Coomassie Blue Dye stained sample and compared the results with a bovine serum albumin standard [67]. A different approach can be found on Condori, et al. (2024) [69] or Cruz, et al. (2023) [89], both of which indirectly quantified protein content by using a nitrogen-to-protein conversion rate obtained from the literature [94].
Notably, microalgal bioprospecting efforts span far beyond major metabolite classes and include singular metabolites such as phenolic compounds [67, 70, 95], flavonoids [70], and lipid, protein or carbohydrate extracts for their potential applications as antimicrobial [70, 96], anticancer [95, 97], antioxidant [67, 70, 95, 98], anti-ageing [95], enzyme inhibiting [95] or prebiotic agents [95]. Additionally, physiological algal processes like bioaccumulation [69, 99, 100], biomineralization [101] phytohormone [102], exopolysaccharide (EPS) [102, 103] or nanoparticle (NP) [104] production are also bioprospected for their industrial applications.
3.3 Bioprospecting: a Herculean endeavour
Despite significant scientific and technological advancements in marine biotechnology [105], the continuous worldwide expansion of the microalgal industry [16] and the rapid development of marine bioprospecting [106], microalgal bioprospecting remains a burdensome endeavour.
To begin with, environmental sampling methodologies are fast and inexpensive [63], but developing a gold-standard guide for best microalgal sampling practice is still to occur [107]. Moreover, sampling location and sample quantities are often determined by convenience, geographic proximity and the presence of a water body. This non-selective bioprospecting overlooks the ecological evolution and natural selection history that shape stress-adapted microalgal biometabolite composition, disregarding the invaluable natural linkage between desired phenotypes and sampling locations [108]. Indiscriminate sampling may yield diverse samples, but technical constraints in post-sampling isolation, culturing, and biometabolite characterization make this approach inefficient. The primary obstacle for bioprospecting pipelines is the current requirement of monoalgal status, a culture condition only achieved after a time-consuming and resource-intensive separation process. Additionally, improper enrichment and isolation workflows can arise taxonomic prevalence and survival biases stemming from enrichment-media selection, microbial competition or, in the case of dinoflagellate presence, to outright predation [109, 110]. Adding to this low-throughput wound is the requirement, a recurrently left desire, for axenic cultures in most current microalgal characterization methodologies [111]. Furthermore, a full methodology guide for the achievement of this tedious, time-consuming and hardly achievable; or impossible [112] axenic outcome is also yet to be created [63].
Similarly ineffective, current microalgal characterization methodologies require a biomass yield in the milligrams [107]. Ideally, this excruciating high biomass yield should be achieved with culture optimization but realistically, bioprospecting efforts do not progress beyond basic discriminations such as fresh or saltwater [70, 84, 86]. This presents a double-edged dilemma as, while rapid growth is a desirable trait, its relevance is diminished if the culture conditions used for its assessment do not match the future culturing parameters required at the prospective application of the strain. Furthermore, the induction of several commonly characterized phenotypic traits; such as lipid production, is largely culture-parameter-dependent [113]. Consequently, a lack of consideration for phenomic plasticity, the ability of an organism to adapt its phenotypic traits to the culture conditions its subject to [114], and this linkage between culture conditions and observable phenes, can result in the erroneous selection of inefficient microalgal isolates or the overlooking of the most efficient ones.
Likewise, the number of desired phenotypic traits and the methodologies employed for their characterization are crucial. Yet, commonly employed characterization methodologies are outdated, low throughput and fail to characterize all relevant parameters [115]. The former and the latter can be exemplified by gravimetric [66, 69, 74] or phenol–sulfuric quantification [67, 69, 74, 86] methodologies. These multi-step, tedious processes requiring milligram quantities of biomass are, on top of that, unable to profile the lipid and carbohydrate species present in the samples respectively [79, 80, 115]. Though they are an improvement, chromatographic techniques are not the silver bullet of microalgal characterization due to their multistep, time-consuming and costly nature, their unforgiving sample quantity requirements and their difficult result standardization across studies [116].
Overall, outdated, inefficient and low throughput culturing, isolation and characterization methodologies (Table 2) [115] compromise the application of efficient workflows and are significant hurdles to the development, application and economical success of microalgal bioprospecting and the expansion of the algal industry.
4 Smart microalgal bioprospecting
Microalgal bioprospecting is currently limited by the employment of low throughput methodologies, a subsequent impediment to the implementation of a biodiverse portfolio of industrially profitable microalgae species. To respond to this impending need, we propose the development of a new wave of high-throughput ''smart'' microalgal bioprospecting workflows founded on ecological considerations, novel approaches and robust methodologies, equipment and techniques.
4.1 From the pond to the laboratory
Sampling and targeting everything, then selecting later is not currently viable. Therefore, smart bioprospecting must aim to enhance the likelihood of isolating only desired microalgal isolates. This must begin with a critical assessment of environmental sampling locations with ecological pressures that are likely to generate the desired microalgal phenotypic traits [17, 62], an approach known as bio-rational collection and screening [17, 62]. Similarly desirable is the application of multivariate approaches that link bio-rational sampling with natural-pressure-imitating stringent laboratory culturing conditions [66, 69, 71, 73]. Combined, these methodologies significantly decrease the pool of ''to-be-characterized'' microalgal strains [62], lowering economic, human and time costs [63] and enhancing throughput. The success of this is exemplified by Royal DSM, a Dutch company that has achieved commercialization and EU novel food approval for a high DHA-producing Schizochytrium sp. strain bioprospected bio-rationally [17, 117, 118].
Despite marketing microalgae's biodiversity, many "bioprospecting" efforts limit their activity to characterizing strains from culture collections [17]. Despite being a source of potentially relevant strains [119], culture collections do not sample bio-rationally. As such, indiscriminate ''culture collection bioprospecting''; coupled with the likely genetic drift, shifts in allele frequencies arisen during population bottlenecks generated by routine serial subculture, and phenotypic changes that a long-term culture is prone to suffer [120, 121], is likely to yield non-competitive strains. Additionally, climatically and microbially ecological obsolescence makes this approach suboptimal when prospective large-scale culturing is a desired bioprospected strain trait. Conversely, local bio-rational sampling offers access to ecologically-competitive isolates [122, 123]. Nevertheless, exotic sampling also holds vast potential. For instance, targeting extremophilic microalgae capable of thriving under cultivation conditions lethal to contaminants is a proven commercially successful approach, as evidenced by Dunaliella salina or Arthospira platensis dominating their respective markets [24, 124].
Combining bio-rational sampling with multivariate approaches and focusing on unique natural [125–127] or man-made [128, 129] ecological niches will enhance both the taxonomical diversity of industrially exploited microalgal species and the discovery of novel compounds with industrial applications [130–132].
4.2 From algal soup to monoalgal culture
Bettering the currently infuriatingly low-throughput enrichment and monoalgal culture isolation [133] requires adaptability to the objectives behind smart bioprospecting. For instance, approaches desiring taxonomical biodiversity should focus on impeding the introduction of unwanted biases. This can be achieved by the fractionation of the original multialgal sample into smaller individual aliquots followed by the enrichment of each individual fraction with multiple distinct culturing medias and conditions. In turn, these differing parameters foster aliquot-specific "biases", which result in differing taxa-specific growth and, even, survival rates. On the contrary, multivariate approaches used to isolate growth under stringent wastewater [134], salinity [135], pH [136, 137], CO2 [138] and light [139] parameters should continue their expansion towards novel selecting parameters such as growth in anaerobic digestates [140] or unfiltered coal-fired flue gas [141].
Regardless, implementing automation and high-throughput cultivation methodologies are increasingly imperative. Multi-well plates [142, 143] and microfluidic platforms [144–146] offer throughputs orders of magnitude above traditional culturing, both in terms of time and in the number of nutrient profiles tested per run [145, 147]. An example is provided in Radzun, et al. (2015) which rapidly optimized the composition and individual concentrations of 12 macro- and microelements under non-limiting CO2 and light conditions in an automated 1, 728 multi-well setup considering the maximum growth rates of 8 different microalgae [147]. Despite these advances, workflow limitations persist due to the small volumes offered by these approaches. Overcoming this also requires automation, which is now implemented in photobioreactors to provide larger culturing volumes. This nascent field, practically unknown to microalgal bioprospecting, is highly adaptable and allows the monitoring of as many culture parameters as sensors exist [148–151] and, as seen in Nguyen, et al. (2018) [150], the adoption of cheap sensor-culturing setups will reduce processing time and costs, reallocating resources to other crucial non-automatable processes.
Nevertheless, no recent technical advances have been as impactful and widely adopted in microalgal bioprospecting as Flow cytometry (FC) and Fluorescence-activated cell sorting (FACS). FC offers rapid and reliable screens of environmental samples through single-cell interrogation by laser interception and detection of scatter light to generate datasets with ''events'' that represent different cell populations. Coupled with a cell sorter, FC becomes FACS, enabling the isolation of desired populations based on specific sets of parameters called "gates". This technology, as demonstrated by Jakob's, et al. (2013) [152], has no less than revolutionised microalgal isolation into monoalgal cultures [63, 133, 153]. FACS is also instrumental in generating monoalgal axenic cultures [63, 152–154]. By interrogating environmental samples for chlorophyll fluorescence, it distinguishes chlorophyll-containing algal cells from bacteria, dead cells, and debris, after which, chlorophyll-positive gating allows monoalgal isolation [111, 152, 155]. Despite its benefits, FACS isolation exposes cells to shear force, electrostatic charges and high-energy lasers. As also seen in Jakob's, et al. (2013), the success of FACS depends on cell parameters like size, shape, abundance and hardiness [152, 154], limiting its universal applicability across all microalgae taxa [63, 154]. This prevents abandoning micromanipulation, the isolation of single cells through aspiration with microcapillaries, a resource-intensive, low-throughput technique requiring highly skilled microscopy users proficient in microalgal morphological identification.
However, an evolution in centrifugation technology holds promise, whereas FACS has limitations. Traditional centrifugation is integral in microalgal studies for tasks like supernatant removal, re-culturing and biomass harvesting [156]. Although density centrifugation has also been proposed to achieve axenic cultures [157], it requires optimization, lacks automation and can reduce cell viability due to shear forces [156]. A promising high-throughput and automatable alternative lies in counterflow centrifugation-based instrumentation [158]. Widely used in cell therapy [158, 159] for its cell concentration, buffer exchange and cell-size based separation capabilities with minimal impact on cell viability [160], the potential of counterflow-centrifugation is yet to be investigated by microalgal researchers. Therefore, counterflow centrifugation could greatly benefit microalgal bioprospecting by enabling size-based separation among various microalgal taxa and their common contaminants [161], facilitating monoalgal axenic isolation.
Regardless, the taxa-wide prevalence of microalgae-attached bacteria [162] makes neither FACS nor counterflow centrifugation an axenic-obtaining silver bullet. Importantly, the reasons and viability behind removing this unique phycosphere must be addressed as, in some cases, this bacterial ''contamination'' is not only not problematic but essential or positive for microalgal development [162, 163]. This is so much so that phycosphere-bioprospecting for the enhanced production of microalgal-bacterial bioproducts or co-cultivation-arisen microalgal productivity improvements is an increasingly developing field [164–167]. Despite this, some workflows still require absolute axenic status and, for that, the employment of ultrasonication; > 20 kHz ultrasonic waves, and its high throughput adaption is needed [155].
After establishing monoalgal cultures, the last pre-characterization step is microalgal identification. Both morphological and traditional molecular identification [168] are not conducive to high-throughput applications. While metabarcoding is preferred for its ability to reduce human error and bias, its independence from taxonomic expertise [168], and its capability to comprehensively assess axenic status or identify phycosphere constituents, there remains a need for improved throughput. As such, the implementation of reliable Artificial Intelligence (AI)—Deep Learning Image Recognition software [168–170] or novel molecular identification technologies [170, 171] is necessary. An example of the latter is Jahn's, et al. (2014) development of a high-throughput metabarcoding 12-well plate setup, employing boiled MiliQ water for algal lysis and an automated sequencing chromatogram analysis methodology [171].
4.3 Bioprospecting into the future
Currently, microalgal characterization methodologies are inefficient, have low throughput [115], and they often require monoalgal cultures. Despite the throughput capabilities of existing and proposed methodologies for achieving this unialgal, axenic or not, state, creating and maintaining multiple monoalgal cultures during bioprospecting is nonsensical, given that most will be discarded during characterization. Could selection be achieved from an enriched multi-taxonomical algal mixture? Moreover, what if a living algal sample were not required?
Culture-independent bioprospecting, has been achieved through metagenomic approaches such as Whole Genome Shotgun [172, 173]. Advances in Next Generation Sequencing (NGS) and global sequencing projects have transformed metagenomic mining, enabling the in silico search for genes encoding biometabolites [172], which becomes commercially viable upon successful gene expression [174]. Nevertheless, microalgal genome sequence databases are currently restricted to a small number of model species, and there is insufficient knowledge of microalgal metabolic pathways [175] and achieving reliable genetic transformation of multiple microalgae species remains unattainable [176]. Together, these current factors limit the application of non-culture dependent microalgal bioprospecting.
Spectroscopic imaging techniques have frequently been proposed as alternative culture-dependent characterization methodologies that enable high-throughput, non-invasive and low-cost characterization [177]. Among these, visible/near infrared, Fourier transform infrared (FTIR), and Raman spectroscopy have garnered significant attention due to their potential in microalgal biometabolite characterization. However, these techniques face several limitations that necessitate extensive optimization and complicate their implementation into high-throughput phenotyping workflows [114]. For instance, Raman spectroscopy requires species-specific optimization [114] and is affected by the background fluorescence of microalgal pigments, while FTIR is affected by spectral interference from water [178]. Altogether, and despite their potential, spectroscopic methodologies are not yet well-suited for high-throughput biometabolite and taxa-wide microalgal bioprospecting.
Another proposed high-throughput approach is Fluorescent Probing (FP), the interrogation of autofluorescing molecules or molecule-specific fluorophores to identify and quantify target metabolites [179]. This approach allows for the assessment of non-monoalgal cultures and has been widely used in lipid bioprospecting, with Nile Red and BODIPY 505/515 being the most common fluorophores for cheap and rapid in situ lipidomic assessment [179, 180]. FP provides throughput levels unimaginable with traditional lipid quantifications when integrated with FACS [180] or microplate workflows [181]. Moreover, various other fluorescent dyes, such as SYTOX Green, a nucleic acid binding fluorophore [182, 183], have extended FP far beyond lipidomics [180, 184, 185]. Although FP presents significant potential for enhancing bioprospecting throughput, challenges persist, including variable fluorophore permeation across different taxa [186], interference with microalgal pigment autofluorescence [180] and the need to establish accurate correlations between fluorescence intensity and target molecule quantity. Investigating novel fluorophores, such as AC-202; a promising alternative to BODIPY [187], could help overcome these challenges and facilitate the transition from traditional low-throughput methodologies [179, 186].
Many of the complications associated with fluorophore-based FP can be mitigated by utilizing microalgae's innate ''fluorophores'': pigments. Pulse Amplitude Modulated (PAM) fluorometry [188], is a non-invasive and high-throughput methodology that offers valuable phenotypic insights by assessing light absorption and photoprotective potentials of microalgal strains [189, 190]. Combining PAM with the optimization of cultivation or stress-induction conditions provides a deeper understanding of the interactions between microalgal photophysiology and cultivation parameters [114, 189, 191]. The Phenoplate, Herdean, et al. (2022), exemplifies the high-throughput capabilities of PAM when integrated with variable cultivation parameters [189, 192], suggesting that further development of similar [142] rapid FP multiparametric workflows could revolutionize microalgal characterization.
Nonetheless, current FP is significantly limited by variable cell wall penetrating abilities and the lack of a diverse array of biometabolite-specific fluorophores or probing methodologies. However, a transformative shift towards nucleic acid or peptide-led and electrophoresis or nanoparticle-assisted methodologies appears imminent. Described elsewhere as biosensors [193], these novel metabolite-specific platforms are not merely aspirational but are already being successfully applied in microalgal research [2, 3, 194, 195]. Central to this approach are Aptamers, small DNA, RNA or peptide chains that specifically bind to biomolecular targets. The Systematic Evolution of Ligands by Exponential Enrichment (SELEX) has evolved into a highly adaptable in vivo or in vitro methodology for aptamer synthesis [196]. SELEX can be performed for a myriad of molecular; or cellular targets [196]. Furthermore, the affordable and versatile nature of nucleic acid and peptide modification facilitates post-SELEX enhancements such as truncation, extension, site-directed mutagenesis and modification or attachment of fluorophores [197] and Quantum Dots, novel inorganic fluorophores [198–201].
Widely applied in the biomedical field [202], aptamer-sensing has been primarily applied in microalgae to detect Harmful Algal Bloom-produced biotoxins [203]. Due to their sensitivity, specific binding, rapid and cost-effective development, and post-development adaptability, aptamers are promising for microalgal metabolite sensing. Several research groups have already proved this potential. For example, Prof. Yoon-E Choi's laboratory (Korea University) has successfully employed single-stranded DNA aptamers for the in vivo sensing of ATP, paramylon and, importantly, β-carotene in the non-cell wall possessing microalgae Euglena gracilis and Ochromonas danica [2, 3, 195, 204, 205]. Furthermore, aptamers targeting biotechnologically relevant molecules, such as the bio-available form of vitamin B12, methylcobalamin [206], and H2O2, a marker of oxidative stress [207], or specific binding sites, such as the plastoquinone binding niche of Photosystem Ⅱ D1 protein [208] have been developed and are poised for implementation across a broader spectrum of microalgae. Aptamer designs targeting biological docking sites, organelle-specific motifs, or even cell-specific motifs [209], represent promising avenues for expanding aptamer applications in diverse biotechnological fields and for enabling smart bioprospecting approaches.
Microalgae's cell walls, cell membranes, and organelle walls still pose significant challenges for intracellular aptamer delivery. Common methods for introducing foreign DNA into microalgae include glass bead agitation, microparticle bombardment, Agrobacterium-based delivery and bacterial conjugation [210, 211]. However, these techniques are not suitable for high-throughput bioprospecting workflows due to their variable efficiency and, in the cases of Agrobacterium delivery or bacterial conjugation [210–212], their non-transient delivery falls under Genetically Modified Organism legislation which would restrict the commercialization of newly bioprospected strains [213]. Despite these challenges, novel intracellular delivery methods are under development [211, 214, 215]. Cell-penetrating peptides, Liposome-mediated delivery and Nanoparticles show promise as alternatives [211], though these emerging approaches require further research to assess their potential, especially with algae. However, even if these methods achieve higher efficiencies, microalgal biodiversity is likely to impede taxa-wide standardized permeation capabilities, a prospective bias that complicates the implementation of these delivery methods for bioprospecting.
In contrast, reversible electroporation, the transient electro-generation of temporary membrane pores, is a reliable and widely used method with a proven record of successful intracellular delivery rates across various microalgae taxa [211, 216]. Electroporation is commonly achieved through Pulsed Electric Fields (PEF), electrical pulses of a voltage in the kV and of a duration in the ms or ns range [217], parameters that require species-specific optimization [216]. Although various methodologies for optimizing electroporation settings exist [216, 218], all electroporation optimization requires monoalgal cultures. Cell size, a parameter that can be selectively managed through upstream methodologies like FACS or counterflow centrifugation, significantly impacts the success of cell permeabilization [219, 220]. Therefore, size-based fractionation of a non-monoalgal sample and its distribution across different electroporation parameter gradients may be essential for achieving efficient taxa-wide intracellular delivery. This approach could be facilitated by commercially available high-throughput, multi-well electroporation platforms or by developing custom in-house setups [221, 222].
Regardless, free-floating aptamers present in a microalgal cytosol would face enzymatic degradation even if successful intracellular delivery is achieved. A solution for this issue is NP conjugation. For example, Prof. Yoon-E Choi's successful aptamer-sensing has been performed through conjugation with Graphene Oxide NPs (GOnS) and Gold NPs (AuNPs) [2, 3, 195, 204, 205]. Interestingly, GOnS provides fluorophore quenching [2, 3, 195, 204, 205], providing a simple yet effective ON–OFF detection platform. Free-floating aptamer-fluorophores are firstly incubated with GOnS to achieve the quenching or OFF state and only after exposure to the aptamer-specific molecule these aptamer-fluorophore complexes detach, subsequently freeing the fluorophore from quenching and achieving the ON state. Furthermore, specific NP designs can also selectively target certain intracellular compartments [223, 224], a transport choice that can also be achieved through NP conjugation with intracellular guiding peptides [225, 226].
5 Conclusion
Microalgal bioprospecting holds transformative potential for advancing diverse microalgal industrial applications and bettering industrial robustness. Yet, current efforts remain hindered by a lack of bio-rational sampling and outdated, low-throughput characterization methodologies. Addressing these gaps demands a shift towards Smart Microalgal Bioprospecting.
The impending abandonment of non-selective sampling requires the integration of bio-rational sampling with expanded use of high-throughput tools such as FACS or implementing novel transdisciplinary approaches such as counterflow centrifugation. In combination with the use of automatable high throughput cultivation platforms such as multi-well plates and microfluidics, this new wave of sampling and laboratory adaptation workflows can improve throughputs, reduce costs and facilitate monoalgal isolation at a scale, an essential stage for accurate microalgal characterization. Moreover, emerging technologies such as fluorescent probing, whether non-invasive PAM or novel biosensor platforms are promising in enabling biometabolite detection. Despite the potential, a further array of biometabolite-specific aptamer or peptide-based biosensors must be developed. Furthermore, cell-penetrating peptides, liposome-mediated delivery, nanoparticles and electroporation technologies require further development to achieve taxa-wide intracellular biosensor delivery.
In addressing these current challenges and exploring emerging technologies with bioprospecting potential, this review aims to aid fellow researchers in the rethinking, developing and implementing of a new wave of smart microalgal bioprospecting efforts.
Notes
Acknowledgement
The authors are grateful to the authors cited in this work for their contributions to the research on microalgal bioprospecting and their application.
Author contributions
Joan Labara Tirado: Conceptualization, Methodology, Writing—Original draft preparation. Andrei Herdean, Peter J. Ralph: Conceptualization, Supervision – Review and Editing.
Data availability
No data was used for the research described in the article.
Declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
-
1.Addicted 04 (Wikipedia User). Wikipedia File Australia on the globe (Australia centered). svg 2014. https://en.m.wikipedia.org/wiki/File:Australia_on_the_globe_(Australia_centered).svg. Accessed 17 Dec 2024. PubMed Google Scholar
-
2.Jin CR, Kim JY, Kim DH, Jeon MS, Choi YE. In vivo monitoring of intracellular metabolite in a microalgal cell using an aptamer/graphene oxide nanosheet complex. ACS Appl Bio Mater 2021;4(6): 5080-9. CrossRef PubMed Google Scholar
-
3.Kim JY, Jin CR, Park J, Kim DG, Kim HS, Choi Y-E. Simultaneous probing of dual intracellular metabolites (ATP and paramylon) in live microalgae using graphene oxide/aptamer nanocomplex. Microchim Acta 2022;189(3): 88. CrossRef PubMed Google Scholar
-
4.Ren L, Chen Y, Li P, Mao Z, Huang P-H, Rufo J, et al. A high-throughput acoustic cell sorter. Lab Chip 2015;15(19): 3870-9. CrossRef PubMed Google Scholar
-
5.Thermo Fisher Scientific INC. CTS Rotea Counterflow Centrifugation System Brochure. https://www.thermofisher.com/au/en/home/clinical/cell-gene-therapy/cell-therapy/cell-therapy-manufacturingsolutions/rotea-counterflow-centrifugation-system/features.html. Accessed 17 Dec 2024. PubMed Google Scholar
-
6.Bhola V, Swalaha F, Ranjith Kumar R, Singh M, Bux F. Overview of the potential of microalgae for CO2 sequestration. Int J Environ Sci Technol 2014;11(7): 2103-18. CrossRef PubMed Google Scholar
-
7.Fabris M, Abbriano RM, Pernice M, Sutherland DL, Commault AS, Hall CC, et al. Emerging technologies in algal biotechnology: toward the establishment of a sustainable, algae-based bioeconomy. Front Plant Sci 2020;11: 279. CrossRef PubMed Google Scholar
-
8.Barsanti L, Gualtieri P. Algae: anatomy, biochemistry, and biotechnology. 3rd ed. Milton: Taylor & Francis Group, 2022. PubMed Google Scholar
-
9.Guiry MD. How many species of algae are there. J Phycol 2012;48(5): 1057-63. CrossRef PubMed Google Scholar
-
10.Park BS, Li Z. Taxonomy and ecology of marine algae. J Mar Sci Eng 2022;10(1): 105. CrossRef PubMed Google Scholar
-
11.Stevenson J. Ecological assessments with algae: a review and synthesis. J Phycol 2014;50(3): 437-61. CrossRef PubMed Google Scholar
-
12.Hopes A, Mock T. Evolution of microalgae and their adaptations in different marine ecosystems. eLS 2015;3: 1. PubMed Google Scholar
-
13.Barati B, Zeng K, Baeyens J, Wang S, Addy M, Gan S-Y, et al. Recent progress in genetically modified microalgae for enhanced carbon dioxide sequestration. Biomass Bioenerg 2021;145: 105927. CrossRef PubMed Google Scholar
-
14.Khan MI, Shin JH, Kim JD. The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb Cell Fact 2018;17(1): 36. CrossRef PubMed Google Scholar
-
15.Mobin S, Alam F. Some promising microalgal species for commercial applications: a review. Energy Procedia 2017;110: 510-7. CrossRef PubMed Google Scholar
-
16.Loke SP. Global market and economic analysis of microalgae technology: status and perspectives. Biores Technol 2022;357: 127329. CrossRef PubMed Google Scholar
-
17.Garbary DJ, Bąk M, Dąbek P, Witkowski A. Abstracts of papers to be presented at the 11th international phycological congress. Phycologia 2017;56(sup4): 1-224. CrossRef PubMed Google Scholar
-
18.Chisti Y. Chapter 2-society and microalgae: understanding the past and present. In: Fleurence A, Fleurence J, editors. Microalgae in health and disease prevention. New York: Academic Press; 2018. PubMed Google Scholar
-
19.West JB. The strange history of atmospheric oxygen. Physiol Rep 2022;10(6): e15214. CrossRef PubMed Google Scholar
-
20.Prihanto A, Jatmiko YD, Nurdiani R, Miftachurrochmah A, Wakayama M. Freshwater microalgae as promising food sources: nutritional and functional properties. Open Microbiol J 2022;16: e2206200. CrossRef PubMed Google Scholar
-
21.Alvarez AL, Weyers SL, Goemann HM, Peyton BM, Gardner RD. Microalgae, soil and plants: a critical review of microalgae as renewable resources for agriculture. Algal Res 2021;54: 102200. CrossRef PubMed Google Scholar
-
22.Abinandan S, Subashchandrabose SR, Venkateswarlu K, Megharaj M. Soil microalgae and cyanobacteria: the biotechnological potential in the maintenance of soil fertility and health. Crit Rev Biotechnol 2019;39(8): 981-98. CrossRef PubMed Google Scholar
-
23.Patel A, Matsakas L, Rova U, Christakopoulos P. A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Biores Technol 2019;278: 424-34. CrossRef PubMed Google Scholar
-
24.Lafarga T, Sánchez-Zurano A, Morillas-España A, Acién-Fernández FG. Extremophile microalgae as feedstock for high-value carotenoids: a review. Int J Food Sci Technol 2021;56(10): 4934-41. CrossRef PubMed Google Scholar
-
25.Varshney P, Mikulic P, Vonshak A, Beardall J, Wangikar PP. Extremophilic micro-algae and their potential contribution in biotechnology. Bioresour Technol 2015;184: 363-72. CrossRef PubMed Google Scholar
-
26.Domozych D, Ciancia M, Fangel JU, Mikkelsen MD, Ulvskov P, Willats WG. The cell walls of green algae: a journey through evolution and diversity. Front Plant Sci 2012;3: 82. CrossRef PubMed Google Scholar
-
27.Jassey VEJ, Walcker R, Kardol P, Geisen S, Heger T, Lamentowicz M, et al. Contribution of soil algae to the global carbon cycle. New Phytol 2022;234(1): 64-76. CrossRef PubMed Google Scholar
-
28.Beardall J, Raven JA. The potential effects of global climate change on microalgal photosynthesis, growth and ecology. Phycologia 2004;43(1): 26-40. CrossRef PubMed Google Scholar
-
29.Claustre H, Legendre L, Boyd PW, Levy M. The oceans' biological carbon pumps: framework for a research observational community approach. Front Mar Sci 2021;8: 780052. CrossRef PubMed Google Scholar
-
30.Kholssi R, Lougraimzi H, Moreno-Garrido I. Effects of global environmental change on microalgal photosynthesis, growth and their distribution. Mar Environ Res 2023;184: 105877. CrossRef PubMed Google Scholar
-
31.Bradbury J. Docosahexaenoic acid (DHA): an ancient nutrient for the modern human brain. Nutrients 2011;3(5): 529-54. CrossRef PubMed Google Scholar
-
32.Crawford MA, Bloom M, Broadhurst CL, Schmidt WF, Cunnane SC, Galli C, et al. Evidence for the unique function of docosahexaenoic acid during the evolution of the modern hominid brain. Lipids 1999;34(S1 Part 1): S39-47. PubMed Google Scholar
-
33.Behrenfeld MJ. Climate-mediated dance of the plankton. Nat Clim Chang 2014;4(10): 880-7. CrossRef PubMed Google Scholar
-
34.Administration NOaA. Carbon dioxide now more than 50% higher than pre-industrial levels. 2022. PubMed Google Scholar
-
35.Petrou K, Kranz SA, Trimborn S, Hassler CS, Ameijeiras SB, Sackett O, et al. Southern Ocean phytoplankton physiology in a changing climate. J Plant Physiol 2016;203: 135-50. CrossRef PubMed Google Scholar
-
36.Beijerinck MW. Culturversuche mit Zoochlorellen, Lichenengonidien und anderen niederen. Algen Bot Ztg 1890;48(725–72): 81-8. PubMed Google Scholar
-
37.Lustig A, Levine AJ. One hundred years of virology. J Virol 1992;66(8): 4629-31. CrossRef PubMed Google Scholar
-
38.Bos L. Beijerinck's work on tobacco mosaic virus: historical context and legacy. Philos Trans R Soc Lond B Biol Sci 1999;354(1383): 675-85. CrossRef PubMed Google Scholar
-
39.García JL, de Vicente M, Galán B. Microalgae, old sustainable food and fashion nutraceuticals. Microb Biotechnol 2017;10(5): 1017-24. CrossRef PubMed Google Scholar
-
40.Sánchez J, Curt MD, Robert N, Fernández J. Chapter two-biomass resources. In: Lago C, Caldés N, Lechón Y, editors. The role of bioenergy in the bioeconomy. Cambridge: Academic Press; 2019. p. 25–111. PubMed Google Scholar
-
41.Weart SR. The idea of anthropogenic global climate change in the 20th century. WIREs Clim Change 2010;1(1): 67-81. CrossRef PubMed Google Scholar
-
42.Fawzy S, Osman AI, Doran J, Rooney DW. Strategies for mitigation of climate change: a review. Environ Chem Lett 2020;18(6): 2069-94. CrossRef PubMed Google Scholar
-
43.Ralph PJ, Pernice M. Save the planet with green industries using algae. PLoS Biol 2023;21(3): e3002061. CrossRef PubMed Google Scholar
-
44.Fuchs W, Rachbauer L, Rittmann SKR, Bochmann G, Ribitsch D, Steger F. Eight up-coming biotech tools to combat climate crisis. Microorganisms 2023;11(6): 1514. CrossRef PubMed Google Scholar
-
45.Hunter P. The potential of molecular biology and biotechnology for dealing with global warming: the biosciences will have to play a leading role in developing new technologies for mitigating the impact of greenhouse gas emissions. EMBO Rep 2016;17(7): 946-8. CrossRef PubMed Google Scholar
-
46.Reisoglu Ş, Aydin S. Microalgae as a promising candidate for fighting climate change and biodiversity loss. 2023. PubMed Google Scholar
-
47.Sadvakasova AK, Kossalbayev BD, Bauenova MO, Balouch H, Leong YK, Zayadan BK, et al. Microalgae as a key tool in achieving carbon neutrality for bioproduct production. Algal Res 2023;72: 103096. CrossRef PubMed Google Scholar
-
48.Singh J, Saxena RC. Chapter 2-an introduction to microalgae: diversity and significance. Amsterdam: Elsevier Inc, 2015: 11-24. PubMed Google Scholar
-
49.Chew KW, Yap JY, Show PL, Suan NH, Juan JC, Ling TC, et al. Microalgae biorefinery: high value products perspectives. Biores Technol 2017;229: 53-62. CrossRef PubMed Google Scholar
-
50.Saeed MU, Hussain N, Shahbaz A, Hameed T, Iqbal HMN, Bilal M. Bioprospecting microalgae and cyanobacteria for biopharmaceutical applications. J Basic Microbiol 2022;62(9): 1110-24. CrossRef PubMed Google Scholar
-
51.Fernandes T, Cordeiro N. Microalgae as sustainable biofactories to produce high-value lipids: biodiversity, exploitation, and biotechnological applications. Mar Drugs 2021;19(10): 573. CrossRef PubMed Google Scholar
-
52.Wu J, Gu X, Yang D, Xu S, Wang S, Chen X, et al. Bioactive substances and potentiality of marine microalgae. Food Sci Nutr 2021;9(9): 5279-92. CrossRef PubMed Google Scholar
-
53.Rizwan M, Mujtaba G, Memon SA, Lee K, Rashid N. Exploring the potential of microalgae for new biotechnology applications and beyond: a review. Renew Sustain Energy Rev 2018;92: 394-404. CrossRef PubMed Google Scholar
-
54.Pulz O, Gross W. Valuable products from biotechnology of microalgae. Appl Microbiol Biotechnol 2004;65(6): 635-48. CrossRef PubMed Google Scholar
-
55.Guiry MD. How many species of algae are there. A reprise. Four kingdoms, 14 phyla, 63 classes and still growing. J Phycol 2024;60(2): 214-28. CrossRef PubMed Google Scholar
-
56.Sharma N, Rai A. Biodiversity and biogeography of microalgae: progress and pitfalls. Environ Rev 2011;19: 1. CrossRef PubMed Google Scholar
-
57.Abida H, Ruchaud S, Rios L, Humeau A, Probert I, De Vargas C, et al. Bioprospecting marine plankton. Mar Drugs 2013;11(11): 4594-611. CrossRef PubMed Google Scholar
-
58.Vuong P, Chong S, Kaur P. The little things that matter: how bioprospecting microbial biodiversity can build towards the realization of United Nations Sustainable development goals. NPJ Biodivers 2022;1(1): 4. CrossRef PubMed Google Scholar
-
59.United Nations DoEaSA. United nations sustainable development goals. 2024. https://sdgs.un.org/goals2024;https://sdgs.un.org/goals. Accessed 10 July 2024. PubMed Google Scholar
-
60.Müller WEG, Batel R, Schröder HC, Müller IM. Traditional and modern biomedical prospecting: part Ⅰ—the history. Evid Based Complement Altern Med 2004;1: 856086. PubMed Google Scholar
-
61.Verma M, Mishra V. An introduction to algal biofuels. In: Srivastava N, Srivastava M, Mishra PK, Gupta VK, editors. Microbial strategies for techno-economic biofuel production. Singapore: Springer; 2020. p. 1–34. PubMed Google Scholar
-
62.Richmond A, Hu Q. Handbook of microalgal culture: applied phycology and biotechnology. Newark: Wiley, 2013. PubMed Google Scholar
-
63.Fernandez-Valenzuela S, Ruvalcaba F, Beltrán-Rocha J, Claudio P, Reyna G. Isolation and culturing axenic microalgae: mini-review. Open Microbiol J 2021;15: 111-9. CrossRef PubMed Google Scholar
-
64.Chiu C-S, Chiu P-H, Yong TC, Tsai H-P, Soong K, Huang H-E, et al. Mechanisms protect airborne green microalgae during long distance dispersal. Sci Rep 2020;10(1): 13984. CrossRef PubMed Google Scholar
-
65.Roque J, Brito Â, Rocha M, Pissarra J, Nunes T, Bessa M, et al. Isolation and characterization of soil cyanobacteria and microalgae and evaluation of their potential as plant biostimulants. Plant Soil 2023;493(1): 115-36. CrossRef PubMed Google Scholar
-
66.Rezaei A, Cheniany M, Ahmadzadeh H, Vaezi J. Evaluation of lipid composition and growth parameters of cold-adapted microalgae under different conditions. BioEnergy Res 2024;17(1): 557-69. PubMed Google Scholar
-
67.Araj-Shirvani M, Honarvar M, Jahadi M, Mizani M. Biochemical profile of Dunaliella isolates from different regions of Iran with a focus on pharmaceutical and nutraceutical potential applications. Food Sci Nutr 2024. CrossRef PubMed Google Scholar
-
68.Ben Ammar FE, Saidane F, Messaoud C, Hamdi M. Screening of efficient microalgae strains isolated from Tunisian ecosystems: Assessment of algal growth rate and added-value bioproducts for biorefinery applications. Biocatal Agric Biotechnol 2024;58: 103140. CrossRef PubMed Google Scholar
-
69.Condori MAM, Condori MM, Gutierrez MEV, Choix FJ, García-Camacho F. Bioremediation potential of the Chlorella and Scenedesmus microalgae in explosives production effluents. Sci Total Environ 2024;920: 171004. CrossRef PubMed Google Scholar
-
70.Grubišić M, Šantek B, Zorić Z, Čošić Z, Vrana I, Gašparović B, et al. Bioprospecting of microalgae isolated from the Adriatic sea: characterization of biomass, pigment, lipid and fatty acid composition, and antioxidant and antimicrobial activity. Molecules 2022;27(4): 1248. CrossRef PubMed Google Scholar
-
71.Badary A, Hidasi N, Ferrari S, Mayfield SP. Isolation and characterization of microalgae strains able to grow on complex biomass hydrolysate for industrial application. Algal Res 2024;78: 103381. CrossRef PubMed Google Scholar
-
72.Katayama T, Takahashi K, Wahid MEA, Yusoff FM, Takahashi K. Picochloropsis malayensis gen. et sp. Nov. (Chlorellales, Chlorophyta), an ammonium tolerant, polyphosphate-accumulating microalga from seawater. Phycol Res 2024. CrossRef PubMed Google Scholar
-
73.Candido C, Cardoso LG, Lombardi AT. Bioprospecting and selection of tolerant strains and productive analyses of microalgae grown in vinasse. Braz J Microbiol 2022;53(2): 845-55. CrossRef PubMed Google Scholar
-
74.Assobhi B, Bouchelta Y, Alsubih M, Alaoui-Sossé B, Bourgeade P, et al. What are the growth kinetics and biochemical compositions of microalgae isolated from diverse aquatic ecosystems in Morocco, France, and Tunisia? Environ Sci Poll Res 2024. CrossRef PubMed Google Scholar
-
75.Aljabri H, Cherif M, Siddiqui SA, Bounnit T, Saadaoui I. Evidence of the drying technique's impact on the biomass quality of Tetraselmis subcordiformis (Chlorophyceae). Biotechnol Biofuels Bioprod 2023;16(1): 85. CrossRef PubMed Google Scholar
-
76.Amouri M, Aziza M, Kaidi F, Abert Vian M, Chemat F, Amrane A, et al. Indigenous microalgae strains characterization for a sustainable biodiesel production. Biotechnol J 2024;19(1): 2300096. CrossRef PubMed Google Scholar
-
77.Sweiss M, Assi S, Barhoumi L, Al-Jumeily D, Watson M, Wilson M, et al. Qualitative and quantitative evaluation of microalgal biomass using portable attenuated total reflectance-Fourier transform infrared spectroscopy and machine learning analytics. J Chem Technol Biotechnol 2024;99(1): 92-108. CrossRef PubMed Google Scholar
-
78.Patel AK, Vadrale AP, Tseng Y-S, Chen C-W, Dong C-D, Singhania RR. Bioprospecting of marine microalgae from Kaohsiung seacoast for lutein and lipid production. Biores Technol 2022;351: 126928. CrossRef PubMed Google Scholar
-
79.Patel A, Antonopoulou I, Enman J, Rova U, Christakopoulos P, Matsakas L. Lipids detection and quantification in oleaginous microorganisms: an overview of the current state of the art. BMC Chem Eng 2019;1(1): 13. CrossRef PubMed Google Scholar
-
80.Han Y, Wen Q, Chen Z, Li P. Review of methods used for microalgal lipid-content analysis. Energy Procedia 2011;12: 944-50. CrossRef PubMed Google Scholar
-
81.Hounslow E, Noirel J, Gilmour DJ, Wright PC. Lipid quantification techniques for screening oleaginous species of microalgae for biofuel production. Eur J Lipid Sci Technol 2017;119(2): 1500469. CrossRef PubMed Google Scholar
-
82.Sun H, Wang Y, He Y, Liu B, Mou H, Chen F, et al. Microalgae-derived pigments for the food industry. Mar Drugs 2023;21(2): 82. CrossRef PubMed Google Scholar
-
83.Pagels F, Pereira RN, Vicente AA, Guedes AC. Extraction of pigments from microalgae and cyanobacteria—a review on current methodologies. Appl Sci 2021;11(11): 5187. CrossRef PubMed Google Scholar
-
84.Khosravinia S, Malekzadeh-Shafaroudi S, Bagheri A, Behdad A, Moshtaghi N. Bioprospecting of ten microalgae species isolated from saline water lake for evaluation of the biodiesel production. BioEnergy Research 2024;17(2): 1090-103. CrossRef PubMed Google Scholar
-
85.Tan KY, Low SS, Manickam S, Ma Z, Banat F, Munawaroh HSH, et al. Prospects of microalgae in nutraceuticals production with nanotechnology applications. Food Res Int 2023;169: 112870. CrossRef PubMed Google Scholar
-
86.Sánchez-Saavedra MP, Castro-Ochoa FY. Bioprospecting for lipid production of eleven microalgae strains for sustainable biofuel production. BioEnergy Res 2024;17(2): 1118-32. CrossRef PubMed Google Scholar
-
87.DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem 1956;28(3): 350-6. CrossRef PubMed Google Scholar
-
88.Maity S, Mallick N. Bioprospecting marine microalgae and cyanobacteria as alternative feedstocks for bioethanol production. Sustain Chem Pharm 2022;29: 100798. CrossRef PubMed Google Scholar
-
89.Cruz JD, Delattre C, Felpeto AB, Pereira H, Pierre G, Morais J, et al. Bioprospecting for industrially relevant exopolysaccharide-producing cyanobacteria under Portuguese simulated climate. Sci Rep 2023;13(1): 13561. CrossRef PubMed Google Scholar
-
90.Mosibo OK, Ferrentino G, Udenigwe CC. Microalgae proteins as sustainable ingredients in novel foods: recent developments and challenges. Foods 2024;13(5): 733. CrossRef PubMed Google Scholar
-
91.Lucakova S, Branyikova I, Hayes M. Microalgal proteins and bioactives for food, feed, and other applications. Appl Sci 2022;12(9): 4402. CrossRef PubMed Google Scholar
-
92.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J biol Chem 1951;193(1): 265-75. CrossRef PubMed Google Scholar
-
93.Shen CH. Chapter 9-quantification and analysis of proteins. In: Shen CH, editor. Diagnostic molecular biology. 2nd ed. Cambridge: Academic Press; 2023. p. 231–57. PubMed Google Scholar
-
94.Lourenço SO, Barbarino E, Lavín PL, Lanfer Marquez UM, Aidar E. Distribution of intracellular nitrogen in marine microalgae: calculation of new nitrogen-to-protein conversion factors. Eur J Phycol 2004;39(1): 17-32. CrossRef PubMed Google Scholar
-
95.Khemiri S, Khelifi N, Messaoud C, Smaali I. Bioprospecting of microalgae for a potential use as enzyme inhibitors, anti-ageing and prebiotic agents. Biocatal Agric Biotechnol 2023;51: 102759. CrossRef PubMed Google Scholar
-
96.Mc Gee D, Archer L, Smyth TJ, Fleming GTA, Touzet N. Bioprospecting and LED-based spectral enhancement of antimicrobial activity of microalgae isolated from the west of Ireland. Algal Res 2020;45: 101704. CrossRef PubMed Google Scholar
-
97.İnan B, Mutlu B, Karaca GA, Koç RÇ, Özçimen D. Bioprospecting Antarctic microalgae as anticancer agent against PC-3 and AGS cell lines. Biochem Eng J 2023;195: 108900. CrossRef PubMed Google Scholar
-
98.Martínez R, García Beltrán A, Kapravelou G, Guzmán A, Lozano A, Gómez-Villegas P, et al. Nutritional and functional assessment of haloarchaea and microalgae from the Andalusian shoreline: promising functional foods with a high nutritional value. J Funct Foods 2024;116: 106194. CrossRef PubMed Google Scholar
-
99.Geng Y, Cui D, Yang L, Xiong Z, Pavlostathis SG, Shao P, et al. Resourceful treatment of harsh high-nitrogen rare earth element tailings (REEs) wastewater by carbonate activated Chlorococcum sp. microalgae. J Hazard Mater 2022;423: 127000. CrossRef PubMed Google Scholar
-
100.Wang H, Liu Z, Cui D, Liu Y, Yang L, Chen H, et al. A pilot scale study on the treatment of rare earth tailings (REEs) wastewater with low C/N ratio using microalgae photobioreactor. J Environ Manage 2023;328: 116973. CrossRef PubMed Google Scholar
-
101.Arumugam K, Mohamad R, Ashari SE, Tan JS, Mohamed MS. Bioprospecting microalgae with the capacity for inducing calcium carbonate biomineral precipitation. Asia-Pac J Chem Eng 2022;17(3): e2767. CrossRef PubMed Google Scholar
-
102.Jose S, Renuka N, Ratha SK, Kumari S, Bux F. Bioprospecting of microalgae from agricultural fields and developing consortia for sustainable agriculture. Algal Res 2024;78: 103428. CrossRef PubMed Google Scholar
-
103.Concórdio-Reis P, David H, Reis MAM, Amorim A, Freitas F. Bioprospecting for new exopolysaccharide-producing microalgae of marine origin. Int Microbiol 2023;26(4): 1123-30. CrossRef PubMed Google Scholar
-
104.Thangadurai D, Sangeetha J, Prasad R. Bioprospecting algae for nanosized materials. 1st ed. Cham: Springer International Publishing AG, 2022. PubMed Google Scholar
-
105.Greco GR, Cinquegrani M. Firms plunge into the sea. Marine biotechnology industry, a first investigation. Front Mar Sci 2016;2: 124. CrossRef PubMed Google Scholar
-
106.Rusyaev SM, Orlov AM. The phenomenon of marine bioprospecting. Biol Bull Rev 2024;14(1): 115-32. CrossRef PubMed Google Scholar
-
107.Ratha SK, Prasanna R. Bioprospecting microalgae as potential sources of "green energy"—challenges and perspectives (review). Appl Biochem Microbiol 2012;48(2): 109-25. CrossRef PubMed Google Scholar
-
108.Beattie AJ, Hay M, Magnusson B, de Nys R, Smeathers J, Vincent JF. Ecology and bioprospecting. Austral Ecol 2011;36(3): 341-56. CrossRef PubMed Google Scholar
-
109.Lee KH, Jeong HJ, Jang TY, Lim AS, Kang NS, Kim J-H, et al. Feeding by the newly described mixotrophic dinoflagellate Gymnodinium smaydae: feeding mechanism, prey species, and effect of prey concentration. J Exp Mar Biol Ecol 2014;459: 114-25. CrossRef PubMed Google Scholar
-
110.Kang HC, Jeong HJ, Park SA, Eom SH, Ok JH, You JH, et al. Feeding by the newly described heterotrophic dinoflagellate Gyrodinium jinhaense: comparison with G dominans and G moestrupii. Mar Biol 2020;167(10): 156. CrossRef PubMed Google Scholar
-
111.Doppler P, Kriechbaum R, Singer B, Spadiut O. Make microalgal cultures axenic again—a fast and simple workflow utilizing fluorescence-activated cell sorting. J Microbiol Methods 2021;186: 106256. CrossRef PubMed Google Scholar
-
112.Pokorny L, Hausmann B, Pjevac P, Schagerl M. How to verify non-presence—the challenge of axenic algae cultivation. Cells 2022;11(16): 2594. CrossRef PubMed Google Scholar
-
113.Rehman M, Kesharvani S, Dwivedi G, Gidwani SK. Impact of cultivation conditions on microalgae biomass productivity and lipid content. Mater Today: Proc 2022;56: 282-90. CrossRef PubMed Google Scholar
-
114.Hoch L, Herdean A, Argyle PA, Ralph PJ. High throughput phenomics for diatoms: challenges and solutions. Prog Oceanogr 2023;216: 103074. CrossRef PubMed Google Scholar
-
115.Selvam J, Mal J, Singh S, Yadav A, Giri BS, Pandey A, et al. Bioprospecting marine microalgae as sustainable bio-factories for value-added compounds. Algal Res 2024;79: 103444. CrossRef PubMed Google Scholar
-
116.Kiani H, Aznar R, Poojary MM, Tiwari BK, Halim R. Chromatographic techniques to separate and identify bioactive compounds in microalgae. Front Energy Res 2022;10: 904014. CrossRef PubMed Google Scholar
-
117.Jacobsen C. Fish oils: composition and health effects. In: Caballero B, Finglas PM, Toldrá F, editors. Encyclopedia of food and health. Oxford: Academic Press; 2016. p. 686–92. PubMed Google Scholar
-
118.Comission EU. Commission Implementing Regulation (EU) 2022/1365 of 4 August 2022 amending Implementing Regulation (EU) 2017/2470 as regards the conditions of use of the novel food Schizochytrium sp. oil rich in DHA and EPA (Text with EEA relevance) Commission implementing regulation (EU) 2022/1365 of 4 August 2022 amending Implementing Regulation (EU) 2017/2470 as regards the conditions of use of the novel food Schizochytrium sp. oil rich in DHA and EPA. 2022. PubMed Google Scholar
-
119.Giudice AL, Rizzo C. Culture collections as hidden sources of microbial biomolecules and biodiversity. Diversity 2020;12(7): 264. CrossRef PubMed Google Scholar
-
120.Foo SC, Mok CY, Ho SY, Khong NMH. Microalgal culture preservation: progress, trends and future developments. Algal Res 2023;71: 103007. CrossRef PubMed Google Scholar
-
121.Chellappan A, Thangamani P, Markose S, Thavasimuthu C, Thangaswamy S, Mariavincent M. Long-term preservation of micro-algal stock for fish hatcheries. Aquac Rep 2020;17: 100329. CrossRef PubMed Google Scholar
-
122.Cheregi O, Ekendahl S, Engelbrektsson J, Strömberg N, Godhe A, Spetea C. Microalgae biotechnology in Nordic countries—the potential of local strains. Physiol Plant 2019;166(1): 438-50. CrossRef PubMed Google Scholar
-
123.Duong VT, Li Y, Nowak E, Schenk PM. Microalgae Isolation and selection for prospective biodiesel production. Energies 2012;5(6): 1835-49. CrossRef PubMed Google Scholar
-
124.Pourkarimi S, Hallajisani A, Alizadehdakhel A, Nouralishahi A, Golzary A. Factors affecting production of beta-carotene from Dunaliella salina microalgae. Biocatal Agric Biotechnol 2020;29: 101771. CrossRef PubMed Google Scholar
-
125.Smith-Bädorf HD, Chuck CJ, Mokebo KR, MacDonald H, Davidson MG, Scott RJ. Bioprospecting the thermal waters of the Roman baths: isolation of oleaginous species and analysis of the FAME profile for biodiesel production. AMB Express 2013;3(1): 9. CrossRef PubMed Google Scholar
-
126.Treves H, Raanan H, Finkel OM, Berkowicz SM, Keren N, Shotland Y, et al. A newly isolated Chlorella sp. from desert sand crusts exhibits a unique resistance to excess light intensity. FEMS Microbiol Ecol 2013;86(3): 373-80. CrossRef PubMed Google Scholar
-
127.Abdullahi ZH, Marselin FN, Khaironizam NIA, Fauzi NFA, Wan Maznah WO. Growth stage-related biomass, pigments, and biochemical composition of Stichococcus bacillaris, Synechococcus sp., and Trentepohlia aurea isolated from Gua Tempurung, a cave in Malaysia. Plant Physiol Biochem 2023;197: 107633. CrossRef PubMed Google Scholar
-
128.Halder N, Goyal D, Aneja RK. Bioprospecting microalgae from sewage water: assessment of biochemicals for biomass utilization. Mol Biotechnol 2023. CrossRef PubMed Google Scholar
-
129.Senhorinho GNA, Laamanen CA, Scott JA. Bioprospecting freshwater microalgae for antibacterial activity from water bodies associated with abandoned mine sites. Phycologia 2018;57(4): 432-9. CrossRef PubMed Google Scholar
-
130.Rinaldi KL, Senhorinho GNA, Laamanen CA, Scott JA. A review of extremophilic microalgae: impacts of experimental cultivation conditions for the production of antimicrobials. Algal Res 2024;78: 103427. CrossRef PubMed Google Scholar
-
131.Rocha LC, de Oliveira JR, Vacondio B, Rodrigues GN, Seleghim MHR, Porto ALM. Bioactive marine microorganisms for biocatalytic reactions in organic compounds. Mar Microbiol 2013. CrossRef PubMed Google Scholar
-
132.Bacellar Mendes LB, Vermelho AB. Allelopathy as a potential strategy to improve microalgae cultivation. Biotechnol Biofuels 2013;6(1): 152. CrossRef PubMed Google Scholar
-
133.Hosseini H, Al-Jabri HM, Moheimani NR, Siddiqui SA, Saadaoui I. Marine microbial bioprospecting: exploitation of marine biodiversity towards biotechnological applications—a review. J Basic Microbiol 2022;62(9): 1030-43. CrossRef PubMed Google Scholar
-
134.Dammak I, Fersi M, Hachicha R, Abdelkafi S. Current insights into growing microalgae for municipal wastewater treatment and biomass generation. Resources 2023;12(10): 119. CrossRef PubMed Google Scholar
-
135.Ishika T, Moheimani NR, Bahri PA, Laird DW, Blair S, Parlevliet D. Halo-adapted microalgae for fucoxanthin production: effect of incremental increase in salinity. Algal Res 2017;28: 66-73. CrossRef PubMed Google Scholar
-
136.Berge T, Daugbjerg N, Hansen PJ. Isolation and cultivation of microalgae select for low growth rate and tolerance to high pH. Harmful Algae 2012;20: 101-10. CrossRef PubMed Google Scholar
-
137.Desjardins SM, Laamanen CA, Basiliko N, Scott JA. Selection and re-acclimation of bioprospected acid-tolerant green microalgae suitable for growth at low pH. Extremophiles 2021;25(2): 129-41. CrossRef PubMed Google Scholar
-
138.Yoo C, Jun S-Y, Lee J-Y, Ahn C-Y, Oh H-M. Selection of microalgae for lipid production under high levels carbon dioxide. Bioresourc Technol 2010;101(1): S71-4. CrossRef PubMed Google Scholar
-
139.Ramanna L, Rawat I, Bux F. Light enhancement strategies improve microalgal biomass productivity. Renew Sustain Energy Rev 2017;80: 765-73. CrossRef PubMed Google Scholar
-
140.Zuliani L, Frison N, Jelic A, Fatone F, Bolzonella D, Ballottari M. Microalgae cultivation on anaerobic digestate of municipal wastewater, sewage sludge and agro-waste. Int J Mol Sci 2016;17(10): 1692. CrossRef PubMed Google Scholar
-
141.Aslam A, Thomas-Hall SR, Mughal TA, Schenk PM. Selection and adaptation of microalgae to growth in 100% unfiltered coal-fired flue gas. Biores Technol 2017;233: 271-83. CrossRef PubMed Google Scholar
-
142.Argyle PA, Hinners J, Walworth NG, Collins S, Levine NM, Doblin MA. A high-throughput assay for quantifying phenotypic traits of microalgae. Front Microbiol 2021;12: 706235. CrossRef PubMed Google Scholar
-
143.Van Wagenen J, Holdt SL, De Francisci D, Valverde-Pérez B, Plósz BG, Angelidaki I. Microplate-based method for high-throughput screening of microalgae growth potential. Biores Technol 2014;169: 566-72. CrossRef PubMed Google Scholar
-
144.Zheng J, Cole T, Zhang Y, Bayinqiaoge YD, Tang SY. An automated and intelligent microfluidic platform for microalgae detection and monitoring. Lab Chip 2024;24(2): 244-53. CrossRef PubMed Google Scholar
-
145.Zheng G, Cui Y, Lu L, Guo M, Hu X, Wang L, et al. Microfluidic chemostatic bioreactor for high-throughput screening and sustainable co-harvesting of biomass and biodiesel in microalgae. Bioact Mater 2023;25: 629-39. PubMed Google Scholar
-
146.Westerwalbesloh C, Brehl C, Weber S, Probst C, Widzgowski J, Grünberger A, et al. A microfluidic photobioreactor for simultaneous observation and cultivation of single microalgal cells or cell aggregates. PLoS ONE 2019;14(4): e0216093. CrossRef PubMed Google Scholar
-
147.Radzun KA, Wolf J, Jakob G, Zhang E, Stephens E, Ross I, et al. Automated nutrient screening system enables high-throughput optimisation of microalgae production conditions. Biotechnol Biofuels 2015;8(1): 65. CrossRef PubMed Google Scholar
-
148.Porras Reyes L, Havlik I, Beutel S. Software sensors in the monitoring of microalgae cultivations. Rev Environ Sci Bio/Technol 2024;23(1): 67-92. CrossRef PubMed Google Scholar
-
149.Tham PE, Ng YJ, Vadivelu N, Lim HR, Khoo KS, Chew KW, et al. Sustainable smart photobioreactor for continuous cultivation of microalgae embedded with Internet of Things. Biores Technol 2022;346: 126558. CrossRef PubMed Google Scholar
-
150.Nguyen BT, Rittmann BE. Low-cost optical sensor to automatically monitor and control biomass concentration in microalgal cultivation. Algal Res 2018;32: 101-6. CrossRef PubMed Google Scholar
-
151.Luttmann R, Bracewell DG, Cornelissen G, Gernaey KV, Glassey J, Hass VC, et al. Soft sensors in bioprocessing: a status report and recommendations. Biotechnol J 2012;7(8): 1040-8. CrossRef PubMed Google Scholar
-
152.Jakob G, Wolf J, Bui T, Posten C, Kruse O, Stephens E, et al. Surveying a diverse pool of microalgae as a bioresource for future biotechnological applications. J Petrol Environ Biotechnol 2013;04: 2. CrossRef PubMed Google Scholar
-
153.Pereira H, Schulze PSC, Schüler LM, Santos T, Barreira L, Varela J. Fluorescence activated cell-sorting principles and applications in microalgal biotechnology. Algal Res 2018;30: 113-20. CrossRef PubMed Google Scholar
-
154.Vu CHT, Lee H-G, Chang YK, Oh H-M. Axenic cultures for microalgal biotechnology: establishment, assessment, maintenance, and applications. Biotechnol Adv 2018;36(2): 380-96. CrossRef PubMed Google Scholar
-
155.Cho D-H, Ramanan R, Kim B-H, Lee J, Kim S, Yoo C, et al. Novel approach for the development of axenic microalgal cultures from environmental samples. J Phycol 2013;49(4): 802-10. CrossRef PubMed Google Scholar
-
156.Soomro RR, Ndikubwimana T, Zeng X, Lu Y, Lin L, Danquah MK. Development of a two-stage microalgae dewatering process—a life cycle assessment approach. Front Plant Sci 2016;7: 113. CrossRef PubMed Google Scholar
-
157.Lee TC, Chan PL, Tam NF, Xu SJ, Lee FW. Establish axenic cultures of armored and unarmored marine dinoflagellate species using density separation, antibacterial treatments and stepwise dilution selection. Sci Rep 2021;11(1): 202. CrossRef PubMed Google Scholar
-
158.Li A, Kusuma GD, Driscoll D, Smith N, Wall DM, Levine BL, et al. Advances in automated cell washing and concentration. Cytotherapy 2021;23(9): 774-86. CrossRef PubMed Google Scholar
-
159.Dargitz CT, Daoudi S, Dunn S, du Jeu XD, Ravinder N. Rotea: a closed and automated instrument for efficient cell isolation, washing and conentration in cell therapy workflows. Cytotherapy 2020;22(5): S200. CrossRef PubMed Google Scholar
-
160.De Stefano JA, Foy JM, Sullivan DW, Dawes SM, Cushion MT, Babcock GF, et al. Fractionation of Pneumocystis carinii developmental stages by counterflow centrifugal elutriation and sequential filtrations. Parasitol Res 1994;80(1): 1-9. CrossRef PubMed Google Scholar
-
161.Kim GY, Son J, Han JI, Park JK. Inertial microfluidics-based separation of microalgae using a contraction-expansion array microchannel. Micromachines (Basel) 2021;12(1): 97. CrossRef PubMed Google Scholar
-
162.Sapp M, Schwaderer AS, Wiltshire KH, Hoppe H-G, Gerdts G, Wichels A. Species-specific bacterial communities in the phycosphere of microalgae. Microb Ecol 2007;53(4): 683-99. CrossRef PubMed Google Scholar
-
163.Samo TJ, Kimbrel JA, Nilson DJ, Pett-Ridge J, Weber PK, Mayali X. Attachment between heterotrophic bacteria and microalgae influences symbiotic microscale interactions. Environ Microbiol 2018;20(12): 4385-400. CrossRef PubMed Google Scholar
-
164.Variem SS, Kizhakkedath VK. Phycosphere associated bacteria; a prospective source of bioactive compounds. Biologia 2021;76(3): 1095-8. CrossRef PubMed Google Scholar
-
165.Santo ÉdE, Ishii M, Pinto UM, Matsudo MC, Carvalho JC. Obtaining bioproducts from the studies of signals and interactions between microalgae and bacteria. Microorganisms 2022;10(10): 2029. CrossRef PubMed Google Scholar
-
166.Perković L, Djedović E, Vujović T, Baković M, Paradžik T, Čož-Rakovac R. Biotechnological enhancement of probiotics through co-cultivation with algae: future or a trend. Mar Drugs 2022;20(2): 142. CrossRef PubMed Google Scholar
-
167.Tandon P, Jin Q. Microalgae culture enhancement through key microbial approaches. Renew Sustain Energy Rev 2017;80: 1089-99. CrossRef PubMed Google Scholar
-
168.Chong JWR, Khoo KS, Chew KW, Vo DV, Balakrishnan D, Banat F, et al. Microalgae identification: Future of image processing and digital algorithm. Bioresourc Technol 2023;369: 128418. CrossRef PubMed Google Scholar
-
169.Otálora P, Guzmán JL, Acién FG, Berenguel M, Reul A. An artificial intelligence approach for identification of microalgae cultures. New Biotechnol 2023;77: 58-67. CrossRef PubMed Google Scholar
-
170.Liu F, Zhang C, Wang Y, Chen G. A review of the current and emerging detection methods of marine harmful microalgae. Sci Total Environ 2022;815: 152913. CrossRef PubMed Google Scholar
-
171.Jahn MT, Schmidt K, Mock T. A novel cost effective and high-throughput isolation and identification method for marine microalgae. Plant Methods 2014;10(1): 26. CrossRef PubMed Google Scholar
-
172.Vuong P, Wise MJ, Whiteley AS, Kaur P. Small investments with big returns: environmental genomic bioprospecting of microbial life. Crit Rev Microbiol 2022;48(5): 641-55. CrossRef PubMed Google Scholar
-
173.Maghembe R, Damian D, Makaranga A, Nyandoro SS, Lyantagaye SL, Kusari S, et al. Omics for bioprospecting and drug discovery from bacteria and microalgae. Antibiotics 2020;9(5): 229. CrossRef PubMed Google Scholar
-
174.Beal J, Goñi-Moreno A, Myers C, Hecht A, de Vicente MDC, Parco M, et al. The long journey towards standards for engineering biosystems. EMBO Rep 2020;21(5): 50521. CrossRef PubMed Google Scholar
-
175.Stavridou E, Karapetsi L, Nteve GM, Tsintzou G, Chatzikonstantinou M, Tsaousi M, et al. Landscape of microalgae omics and metabolic engineering research for strain improvement: an overview. Aquaculture 2024;587: 740803. CrossRef PubMed Google Scholar
-
176.Maréchal E. Grand challenges in microalgae domestication. Front Plant Sci 2021;12: 764573. CrossRef PubMed Google Scholar
-
177.Liu J-Y, Zeng L-H, Ren Z-H. Recent application of spectroscopy for the detection of microalgae life information: a review. Appl Spectrosc Rev 2020;55(1): 26-59. CrossRef PubMed Google Scholar
-
178.Podevin M, Fotidis IA, Angelidaki I. Microalgal process-monitoring based on high-selectivity spectroscopy tools: status and future perspectives. Crit Rev Biotechnol 2018;38(5): 704-18. CrossRef PubMed Google Scholar
-
179.Mutanda T, Ramesh D, Karthikeyan S, Kumari S, Anandraj A, Bux F. Bioprospecting for hyper-lipid producing microalgal strains for sustainable biofuel production. Biores Technol 2011;102(1): 57-70. CrossRef PubMed Google Scholar
-
180.Hyka P, Lickova S, Přibyl P, Melzoch K, Kovar K. Flow cytometry for the development of biotechnological processes with microalgae. Biotechnol Adv 2013;31(1): 2-16. CrossRef PubMed Google Scholar
-
181.Katayama T, Rahman NA, Khatoon H, Kasan NA, Nagao N, Yamada Y, et al. Bioprospecting of tropical microalgae for high-value products: n-3 polyunsaturated fatty acids and carotenoids. Aquac Rep 2022;27: 101406. CrossRef PubMed Google Scholar
-
182.Sato M, Murata Y, Mizusawa M, Iwahashi H, Oka S. A simple and rapid dual-fluorescence viability assay for microalgae. Microbiol Cult Coll 2004;20: 53-9. PubMed Google Scholar
-
183.Machado MD, Soares EV. Development of a short-term assay based on the evaluation of the plasma membrane integrity of the alga Pseudokirchneriella subcapitata. Appl Microbiol Biotechnol 2012;95(4): 1035-42. CrossRef PubMed Google Scholar
-
184.Reza AM, Rakhi SF, Zhu X, Tang Y, Qin J. Visualising the emerging platform of using microalgae as a sustainable bio-factory for healthy lipid production through biocompatible AIE probes. Biosensors 2022;12(4): 208. CrossRef PubMed Google Scholar
-
185.Archer SD, Posman KM, DeStefano J, Harrison AO, Ladina A, Cheff EA, et al. Fluorescent detection of bromoperoxidase activity in microalgae and planktonic microbial communities using aminophenyl fluorescein. Front Mar Sci 2019;6: 68. CrossRef PubMed Google Scholar
-
186.Schmidt J. Development and evaluation of microalgae screening procedures and cultivation systems for biofuel applications: Technische Universität München; 2018. PubMed Google Scholar
-
187.Harchouni S, Field B, Menand B. AC-202, a highly effective fluorophore for the visualization of lipid droplets in green algae and diatoms. Biotechnol Biofuels 2018;11(1): 120. CrossRef PubMed Google Scholar
-
188.White S, Anandraj A, Bux F. PAM fluorometry as a tool to assess microalgal nutrient stress and monitor cellular neutral lipids. Biores Technol 2011;102(2): 1675-82. CrossRef PubMed Google Scholar
-
189.Herdean A, Sutherland DL, Ralph PJ. Phenoplate: an innovative method for assessing interacting effects of temperature and light on non-photochemical quenching in microalgae under chemical stress. New Biotechnol 2022;66: 89-96. CrossRef PubMed Google Scholar
-
190.Solovchenko A, Lukyanov A, Vasilieva S, Lobakova E. Chlorophyll fluorescence as a valuable multitool for microalgal biotechnology. Biophys Rev 2022;14(4): 973-83. CrossRef PubMed Google Scholar
-
191.Solovchenko AE, Vasilieva SG, Zaitsev P, Lukyanov AA, Skripnikova EV, Antal TK. Approaches to rapid screening of pharmaceutical xenobiotic effects on microalgae via monitoring of photosynthetic apparatus condition. J Appl Phycol 2022;34(1): 353-61. CrossRef PubMed Google Scholar
-
192.Herdean A, Hall C, Hughes DJ, Kuzhiumparambil U, Diocaretz BC, Ralph PJ. Temperature mapping of non-photochemical quenching in Chlorella vulgaris. Photosynth Res 2023;155(2): 191-202. CrossRef PubMed Google Scholar
-
193.Patwari P, Pruckner F, Fabris M. Biosensors in microalgae: a roadmap for new opportunities in synthetic biology and biotechnology. Biotechnol Adv 2023;68: 108221. CrossRef PubMed Google Scholar
-
194.Maeno T, Uzawa T, Kono I, Okano K, Iino T, Fukita K, et al. Targeted delivery of fluorogenic peptide aptamers into live microalgae by femtosecond laser photoporation at single-cell resolution. Sci Rep 2018;8(1): 8271. CrossRef PubMed Google Scholar
-
195.Kim JY, Lee M, Kim KY, Kim HS, Oh B, Son J, et al. Uptake of spherical nucleic acid (SNA) in Ochromonas danica: a new potential biotechnological tool. Algal Res 2024;78: 103385. CrossRef PubMed Google Scholar
-
196.Kohlberger M, Gadermaier G. SELEX: critical factors and optimization strategies for successful aptamer selection. Biotechnol Appl Biochem 2022;69(5): 1771-92. CrossRef PubMed Google Scholar
-
197.Yu H, Zhu J, Shen G, Deng Y, Geng X, Wang L. Improving aptamer performance: key factors and strategies. Microchim Acta 2023;190(7): 255. CrossRef PubMed Google Scholar
-
198.Li J, Wu H, Santana I, Fahlgren M, Giraldo JP. Standoff optical glucose sensing in photosynthetic organisms by a quantum dot fluorescent probe. ACS Appl Mater Interfaces 2018;10(34): 28279-89. CrossRef PubMed Google Scholar
-
199.Qian ZS, Shan XY, Chai LJ, Chen JR, Feng H. A fluorescent nanosensor based on graphene quantum dots–aptamer probe and graphene oxide platform for detection of lead (Ⅱ) ion. Biosens Bioelectron 2015;68: 225-31. CrossRef PubMed Google Scholar
-
200.Giardi MT, Zappi D, Turemis M, Varani G, Lo Celso F, Barone G, et al. Quantum dots functionalised artificial peptides bioinspired to the D1 protein from the photosystem Ⅱ of Chlamydomonas reinhardtii for endocrine disruptor optosensing. Talanta 2021;224: 121854. CrossRef PubMed Google Scholar
-
201.Jin D, Xi P, Wang B, Zhang L, Enderlein J, van Oijen AM. Nanoparticles for super-resolution microscopy and single-molecule tracking. Nat Methods 2018;15(6): 415-23. CrossRef PubMed Google Scholar
-
202.Ștefan G, Hosu O, De Wael K, Lobo-Castañón MJ, Cristea C. Aptamers in biomedicine: selection strategies and recent advances. Electrochim Acta 2021;376: 137994. CrossRef PubMed Google Scholar
-
203.Cunha I, Biltes R, Sales M, Vasconcelos V. Aptamer-based biosensors to detect aquatic phycotoxins and cyanotoxins. Sensors (Basel) 2018;18(7): 2367. CrossRef PubMed Google Scholar
-
204.Kim JY, Oh J-J, Kim DH, Park J, Kim HS, Choi Y-E. Rapid and accurate quantification of paramylon produced from Euglena gracilis using an ssDNA aptamer. J Agric Food Chem 2020;68(1): 402-8. CrossRef PubMed Google Scholar
-
205.Kim JY, Jin CR, Kim HS, Park J, Choi Y-E. Fluorogenic, "on-off" nanosensor based on dual-quenching effect for imaging intracellular metabolite of various microalgae. Biosens Bioelectron 2022;198: 113839. CrossRef PubMed Google Scholar
-
206.Kumudha A, Selvakumar S, Dilshad P, Vaidyanathan G, Thakur MS, Sarada R. Methylcobalamin—a form of vitamin B12 identified and characterised in Chlorella vulgaris. Food Chem 2015;170: 316-20. CrossRef PubMed Google Scholar
-
207.Wu H, Nißler R, Morris V, Herrmann N, Hu P, Jeon S-J, et al. Monitoring plant health with near-infrared fluorescent H2O2 nanosensors. Nano Lett 2020;20(4): 2432-42. CrossRef PubMed Google Scholar
-
208.Scognamiglio V, Stano P, Polticelli F, Antonacci A, Lambreva MD, Pochetti G, et al. Design and biophysical characterization of atrazine-sensing peptides mimicking the Chlamydomonas reinhardtii plastoquinone binding niche. Phys Chem Chem Phys 2013;15(31): 13108-15. CrossRef PubMed Google Scholar
-
209.Liu F, Zhang C, Duan Y, Ma J, Wang Y, Chen G. In vitro selection and characterization of a DNA aptamer targeted to Prorocentrum minimum-a common harmful algae. Sci Total Environ 2022;830: 154771. CrossRef PubMed Google Scholar
-
210.Gutiérrez S, Lauersen KJ. Gene delivery technologies with applications in microalgal genetic engineering. Biology (Basel) 2021;10(4): 265. PubMed Google Scholar
-
211.Gutiérrez S, Lauersen KJ. Gene delivery technologies with applications in microalgal genetic engineering. Biology 2021;10(4): 265. CrossRef PubMed Google Scholar
-
212.Ortiz-Matamoros MF, Villanueva MA, Islas-Flores T. Genetic transformation of cell-walled plant and algae cells: delivering DNA through the cell wall. Brief Funct Genomics 2017;17(1): 26-33. CrossRef PubMed Google Scholar
-
213.Grama SB, Liu Z, Li J. Emerging trends in genetic engineering of microalgae for commercial applications. Mar Drugs 2022;20(5): 285. CrossRef PubMed Google Scholar
-
214.Mou Q, Xue X, Ma Y, Banik M, Garcia V, Guo W, et al. Efficient delivery of a DNA aptamer-based biosensor into plant cells for glucose sensing through thiol-mediated uptake. Sci Adv 2022;8(26): eabo0902. CrossRef PubMed Google Scholar
-
215.Hyman JM, Geihe EI, Trantow BM, Parvin B, Wender PA. A molecular method for the delivery of small molecules and proteins across the cell wall of algae using molecular transporters. Proc Natl Acad Sci 2012;109(33): 13225-30. CrossRef PubMed Google Scholar
-
216.Muñoz CF, de Jaeger L, Sturme MHJ, Lip KYF, Olijslager JWJ, Springer J, et al. Improved DNA/protein delivery in microalgae—a simple and reliable method for the prediction of optimal electroporation settings. Algal Res 2018;33: 448-55. CrossRef PubMed Google Scholar
-
217.Geada P, Rodrigues R, Loureiro L, Pereira R, Fernandes B, Teixeira JA, et al. Electrotechnologies applied to microalgal biotechnology—applications, techniques and future trends. Renew Sustain Energy Rev 2018;94: 656-68. CrossRef PubMed Google Scholar
-
218.Garcia PA, Ge Z, Moran JL, Buie CR. Microfluidic screening of electric fields for electroporation. Sci Rep 2016;6(1): 21238. CrossRef PubMed Google Scholar
-
219.Agarwal A, Zudans I, Weber EA, Olofsson J, Orwar O, Weber SG. Effect of cell size and shape on single-cell electroporation. Anal Chem 2007;79(10): 3589-96. CrossRef PubMed Google Scholar
-
220.Azencott HR, Peter GF, Prausnitz MR. Influence of the cell wall on intracellular delivery to algal cells by electroporation and sonication. Ultrasound Med Biol 2007;33(11): 1805-17. CrossRef PubMed Google Scholar
-
221.Patino CA, Pathak N, Mukherjee P, Park SH, Bao G, Espinosa HD. Multiplexed high-throughput localized electroporation workflow with deep learning-based analysis for cell engineering. Sci Adv 2022;8(29): eabn7637. CrossRef PubMed Google Scholar
-
222.Guignet EG, Meyer T. Suspended-drop electroporation for high-throughput delivery of biomolecules into cells. Nat Methods 2008;5(5): 393-5. CrossRef PubMed Google Scholar
-
223.Kwak S-Y, Lew TTS, Sweeney CJ, Koman VB, Wong MH, Bohmert-Tatarev K, et al. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat Nanotechnol 2019;14(5): 447-55. CrossRef PubMed Google Scholar
-
224.Giraldo JP, Landry MP, Faltermeier SM, McNicholas TP, Iverson NM, Boghossian AA, et al. Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat Mater 2014;13(4): 400-8. CrossRef PubMed Google Scholar
-
225.Santana I, Jeon S-J, Kim H-I, Islam MR, Castillo C, Garcia GFH, et al. Targeted carbon nanostructures for chemical and gene delivery to plant chloroplasts. ACS Nano 2022;16(8): 12156-73. CrossRef PubMed Google Scholar
-
226.Santana I, Wu H, Hu P, Giraldo JP. Targeted delivery of nanomaterials with chemical cargoes in plants enabled by a biorecognition motif. Nat Commun 2020;11(1): 2045. CrossRef PubMed Google Scholar
Copyright information
© The Author(s) 2025.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.






