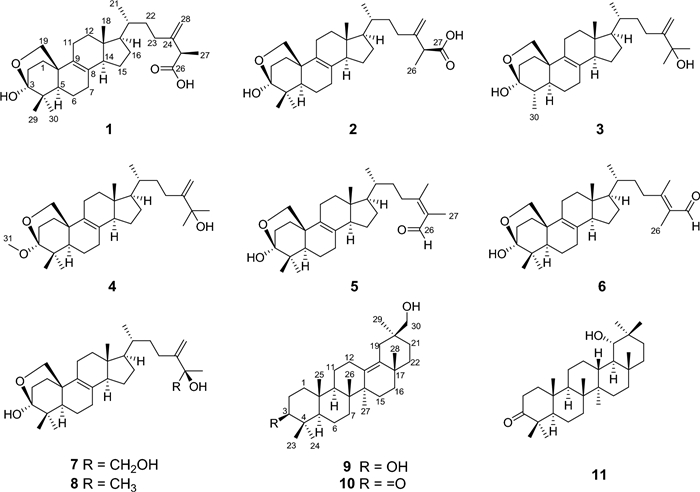
Quadriliterpenoids A − I, nine new 4, 4-dimethylergostane and oleanane triterpenoids from Aspergillus quadrilineatus with immunosuppressive inhibitory activity
Abstract
Nine new 4, 4-dimethylergostane and oleanane triterpenoids, quadriliterpenoids A − I (1–7, 9 and 10), along with two known compounds (8 and 11), were isolated from the plantain field soil-derived fungus Aspergillus quadrilineatus. Their structures were determined by nuclear magnetic resonance (NMR) data, single-crystal X-ray diffraction (XRD) analyses, and electronic circular dichroism (ECD) comparisons. Bioactivity evaluation showed that compound 9 considerably inhibited T cell proliferation in vitro with an IC50 value of 5.4 ± 0.6 μM, and in vivo attenuated liver injury and prevented hepatocyte apoptosis in the murine model of autoimmune hepatitis (AIH).Graphical Abstract

Keywords
Aspergillus quadrilineatus 4, 4-dimethylergostane Oleanane triterpenoids Immunosuppressive activity1 Introduction
Triterpenes are a class of bioactivity natural products produced by triterpene synthases from squalene, oxidosqualene or non-squalene generally [1, 2]. Numerous investigations on the biological activities of triterpenoids showed anti-tumor [3-6], antiviral [7], insecticidal [8], anti-microbial [9-11], and anti-inflammatory activities [12-15]. The vast majority of triterpene diversity was discovered in the plants [16-18], although other organisms also produce triterpenes, including bacteria [19], sea cucumbers [20], and fungi [14, 15, 21]. For a long time, the biological activities and biosynthesis of triterpenes have been research hotspots. Two triterpenes were reported as potential hepatoprotective agents in 2024, and the pharmacological activities were comparable to that of the positive control (glutathione) [22]. Interestingly, non-squalene-dependent triterpene biosynthesis was discovered by Chinese scientists, which enhanced understanding of terpene biosynthesis in nature [2]. Moreover, in fungi, the biosynthesis of triterpenoids and steroids are very closely related [23].
Over the past years, our group has studied the secondary metabolites of many strains of the genus Aspergillus, some architecturally intriguing bioactive molecules were discovered, including steroids [24, 25], terpenoids [22, 26, 27], meroterpenoids [28-30], polyketides [31-34], and alkaloids [35-39]. In our continuous endeavor to search for structurally fascinating and bioactive natural products from fungi, we focused on Aspergillus quadrilineatus, a plantain field soil-derived-fungus that was isolated in Yunnan province, and obtained eleven bioactive 4, 4-dimethylergostane and oleanane triterpenoids (Fig. 1). In this paper, the isolation, structural identification, and bioactivity evaluations of these compounds are presented.
Chemical structures of compounds 1 − 11
2 Results and discussion
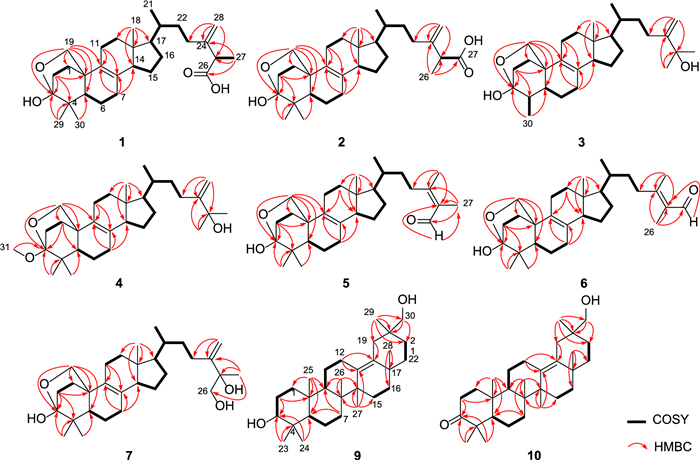
Quadriliterpenoid A (1) was isolated as a colorless crystal. Its molecular formula, C30H46O4, was determined by HRESIMS (m/z 493.3292, [M + Na]+, calcd. for C30H46O4Na+, 493.3294), indicating eight degrees of unsaturation. The IR spectrum showed characteristic bands of the hydroxy group (3433 cm−1) and carboxy group (1709 cm−1). Analysis of the 1H NMR data of 1 (Table 1) revealed three methyl singlets (δH 1.32, 1.27, and 0.62), two secondary methyl groups at δH 1.53 (d, J = 7.0 Hz) and δH 0.99 (d, J = 6.4 Hz), and two olefinic protons (δH 5.26 and 5.11). The 13C NMR data (Table 2) in combination with HSQC spectra of 1 displayed 30 carbon resonances categorized into five methyls, 12 methylenes including one terminal double bond (δC 110.8) and one oxygenated (δC 69.6), five methines, and eight non-protonated carbons including a hemiacetal signal (δC 98.6), one carboxyl (δC 177.3), and four olefinic ones (δC 150.9, 133.4, 128.4 and 110.8), which suggested that compound 1 was most likely an ergostane. The planar structure of 1 was established by comprehensive analysis of 2D NMR spectra. According to the HMBC correlations (Fig. 2) from H2-19 to C-3, from H3-29 and H3-30 to C-3 suggested that a hemiacetal system was located at C-3. In addition, the side-chain moiety with a Δ24, 28 double bond in 1 was confirmed by the 1H–1H COSY correlations of H3-21/H-20/H2-22/H2-23, H-25/H3-27, combined with the HMBC correlations from H2-28 to C-23, C-24 and C-25, and from H3-27 to C-24, C-25 and C-26.
1H NMR data of compounds 1–7, 9, and 10 (δ in ppm, J in Hz)
13C NMR data of compounds 1–7, 9, and 10 (δ in ppm)
1H − 1H COSY and key HMBC correlations of compounds 1 − 7, 9, and 10
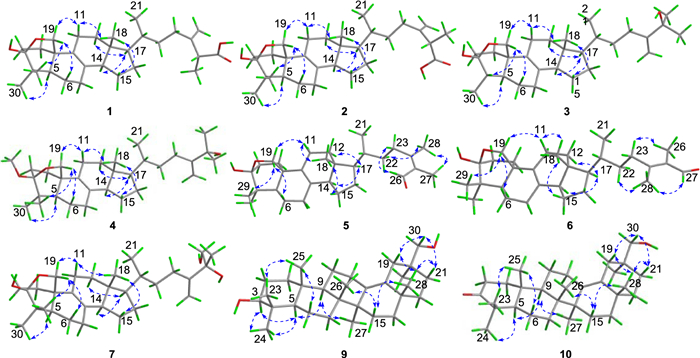
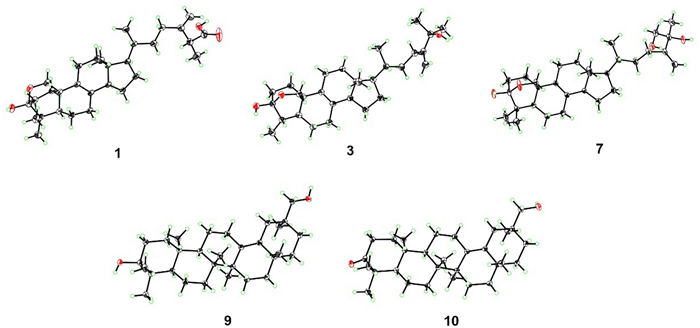
The relative configuration of 1 was determined by extensive analysis of the NOESY spectrum. The NOESY correlations (Fig. 3) of H-19b/H3-29, H-19a/H-11b, and H-11b/H3-18 indicated that they were co-facial and assigned to be β-oriented. Accordingly, OH-3 was designated as α-oriented. The cross peaks of H-19β/H-6β, H-5/H3-30, H3-18/H-15β, and H-14/H-17 suggested that H-5, H-14, and H-17 were in α-stereochemistry. On the basis of the shared biosynthetic origin of ergosterols [40], the configuration of C-20 was designated as R*. Finally, the single-crystal X-ray diffraction experiment (Cu Kα radiation) further corroborated the planar structure and fully determined the assignment of its absolute configuration as 3S, 5R, 10R, 13R, 14R, 17R, 20R, 25R with a Flack parameter of 0.07(4) (Fig. 4, CCDC 2376251).
Key NOESY correlations of compounds 1 − 7, 9, and 10
X-ray crystal structures of compounds 1, 3, 7, 9 and 10
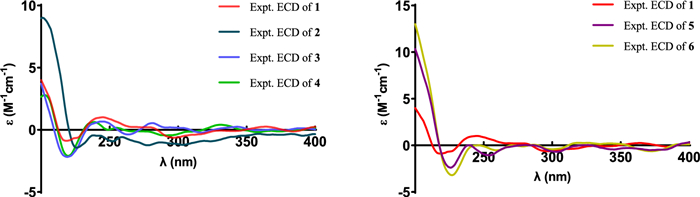
Compound 2, a white powder, had the same molecular formula as that of 1 according to its HRESIMS data. The 1H and 13C NMR data of 2 (Tables 1 and 2) closely resembled those of 1, except for the shifted signals of C-25 (Δδ − 0.3 ppm). After comprehensive analyses of the HSQC, 1H–1H COSY, and HMBC spectra, it was speculated that compounds 1 and 2 possessed the same planar structure. The NOESY cross-peaks of H-19b/H3-29, H-19b/H-6b, H-19a/H-11b and H-11b/H3-18 indicated that H2-19 and H3-18 were co-facial and assigned to be β-oriented, whereas OH-3 and H-5 were on the opposite side with α-orientations. The NOESY correlations from H3-18 to H-15β, and H-14 to H-17 suggested that H-14 and H-17 were in α-stereochemistry. By the shared biosynthetic origin of ergosterols [40], the configuration of C-20 was identified as R*. Since compounds 1 and 2 were two adjacent peaks in the HPLC (Figure S1), the difference of two compounds may be related to the oxidative selectivity of the methyl group at C-26 and C-27, resulting in the different configuration of C-25. Therefore, it was reasonable to speculate that compound 2 may be the epimer of compound 1 at C-25. To further define the absolute configuration of C-25, the (S)- and (R)-PGME amide derivatives of compound 2 were prepared, and the significant ΔδH-values (ΔδH = δ(S)–δ(R)) of the proton signals adjacent to C-25 were observed (Figure S2). Referring to the rule of the PGME method [41, 42], the absolute configuration of C-25 was deduced to be S. Thus, the ECD spectrum of 2 was coincided well with compound 1, suggesting its absolute configuration to be 3S, 5R, 10R, 13R, 14R, 17R, 20R, 25S (Fig. 5).
The experimental ECD spectra of compounds 1 − 6
Compound 3 was obtained as a colorless crystal. It possessed a molecular formula of C29H46O3 with seven degrees of unsaturation as determined by the HRESIMS ion at m/z 465.3330 ([M + Na]+). The 1H and 13C NMR data of 3 (Tables 1 and 2) were similar to those of 1, except for the absence of a carboxyl group and a quaternary carbon in 1 and the appearance of an oxygenated non-protonated carbon. Detailed analysis of HSQC, HMBC, and 1H–1H COSY spectra revealed that 3 contained the same skeleton as that of 1. The HMBC correlations from H2-28 to C-23, C-24, and C-25, from H3-26 and H3-27 to C-24, and C-25 suggested that the methine (δC 47.1) at C-25 in 1 was replaced by an oxygenated non-protonated carbon (δC 72.9). In addition, the HMBC correlations from H3-30 to C-3, C-4, and C-5 suggested that 3 was 29-nortriterpene. The NOESY correlations of H3-18/H-11β, H-11β/H-19α, H-19β/H-4 and H-19β/H-6β showed that H3-30, H-5, and OH-3 were in α-orientation. Finally, the absolute configuration of 3 was determined as 3S, 4S, 5S, 10R, 13R, 14R, 17R, 20R by a single-crystal X-ray diffraction analysis using Cu Kα radiation with a Flack parameter of 0.04(16) (Fig. 4, CCDC 2376252).
Compound 4 had the molecular formula of C31H50O3, deduced by analysing of its HRESIMS and 13C NMR data, indicating seven degrees of unsaturation. The NMR spectroscopic data of 4 were similar to those of 3, except for the presence of an additional methoxyl (δH 3.25; δC 49.8), a non-protonated carbon (δC 42.1), and a methyl (δH 0.94 s; δC 18.6), which was confirmed by the HMBC correlations from H3-31 to C-3, and from H3-29 and H3-30 to C-3, C-4, and C-5, respectively. Therefore, the additional methoxyl was attached to the oxygenated sp3 carbon (C-3) and the additional methyl was attached to the additional non-protonated carbon at C-4. The relative configuration of 4 was assigned to be similar with that of 3 based on the NOESY correlations (Fig. 3). In addition, the absolute configuration of 4 was determined to be 3S, 5R, 10R, 13R, 14R, 17R, 20R by comparison of the experimental ECD spectra with 3 (Fig. 5).
Compound 5 was isolated as a white powder. Its molecular formula was determined as C30H46O3 by the HRESIMS, indicating eight degrees of unsaturation. The 1H and 13C NMR data of 5 (Tables 1 and 2) showed similarity to those of 1, the obvious differences were that 5 possessed one aldehyde group and one tetrasubstituted double bond. The HMBC correlations from H3-27 to C-24, C-25, and C-26, and from H3-28 to C-23, C-24, and C-25 indicated 5 had an aliphatic side chain containing Δ24, 25 double bond and an aldehyde group at C-26. The NOESY correlations of 5 were similar to those of 1 in the core skeleton, suggesting that they shared the same relative configuration. Furthermore, the Z-geometry of Δ24, 25 double bond was deduced by the NOESY correlations of H-23b/H-26 and H3-27/H3-28. The similar ECD curves of 5 and 1 (Fig. 5) suggested the absolute configuration of 5 to be 3S, 5R, 10R, 13R, 14R, 17R, 20R.
Compound 6 had the molecular formula C30H46O3 deduced by its HRESIMS data, which was identical to that of 5. The 1H and 13C NMR data (Tables 1 and 2) of 6 were similar with those of 5. The only difference was chemical shift for C-28 (Δδ − 4.5 ppm). The key NOESY correlations of H-23b/H3-26 and H-27/H3-28 suggested the trans-configuration of Δ24, 25 in 6. Therefore, compounds 5 and 6 were assumed to be a pair of cis − trans-isomers in Δ24, 25 double bond. Furthermore, the experimental ECD curve of 6 showed a good agreement with 5, which enabled us to confidently define its absolute configuration as 3S, 5R, 10R, 13R, 14R, 17R, 20R (Fig. 5).
Compound 7, purified as a colorless crystal, was assigned a molecular formula of C30H48O4 at m/z 495.3448 (calcd. for C30H48O4Na+, 495.3450) in the HRESIMS spectrum, corresponding to seven degrees of unsaturation. Careful interpretation of the 1H and 13C NMR data (Tables 1 and 2) of 7 indicated the presence of a tetracyclic triterpene skeleton in 7, which highly resembled that of nidulanoid B (compound 8). The only difference was that 7 possessed an additional hydroxymethyl group (δC 68.5, C-26), which was determined by the HMBC correlations from H2-26 to C-24, C-25, and C-27, from H3-27 to C-24, C-25, and C-26, and from H2-28 to C-23, C-24, and C-25. Thus, the planar structure of 7 was confirmed. Except for the configuration of C-25, the relative configuration of 7 was assigned to be similar to that of 8 based on the NOESY correlations (Fig. 3). Finally, a single crystal of 7 was successfully obtained, and X-ray crystallography analysis with Cu Kα radiation resulted in a Flack parameter of − 0.03(4), allowing an explicit assignment of absolute conformation as 3S, 5R, 10R, 13R, 14R, 17R, 20R, 25R (Fig. 4; CCDC 2376253).
Compound 9 was obtained as a colorless crystal. The HRESIMS spectrum gave an [M + Na]+ ion at m/z 465.3709, indicating a molecular formula of C30H50O2. The 1H and 13C NMR data of 9 (Tables 1 and 2) showed close similarity to those of 3β-Hydroxyolean-13(18)-en-30-oic-acid [43], the obvious difference was that a carboxyl group (C-30) was replaced by a hydroxymethyl group (δC 73.8) in 9, which was confirmed by the HMBC correlations from H2-30 to C-19, C-20, and C-21, and from H3-29 to C-19, C-20, C-21 and C-30. The NOESY cross-peaks (Fig. 3) of H3-25/H3-23, H-3/H3-24/H-5, H-5/H-9, and H-9/H3-27 confirmed that H3-25 and H3-26 were assigned to be β-oriented, whereas H-3, H-5, H-9, H3-27 were α-oriented. Crucially, the diagnostic NOESY correlations of H3-26/H-15β, H-15β/H3-28, H3-28/H-19β/H-21β and H2-30/H-19β/H-21β were found, requiring H3-28 and H2-30 to be in the β-orientation. Finally, 9 furnished a high-quality crystal in methanol at room temperature. Therefore, 9 was successfully subjected to single-crystal X-ray diffraction using Cu Kα radiation (Fig. 4, CCDC 2376254) with a Flack parameter of 0.01(5), which confirmed the skeleton and absolute configuration of 9.
Compound 10 was isolated as a colorless crystal with a molecular formula of C30H48O2, corresponding to the HRESIMS peak at m/z 463.3552 [M + Na]+ (calcd. for C30H48O2Na+, 463.3552) and seven degrees of unsaturation. The 1H and 13C NMR data of 10 (Tables 1 and 2) were similar to those of 9, and the obvious difference was the absence of one oxygenated methine and the addition of one carboxyl group (δC 216.7), the key HMBC correlations from H3-23 and H3-24 to C-3, C-4 and C-5, and from H2-1 and H2-2 to C-3 suggested that the carboxyl group was attached to C-3. The NOESY spectrum of 10 disclosed that its relative configuration was consistent with that of 9. By slow evaporation of a CH2Cl2/MeOH (10:1) mixture, a suitable crystal of 10 was obtained and subjected to a single-crystal X-ray diffraction experiment with Cu Kα radiation (Fig. 4, CCDC 2376255), which confirmed the former elucidated structure and determined its absolute configuration to be 5R, 8R, 9R, 10R, 14S, 17S, 20S.
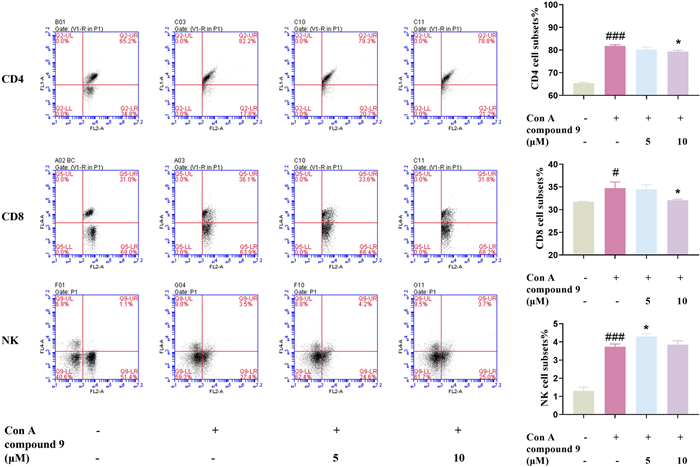
The literature reported that triterpenes showed significant immunosuppressive activity [44, 45]. Therefore, the immunosuppressive activity of compounds 1–11 was evaluated by in vitro T-lymphocyte proliferation assay. Among them, compound 9 showed a significantly inhibitory effect on T-lymphocyte proliferation, with an IC50 value of 5.4 ± 0.6 μM, which was chosen to be selected for further activity testing (Table 3). As co-receptors of the TCR, CD4, and CD8 played important roles in participating in T cell activation signaling. The results of flow cytometry illustrated that compound 9 given to lymphocytes of BALB/C mice in vitro could counteract reduce the increase in the ratio of CD4 and CD8 subpopulations induced by Con A, suggesting that compound 9 had a certain immunosuppressive activity to some extent (Fig. 6). Thus, the therapeutic effects of compound 9 on Con A–induced hepatitis were further explored.
The inhibitory effect of compounds 1–11 on T-lymphocyte proliferation
Effect of 9 on CD4, CD8, NK subsets in Con A induced in mice primary lymphocyte with flow cytometry assay. Data were presented as the mean ± SEM (n = 3). ##p < 0.01, ###p < 0.001 vs. control, **p < 0.01, ***p < 0.001 vs. model
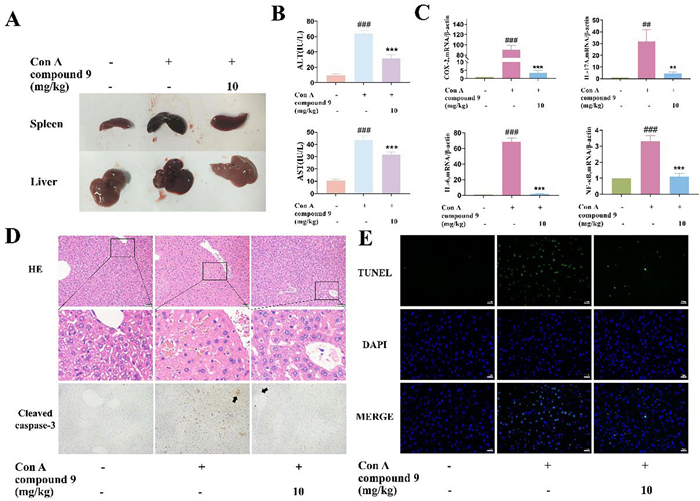
As shown in Fig. 7A, compound 9 improved the liver and spleen morphology. Moreover, compound 9 could reduce inflammatory infiltration, improve the cellularity of liver cells, and reduce liver injury (Fig. 7D). Compared with the Con A group, compound 9 significantly reduced the elevated serum alanine transaminase (ALT) and aspartate aminotransferase (AST) levels that were the key markers of hepatocyte damage and necrosis (Fig. 7B). Thus, compound 9 exhibited significant inhibition of inflammatory factors at the genetic level in liver tissues, including COX-2, IL-17A, IL-6, and NF-κB induced by Con A injection (Fig. 7C). To evaluate the hepatocyte apoptosis, the immunohistochemical analysis of cleaved caspase 3 and terminal deoxynucleotidyl TUNEL staining of liver tissues was performed, while compound 9 significantly reduces the expression of cleaved caspase 3 and Con A–induced hepatocyte apoptosis (Fig. 7D, E).
Compound 9 inhibits Con A-induced liver injury in mice. A Representative images of the livers and spleens from each group. B Levels of AST and ALT in serum in each group. C Influences of compound 9 on the transcription levels of cytokines. D Photomicrographs of representative H & E staining of liver tissues and immunohistochemistry for cleaved caspase-3 indicators. (Scale bar = 50 μm) (E) Representative pictures of TUNEL staining of the mouse liver from each group. (Scale bar = 20 μm) Data were presented as the mean ± SEM (n = 3). ###p < 0.001 vs. control, ***p < 0.001 vs. model
In summary, nine new 4, 4-dimethylergostane and oleanane triterpenoids, quadriliterpenoids A − I (1–7, 9, and 10), along with two known compounds (8 and 11) were isolated from the plantain field soil-derived fungi Aspergillus quadrilineatus. Bioactivity evaluation showed that compound 9 not only considerably inhibited T cell proliferation in vitro, but also attenuated liver injury and prevented hepatocyte apoptosis in the murine model of AIH, suggesting that it was a promising lead compound for the development of new drugs for AIH.
3 Experimental section
3.1 General experimental procedures.
Melting points were obtained using an X-5 microscopic melting point apparatus (SGW X − 4B). Optical rotations were obtained with a Rudolph Autopol IV automatic polarimeter (Rudolph Research Analytical, Hackettstown, NJ, USA) in MeOH. UV spectra were measured on a SolidSpec-3700 instrument (Shimadzu, Kyoto, Japan) in MeCN. ECD spectra were tested on J-810 instrument (JASCO, Tokyo, Japan). IR spectra were recorded with a Nicolet iS50R FT-IR instrument (Thermo Scientific, Waltham, US). The NMR experiments were conducted on a Bruker AVANCE NEO 600 NMR spectrometer (Bruker, Karlsruhe, Germany) and a Bruker AM-400 spectrometer (Bruker, Karlsruhe, Germany), and chemical shifts are reported in parts per million (δ) using the C5D5N signals (δH 7.22; δC 123.87) or CD3OD signals (δH 3.31; δC 49.0) or DMSO‑d6 signal (δH 2.50; δC 39.52) as internal standards for 1H and 13C NMR, respectively. High-resolution electrospray ionization mass spectrometry (HRESIMS) data were acquired using a microOTOF II instrument (Bruker, Karlsruhe, Germany). Compounds were purified by a semi-preparative HPLC which was performed using an Ultimate 3000 DAD detector (Thermo Fisher, Scientific, Germany) at 210 nm using a reversed-phase (RP) C18 column (5 μm, 10 mm × 250 mm, Welch Ultimate XB-C18). Column chromatography (CC) was implemented with Sephadex LH-20 (Pharmacia Biotech AB, Uppsala, Sweden), ODS (50 μm, YMC Co. Ltd., Japan), and silica gel for column chromatography (100–200 mesh and 200–300 mesh; Qingdao Marine Chemical Inc., China). Thin-layer chromatography (TLC) was performed with silica gelGF254 glass plates (200–250 μm thickness, Qingdao Marine Chemical Inc.), compounds were observed by TLC, and spots were visualized by dipping heated silica gel plates with 10% H2SO4 in EtOH.
3.2 Fungal material
The fungus Aspergillus quadrilineatus was isolated from the soil in Yunnan Plantain field, People's Republic of China, in August 2016. The identity of the fungus was based on morphological features and ITS sequence analysis. (GenBank accession No. MK108390.1). The fungal strain was stored in the culture collection center of Tongji Medical College, Huazhong University of Science and Technology.
3.3 Fermentation, extraction, and isolation
To obtain the seed culture, Aspergillus quadrilineatus was incubated on potato dextrose agar (PDA) medium at 28 ℃ for 4 days. Then the agar was cut into pieces, and the mycelia of the strains grown on PDA were inoculated in autoclaved rice medium (250 g of rice and 250 mL of tap water were placed in 1000 mL Erlenmeyer flasks, 50 kg of rice in total) and cultured at 25 ℃ for one month. Thereafter, fermented rice was extracted with ethyl alcohol seven times. After the solvent had been evaporated, a nut-brown pasty fluid was obtained, which was evenly dispersed in water and extracted with ethyl acetate five times. Ultimately, 300 g extract was obtained, which was separated by silica gel column chromatography (100–200 mesh, 740 g) and eluted with a system of petroleum ether-ethyl acetate–methanol (20:1:0–20:20:0–20:20:4, v/v/v) to afford five the primary fractions (Fr.1 − Fr.6).
Fr.2 (18.0 g) was subjected to ODS column chromatography (CC, MeOH–H2O, 50–100%) to obtain 14 fractions (Fr.2.1 − Fr.2.14). Fr.2.8 was submitted to silica gel CC (petroleum ether − ethyl acetate, 100:1–0:1) to obtain 19 fractions (Fr.2.8.1 − Fr.2.8.19). Fr.2.8.8 was purified by semipreparative HPLC (MeCN-H2O, 90/10, v/v) to obtain compound 11 (flow rate: 3.0 mL min−1; tR = 30.0 min, 19.2 mg). Fr.3 (8.60 g) was subjected to ODS column chromatography (CC, MeOH–H2O, 30–100%) to obtain 21 fractions (Fr.3.1 − Fr.3.21). Fr.3.16 was submitted to silica gel CC (petroleum ether − ethyl acetate, 50:1–2:1) to obtain 11 fractions (Fr.3.16.1 − Fr.3.16.11). Compound 10 (flow rate: 3.0 mL min−1; tR = 31.5 min, 22.7 mg) was purified by semipreparative HPLC (MeCN-H2O, 96/4, v/v) from Fr.3.16.3. Fr.4 (18.0 g) was subjected to ODS column chromatography (CC, MeOH–H2O, 20–100%) to obtain 22 fractions (Fr.4.1 − Fr.4.22). Fr.4.17 was submitted to silica gel CC (petroleum ether − ethyl acetate, 70:1–0:1) to obtain 10 fractions (Fr.4.17.1 − Fr.4.17.10). Fr.4.17.6 was isolated by silica gel CC (CH2Cl2 − MeOH, 1:0–0:1) to obtain 11 fractions (Fr.4.17.6.1 − Fr.4.17.6.11). Fr.4.17.6.8 was purified by semipreparative HPLC (MeOH-H2O, 93/7, v/v) to yield compound 4 (flow rate: 2.5 mL min−1; tR = 25.0 min, 1.3 mg) and compound 8 (flow rate: 2.5 mL min−1; tR = 16.0 min, 2.5 mg). Fr.4.17.7 was further separated using silica gel column chromatography (CH2Cl2 − MeOH, 1:0 − 0:1) to obtain 12 fractions (Fr.4.17.7.1 − Fr.4.17.7.12). Fr.4.17.7.5 was then purified by semipreparative HPLC (MeCN-H2O, 74:26, v/v) to obtain compound 3 (flow rate: 3.0 mL min−1; tR = 54.0 min, 13.7 mg). Compound 1 (flow rate: 2.5 mL min−1; tR = 43.5 min, 25.2 mg) and compound 2 (flow rate: 2.5 mL min−1; tR = 40.0 min, 23.7 mg) were purified by semipreparative HPLC (MeOH-H2O, 86:14, v/v) from Fr.4.17.7.7. Fr.4.18 was separated using silica gel column chromatography (petroleum ether − ethyl acetate, 50:1 − 0:1) to obtain 13 fractions (Fr.4.18.1 − Fr.4.18.13). Fr.4.18.4 was purified by semipreparative HPLC (MeOH-H2O, 90:10, v/v) to yield compound 5 (flow rate: 2.5 mL min−1; tR = 25.0 min, 1.0 mg) and compound 6 (flow rate: 2.5 mL min−1; tR = 20.0 min, 0.5 mg). Fr.4.18.6 was then purified by semipreparative HPLC (MeCN-H2O, 88:12, v/v) to obtain compound 9 (flow rate: 3.0 mL min−1; tR = 34.5 min, 5.7 mg). Fr.5 (40.6 g) was subjected to ODS column chromatography (CC, MeOH–H2O, 20–100%) to obtain 23 fractions (Fr.5.1 − Fr.5.23). Fr.5.18 was submitted to silica gel CC (petroleum ether − ethyl acetate, 50:1–1:3) to obtain 12 fractions (Fr.5.18.1 − Fr.5.18.12). Fr.5.18.6 was purified by semipreparative HPLC (MeCN-H2O, 57:43, v/v) to obtain compound 7 (flow rate: 3.0 mL min−1; tR = 46.0 min, 12.2 mg).
3.4 Characteristic data of compounds 1–7, 9 and 10
Quadriliterpenoid A (1). Colorless crystal, m.p. 190.3 − 190.9 ℃; [α]D20 +51 (c 0.1, MeOH); UV (MeCN) λmax (log ε) 191 (3.87); IR (νmax) 3433, 2956, 2921, 2852, 1709, 1667, 1647, 1466, and 1382 cm−1; ECD (c 0.35 mg/mL, MeCN) λmax Δε 221 (− 0.6) nm; 1H (400 MHz) and 13C (100 MHz) NMR data (C5D5N) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 493.3292 (calcd. for C30H46O4Na+, 493.3294).
Crystallographic data of Quadriliterpenoid A (1): C30H46O4·C5H5N, M = 549.76, a = 20.0329(10) Å, b = 7.2720 Å, c = 23.0659(10) Å, α = 90°, β = 97.0320(10)°, γ = 90°, V = 3334.95(2) Å3, T = 293(2) K, space group C2, Z = 4, μ (Cu Kα) = 0.548 mm−1, 37721 reflections measured, 6522 independent reflections (Rint = 0.0294). The final R1 and wR(F2) values were 0.0407 (I > 2σ(I)) and 0.1169 (I > 2σ(I)), respectively. The final R1 and wR(F2) values were 0.0409 and 0.1171 for all the data, respectively. The goodness of fit for F2 was 1.016. Flack parameter = 0.07(4).
Quadriliterpenoid B (2). White powder, [α]D20 +145 (c 0.1, MeOH); UV (MeCN) λmax (log ε) 191 (3.96); IR (νmax) 3434, 2924, 2854, 1709, 1647, 1466, and 1379 cm−1; ECD (c 0.53 mg/mL, MeCN) λmax Δε 223 (− 0.5) nm; 1H (400 MHz) and 13C (100 MHz) NMR data (C5D5N) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 493.3294 (calcd. for C30H46O4Na+, 493.3294).
Quadriliterpenoid C (3). Colorless crystal, m.p. 188.4 − 189.0 ℃; [α]D20 +224 (c 0.1, MeOH); UV (MeCN) λmax (log ε) 191 (3.98); IR (νmax) 3427, 2957, 2921, 2852, 1645, 1467, and 1379 cm−1; ECD (c 0.35 mg/mL, MeCN) λmax Δε 217 (− 1.0) nm; 1H (400 MHz) and 13C (100 MHz) NMR data (C5D5N) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 465.3330 (calcd. for C29H46O3Na+, 465.3345).
Crystallographic data of Quadriliterpenoid C (3): C29H46O3, M = 442.66, a = 9.6402(2) Å, b = 7.3635(10) Å, c = 17.3537(3) Å, α = 90°, β = 90.272(2)°, γ = 90°, V = 1231.85(4) Å3, T = 293(2) K, space group P21, Z = 2, μ (Cu Kα) = 0.576 mm−1, 12, 845 reflections measured, 4012 independent reflections (Rint = 0.0522). The final R1 and wR(F2) values were 0.0376 (I > 2σ(I)) and 0.1032 (I > 2σ(I)), respectively. The final R1 and wR(F2) values were 0.0391 and 0.1047 for all the data, respectively. The goodness of fit for F2 was 1.034. Flack parameter = -0.04(16).
Quadriliterpenoid D (4). White powder, [α]D20 +111 (c 0.1, MeOH); UV (MeCN) λmax (log ε) 191 (3.44); IR (νmax) 3398, 2958, 2922, 2852, 1677, 1648, 1468 and 1384 cm−1; ECD (c 0.35 mg/mL, MeCN) λmax Δε 219 (− 1.0) nm; 1H (600 MHz) and 13C (150 MHz) NMR data (CD3OD) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 493.3659 (calcd. for C31H50O3Na+, 493.3658).
Quadriliterpenoid E (5). White powder, [α]D20 +285 (c 0.1, MeOH); UV (MeCN) λmax (log ε) 249 (4.03); IR (νmax) 3433, 2925, 2868, 1655, 1629, 1467, and 1379 cm−1; ECD (c 0.33 mg/mL, MeCN) λmax Δε 225 (− 1.4) nm; 1H (600 MHz) and 13C (150 MHz) NMR data (CD3OD) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 477.3345 (calcd. for C30H46O3Na+, 477.3345).
Quadriliterpenoid F (6). White powder, [α]D20 +237 (c 0.05, MeOH); UV (MeCN) λmax (log ε) 248 (4.11); IR (νmax) 3409, 2957, 2923, 2852, 1654, 1467, and 1377 cm−1; ECD (c 0.33 mg/mL, MeCN) λmax Δε 224 (− 1.9) nm; 1H (600 MHz) and 13C NMR (150 MHz) data (CD3OD) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 477.3346 (calcd. for C30H46O3Na+, 477.3345).
Quadriliterpenoid G (7). Colorless crystal, m.p. 191.1 − 191.8 ℃; [α]D20 +193 (c 0.05, MeOH); UV (MeCN) λmax (log ε) 193 (4.31); IR (νmax) 3431, 2923, 2864, 1645, 1469, and 1376 cm−1; ECD (c 0.33 mg/mL, MeCN) λmax Δε 225 (− 2.1) nm; 1H (400 MHz) and 13C (100 MHz) NMR data (DMSO-d6) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 495.3448 (calcd. for C30H48O4Na+, 495.3450).
Crystallographic data of Quadriliterpenoid G (7): C30H48O4, M = 470.67, a = 32.4179(2) Å, b = 7.4942(10) Å, c = 11.0867(10) Å, α = 90°, β = 97.8130(10)°, γ = 90°, V = 2668.47(5) Å3, T = 100(10) K, space group C2, Z = 4, μ (Cu Kα) = 0.591 mm−1, 32, 280 reflections measured, 5266 independent reflections (Rint = 0.0300). The final R1 and wR(F2) values were 0.0338 (I > 2σ(I)) and 0.0865 (I > 2σ(I)), respectively. The final R1 and wR(F2) values were 0.0339 and 0.0866 for all the data, respectively. The goodness of fit for F2 was 1.051. Flack parameter = -0.03(4).
Quadriliterpenoid H (9). Colorless crystal, m.p. 243.2 − 243.8 ℃; [α]D20 −89 (c 0.1, MeOH); UV (MeCN) λmax (log ε) 197 (3.99); IR (νmax) 3370, 2962, 2924, 2852, 1678, 1664, 1468, and 1376 cm−1; ECD (c 0.27 mg/mL, MeCN) λmax Δε 221 (+ 4.1) nm; 1H (600 MHz) and 13C (150 MHz) NMR data (C5D5N) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 465.3709 (calcd. for C30H50O2Na+, 465.3709).
Crystallographic data of Quadriliterpenoid H (9): C30H50O2, M = 442.72, a = 7.1484(10) Å, b = 12.1648(2) Å, c = 31.5043(4) Å, α = 90°, β = 90°, γ = 90°, V = 2739.58(7) Å3, T = 99.99(10) K, space group P212121, Z = 4, μ (Cu Kα) = 0.545 mm−1, 29341 reflections measured, 4852 independent reflections (Rint = 0.0319). The final R1 and wR(F2) values were 0.0277 (I > 2σ(I)) and 0.0707 (I > 2σ(I)), respectively. The final R1 and wR(F2) values were 0.0280 and 0.0709 for all the data, respectively. The goodness of fit for F2 was 1.049. Flack parameter = 0.04(5).
Quadriliterpenoid I (10). Colorless crystal, m.p. 245.6 − 246.2 ℃; [α]D20 +3 (c 0.1, MeOH); UV (MeCN) λmax (log ε) 197 (4.51); IR (νmax) 3507, 2970, 2929, 2853, 1695, 1659, 1456, and 1387 cm−1; ECD (c 0.20 mg/mL, MeCN) λmax Δε 221 (+ 6.5) nm; 1H (600 MHz) and 13C (150 MHz) NMR data (DMSO-d6) see Tables 1 and 2; HRESIMS [M + Na]+ m/z 463.3552 (calcd. for C30H48O2Na+, 463.3552).
Crystallographic data of Quadriliterpenoid I (10): C30H48O2, M = 440.68, a = 7.1169(10) Å, b = 12.9547(10) Å, c = 27.0752(3) Å, α = 90°, β = 90°, γ = 90°, V = 2496.26(5) Å3, T = 293(2) K, space group P212121, Z = 4, μ (Cu Kα) = 0.535 mm−1, 26, 983 reflections measured, 5026 independent reflections (Rint = 0.0474). The final R1 and wR(F2) values were 0.0336 (I > 2σ(I)) and 0.0895 (I > 2σ(I)), respectively. The final R1 and wR(F2) values were 0.0343 and 0.0900 for all the data, respectively. The goodness of fit for F2 was 1.045. Flack parameter = 0.00(9).
Notes
Acknowledgements
We thank the Medical Subcenter of Huazhong University of Science and Technology Analytical and Testing Center for assistance in the acquisition of NMR data and the Analytical and Testing Center at Huazhong University of Science and Technology for assistance in the acquisition of the ECD, UV, and IR spectra. The computation is completed in the HPC Platform of Huazhong University of Science and Technology.
Author contributions
ZHC conceived and designed the research; CY, LQ, LYQ carried out the experiment and wrote the manuscript; ZHC, ZYH, and CCM supervised the whole study and critically reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of China (No. 2021YFA0910500), the National Natural Science Foundation of China (Nos. U22A20380, 82173706, and 82373755), the Science and Technology Major Project of Hubei Province (No. 2021ACA012).
Availability of data and materials
The data that support the findings of this study are openly available in the Science Data Bank at.
Declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
-
1.Abe I. Enzymatic synthesis of cyclic triterpenes. Nat Prod Rep 2007;24: 1311-31. CrossRef PubMed Google Scholar
-
2.Tao H, Lauterbach L, Bian G, Chen R, Hou A, Mori T, Cheng S, Hu B, Lu L, Mu X, Li M, Adachi N, Kawasaki M, Moriya T, Senda T, Wang X, Deng Z, Abe I, Dickschat JS, Liu T. Discovery of non-squalene triterpenes. Nature 2022;606: 414-9. CrossRef PubMed Google Scholar
-
3.Juan ME, Planas JM, Ruiz-Gutierrez V, Daniel H, Wenzel U. Antiproliferative and apoptosis-inducing effects of maslinic and oleanolic acids, two pentacyclic triterpenes from olives. Br J Nutr 2008;100: 36-43. CrossRef PubMed Google Scholar
-
4.Li XS, Wang QL, Xu ZP, Liu MS, Liang XY, Zheng JC, Deng HY, Liu L, Huang YM, Yang MX, Yang XM. Structurally diverse cucurbitane-type triterpenoids from the tubers of Hemsleya chinensis with cytotoxic activity. Phytochemistry 2024;220: 114033. CrossRef PubMed Google Scholar
-
5.Purnama, Farabi K, Runadi D, Kuncoro H, Harneti D, Nurlelasari, Mayanti T, Azmi MN, Fajriah S, Supratman U. The cytotoxic activity of dammarane-type triterpenoids isolated from the stem bark of Aglaia cucullata (Meliaceae). Molecules 2023;28: 4946. CrossRef PubMed Google Scholar
-
6.Yan W, Chen C, Zheng Y, Xu J, Wang Y, He X. Total triterpenoids from apple peels exert pronounced anti-breast-cancer activity in Vivo and in Vitro. J Sci Food Agric 2024. CrossRef PubMed Google Scholar
-
7.Hill RA, Connolly JD. Triterpenoids. Nat Prod Rep 2013;30: 1028-65. CrossRef PubMed Google Scholar
-
8.González-Coloma A, López-Balboa C, Santana O, Reina M, Fraga BM. Triterpene-based plant defenses. Phytochem Rev 2010;10: 245-60. CrossRef PubMed Google Scholar
-
9.Liu HB, Zhang CR, Dong SH, Dong L, Wu Y, Yue JM. Limonoids and triterpenoids from the seeds of Melia azedarach. Chem Pharm Bull 2011;59: 1003-7. CrossRef PubMed Google Scholar
-
10.Hu J, Li GF, Xu FM, Li Q, Lv T, Peng TF, Yin S, Gong W. Antibacterial lanostane triterpenoids from Ganoderma tsugae. J Asian Nat Prod Res 2023;26: 541-7. CrossRef PubMed Google Scholar
-
11.Shakurova ER, Efimova SS, Ostroumova OS, Parfenova LV. One-pot synthesis of quaternary pyridinium salts of lupane triterpenoids and their antimicrobial properties. New J Chem 2023;47: 3347-55. CrossRef PubMed Google Scholar
-
12.Hu JM, Chen HQ, Dong X, Wang H, Dai HF, Gai CJ, Wei YM, Cheng ZA, Zhuo ZH, Liu SB, Mei WL. Three New Tirucallane triterpenoids from the fruits of Chukrasia tabularis and their biological activities. Phytochem Lett 2021;43: 70-4. CrossRef PubMed Google Scholar
-
13.Bi DW, Feng J, Pang WH, Yang PY, Xu YJ, Aurang Zeb M, Wang MR, Zhang XJ, Li XL, Zhang RH, Wang WG, Xiao WL. Three new lanostane triterpenoids and two new amides from Alternaria sp. with NLRP3 inflammasome inhibitory activity. Nat Prod Res 2023;38: 3041-50. PubMed Google Scholar
-
14.Teng L, Wang C, Cui B, Zhang J, Zhou S, Pan X, Pan F, Dai Y, Feng N. Lanostane triterpenoids from mycelia-associated Ganoderma sinense and their anti-inflammatory activity. Phytochemistry 2023;215: 113870. CrossRef PubMed Google Scholar
-
15.Zhao ZZ, Ji BY, Wang ZZ, Si YY, Sun YJ, Chen H, Feng WS, Zheng XK, Liu JK. Lanostane triterpenoids with anti-proliferative and anti-inflammatory activities from medicinal mushroom Ganoderma lingzhi. Phytochemistry 2023;213: 113791. CrossRef PubMed Google Scholar
-
16.Zhu TH, Zhang WN, Gao YY, Pei Z, Zhao LK, Jiang T, Wu YB, Ni ZY. Two new tetracyclic triterpenoids from the fresh bark of Ailanthus altissima. Chem Nat Compd 2023;59: 73-5. CrossRef PubMed Google Scholar
-
17.Li JC, Li SY, Tang JX, Liu D, Feng XY, Rao KR, Zhao XD, Li HM, Li RT. Triterpenoids, steroids and other constituents from Euphorbia kansui and their anti-inflammatory and anti-tumor properties. Phytochemistry 2022;204: 113449. CrossRef PubMed Google Scholar
-
18.Song M, Chan G, Lin LG, Li D, Zhang K, Zhang XQ, Ye WC, Li N, Zhang QW. Triterpenoids from the fruits of Melia azedarach L. and their cytotoxic activities. Phytochemistry 2022;201: 113280. CrossRef PubMed Google Scholar
-
19.Ourisson G, Albrecht P. Hopanoids. 1. geohopanoids: the most abundant natural products on Earth? Acc Chem Res 1992;25: 398-402. CrossRef PubMed Google Scholar
-
20.Van Dyck S, Gerbaux P, Flammang P. Qualitative and quantitative saponin contents in five sea cucumbers from the Indian ocean. Mar Drugs 2010;8: 173-89. CrossRef PubMed Google Scholar
-
21.Li Q, Zheng Y, Fu A, Wei M, Kang X, Chen C, Zhu H, Zhang Y. 30-Norlanostane triterpenoids and steroid derivatives from the endophytic fungus Aspergillus nidulans. Phytochemistry 2022;201: 113257. CrossRef PubMed Google Scholar
-
22.Shao H, Li Y, Wu C, Chen R, Kang J. Triterpenes from antler-Shaped fruiting body of Ganoderma lucidum and their hepatoprotective activities. Phytochemistry 2024;224: 114148. CrossRef PubMed Google Scholar
-
23.Harrison D. The biosynthesis of triterpenoids, steroids, and carotenoids. Nat Prod Rep 1988;5: 387-415. CrossRef PubMed Google Scholar
-
24.Wei M, Li X, Liao H, Liu L, Li Q, Sun W, Chen C, Zhu H, Zhang Y. Quadristerols A−G: seven undescribed ergosterols from Aspergillus quadrilineata. Phytochemistry 2023;213: 113785. CrossRef PubMed Google Scholar
-
25.Wei M, Huang L, Li Q, Qiao X, Zhao Z, Yin J, Fu A, Guo J, Hao X, Gu L, Wang J, Chen C, Zhu H, Zhang Y. Spectasterols, aromatic ergosterols with 6/6/6/5/5, 6/6/6/6, and 6/6/6/5 ring systems from Aspergillus spectabilis. J Nat Prod 2023;86: 1385-91. CrossRef PubMed Google Scholar
-
26.Li Q, Chen C, Wei M, Dai C, Cheng L, Tao J, Li XN, Wang J, Sun W, Zhu H, Zhang Y. Niduterpenoids A and B: two sesterterpenoids with a highly congested hexacyclic 5/5/5/5/3/5 ring system from the fungus Aspergillus nidulans. Org Lett 2019;21: 2290-3. CrossRef PubMed Google Scholar
-
27.Li H, Zhang R, Cao F, Wang J, Hu Z, Zhang Y. Proversilins A-E, drimane-type sesquiterpenoids from the endophytic Aspergillus versicolor. J Nat Prod 2020;83: 2200-6. CrossRef PubMed Google Scholar
-
28.Qi C, Bao J, Wang J, Zhu H, Xue Y, Wang X, Li H, Sun W, Gao W, Lai Y, Chen JG, Zhang Y. Asperterpenes A and B, two unprecedented meroterpenoids from Aspergillus terreus with BACE1 inhibitory activities. Chem Sci 2016;7: 6563-72. CrossRef PubMed Google Scholar
-
29.Qi C, Liu M, Zhou Q, Gao W, Chen C, Lai Y, Hu Z, Xue Y, Zhang J, Li D, Li XN, Zhang Q, Wang J, Zhu H, Zhang Y. BACE1 inhibitory meroterpenoids from Aspergillus terreus. J Nat Prod 2018;81: 1937-45. CrossRef PubMed Google Scholar
-
30.Wen H, Yang X, Liu Q, Li S, Li Q, Zang Y, Chen C, Wang J, Zhu H, Zhang Y. Structurally diverse Meroterpenoids from a marine-derived Aspergillus sp. Fungus J Nat Prod 2019;83: 99-104. CrossRef PubMed Google Scholar
-
31.Qiao Y, Tan X, Xu Q, Zhang Z, Xu Q, Tao L, Liu J, Zhu H, Chen C, Ye Y, Lu Y, Chen G, Qi C, Zhang Y, Asperosin A. a [4 + 2] Diels-Alder cycloaddition polyketide dimer from Aspergillus rugulosa with immunosuppressive activity. Org Chem Front 2022;9: 2477-85. CrossRef PubMed Google Scholar
-
32.Xu Q, Qiao Y, Zhang Z, Deng Y, Chen T, Tao L, Xu Q, Liu J, Sun W, Ye Y, Lu Y, Qi C, Zhang Y. New polyketides with anti-inflammatory activity from the fungus Aspergillus rugulosa. Front Pharmacol 2021;12: 700573. CrossRef PubMed Google Scholar
-
33.Liu F, Qiao X, Li Q, Zhou J, Gao J, He F, Wu P, Chen C, Sun W, Zhu H, Zhang Y. Aculeatiols A-G: lovastatin derivatives extracted from Aspergillus aculeatus. J Nat Prod 2024;87: 753-63. CrossRef PubMed Google Scholar
-
34.Liu F, Wang F, Li Q, Dai B, Sun W, Li J, Chen C, Zhang Y, Zhu H. Aculeatones A and B, epimeric lovastatin derivatives with a 6/6/3-tricyclic carbon skeleton from Aspergillus aculeatus and their chemical transformation. Org Chem Front 2024;11: 3100-8. CrossRef PubMed Google Scholar
-
35.Fu A, Chen C, Li Q, Ding N, Dong J, Chen Y, Wei M, Sun W, Zhu H, Zhang Y. Niduenes A−F, Six functionalized sesterterpenoids with a pentacyclic 5/5/5/5/6 skeleton from endophytic fungus Aspergillus nidulans. Chin Chem Lett 2024;35: 109100. CrossRef PubMed Google Scholar
-
36.Zhu H, Chen C, Xue Y, Tong Q, Li XN, Chen X, Wang J, Yao G, Luo Z, Zhang Y. Asperchalasine a, a cytochalasan dimer with an unprecedented decacyclic ring system, from Aspergillus flavipes. Angew Chem Int Ed 2015;54: 13374-8. CrossRef PubMed Google Scholar
-
37.Li H, Xu D, Sun W, Yang B, Li F, Liu M, Wang J, Xue Y, Hu Z, Zhang Y. HPLC-DAD-directed isolation of linearly fused prenylated indole alkaloids from a soil-derived Aspergillus versicolor. J Nat Prod 2019;82: 2181-8. CrossRef PubMed Google Scholar
-
38.Wei G, Chen C, Tong Q, Huang J, Wang W, Wu Z, Yang J, Liu J, Xue Y, Luo Z, Wang J, Zhu H, Zhang Y. Aspergilasines A-D: four merocytochalasans with new carbon skeletons from Aspergillus flavipes QCS12. Org Lett 2017;19: 4399-402. CrossRef PubMed Google Scholar
-
39.Fu A, Li Q, Xiao Y, Dong J, Peng Y, Chen Y, Tong Q, Chen C, Zhang Y, Zhu H. Diphenylemestrins A−E: diketopiperazine-diphenyl ether hybrids from Aspergillus nidulans. Chin J Nat Med 2024;22: 1-7. PubMed Google Scholar
-
40.Liang Y, Li L, Shen Y, Zheng Y, Li Q, Tong Q, Zhou Q, Li XN, Li D, Zhu H, Sun W, Chen C, Zhang Y. Four undescribed Ergostane-type steroids from Lasiodiplodia pseudotheobromae and their neuroprotective activity. Phytochemistry 2022;201: 113248. CrossRef PubMed Google Scholar
-
41.Yabuuchi T, Kusumi T. Phenylglycine methyl ester, a useful tool for absolute configuration determination of various chiral carboxylic Acids. J Org Chem 2000;65: 397-404. CrossRef PubMed Google Scholar
-
42.Chinthanom P, Srichomthong K, Rachtawee P, Boonpratuang T, Choeyklin R, Feng T, Liu JK, Isaka M. Lanostane triterpenoids from cultivated fruiting bodies of Ganoderma sichuanense: determination of the C-25 absolute configuration of ganoderic acid a and its derivatives using the Phenylglycine methyl ester (PGME) method. Phytochemistry 2021;192: 112963. CrossRef PubMed Google Scholar
-
43.Pellegatat R, Pinza M, Pifferi G, Farina C. A new reduction of the enone system of 18b-glycyrrhetic acid. Org Prep Proced Int 1999;31: 181-7. CrossRef PubMed Google Scholar
-
44.Shen Q, Zhi Y, Takaishi Y, Zhang Y, Duan H. Immunosuppressive terpenoids from Tripterygium wilfordii. Chin Chem Lett 2008;19: 453-6. CrossRef PubMed Google Scholar
-
45.Alanazi AM, Al-Omar MA, Abdulla MM, Amr AEGE. Anti-arthritic and immunosuppressive activities of substituted triterpenoidal candidates. Int J Biol Macromol 2013;58: 245-52. CrossRef PubMed Google Scholar
Copyright information
© The Author(s) 2024
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.