Six New 3, 5-Dimethylcoumarins from Chelonopsis praecox, Chelonopsis odontochila and Chelonopsis pseudobracteata
Abstract
Ten 3, 5-dimethylcoumarins (1–6 and 8‒11) involving six new ones (1–6), together with a known 3-methylcoumarin (7), were isolated from the aerial parts of three Chelonopsis plants, C. praecox, C. odontochila, and C. pseudobracteata. The structures of the new compounds were determined by extensive HRESIMS, 1D and 2D NMR spectroscopic analyses. According to the substitution at C-5, these coumarins were classified into 5-methyl, 5-hydroxymethyl, 5-formyl, and 5-nor types. All the isolates were assayed for their inhibition on α-glucosidase, protein tyrosine phosphatase 1B, and T-cell protein tyrosine phosphatase in vitro.Graphical Abstract
Keywords
3, 5-Dimethylcoumarins 3-Methylcoumarin Chelonopsis Enzyme inhibition1 Introduction
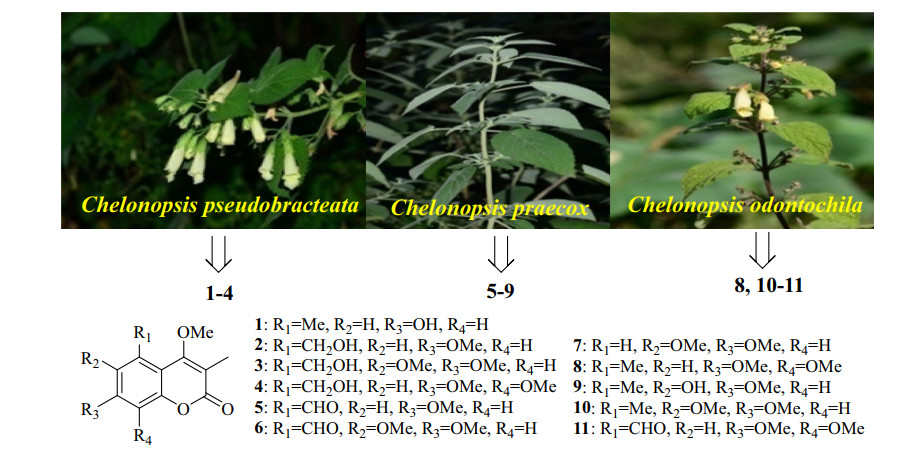
Coumarins with the benzo-α-pyrone core are widely distributed in plant kingdom and show a wide range of biological activities, including antimicrobial, antiviral, antidiabetic antiinflammatory, and antihypertensive activities, etc. [1]. Structurally, coumarins can be divided into simple coumarins, C-substituted coumarins, miscellaneous coumarins, biscoumarins, and triscoumarins. Besides hydroxy and methoxy groups, isopentenyl related C5-groups are the most common substituents present in coumarins, which are generally located at C-3, C-6, or C-8 positions by C–C linkage [2-4]. The methyl substituent in coumarin is very unusual, and only limited coumarins with the methylation at C-3, C-5, or C-6 positions have been reported. Currently, tens of 3, 5-dimethylcoumarins have been isolated from Clutia lanceolata [5], Clutia abyssinica [6], Juniperus sabina [7], Leucas inflata [8], and Sideritis pullulans [9], but never from Chelonopsis plants. Our previous investigation on Chelonopsis plants yielded a series of diterpenoids with α-glucosidase inhibitory activity, i.e., ten ent-kauranes from C. praecox [10], and 13 ent-labdanes and 11 ent-kauranes from C. odontochila [11]. As a continuous search for antidiabetic candidates from natural sources [12-16], ten 3, 5-dimethylcoumarins (1–6 and 8‒11) involving six new ones and one known 3-methylcoumarin (7) were first isolated from three Chelonopsis plants (Fig. 1). Herein, we report their isolation, structural elucidation, and enzymatic inhibition on α-glucosidase, protein tyrosine phosphatase 1B (PTP1B), and T-cell protein tyrosine phosphatase (TCPTP).
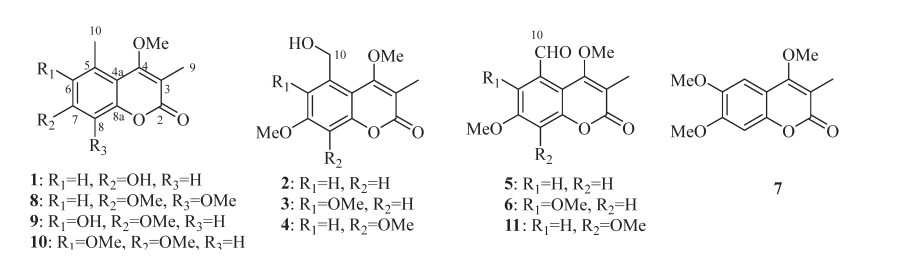
Chemical structures of compounds 1–11
2 Results and Discussion
2.1 Structural Elucidation
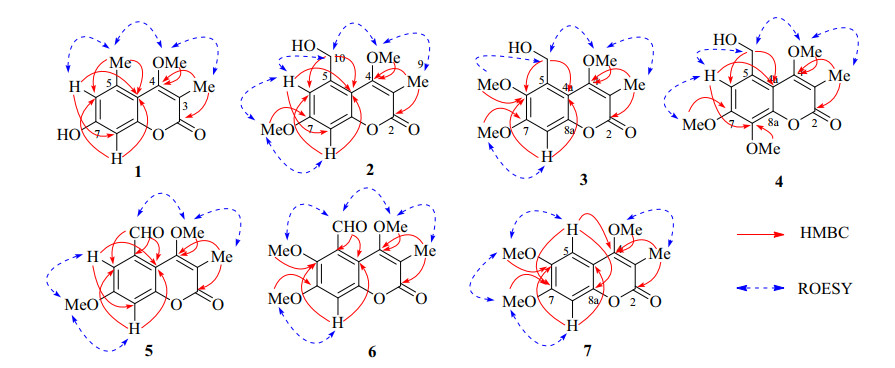
Compound 1 had a chemical composition of C12H12O4 deduced by the [M + H]+ ion at m/z 221.0809, accounting for seven indices of hydrogen deficiency. The UV spectrum showed characteristic absorption at λmax 321 nm for coumarins. The IR absorptions at 3204, 1681, 1610, and 1454 cm-1 were indicative for the presence of hydroxyl, carbonyl, and aromatic functionalities. In the 1H NMR spectrum, two meta-coupled aromatic protons at δH 6.93 (J = 2.4 Hz) and 6.87 (J = 2.4 Hz), one methoxy at δH 3.64, and two singlet methyls at δH 2.56 and 2.10 were well recognized (Table 1). The 13C NMR spectrum displayed 12 carbons comprising one carbonyl carbon, eight olefinic carbons, one methoxy, and two methyls. The 1H and 13C NMR data of 1 showed high resemblance with 6-hydroxy-3, 5-dimethyl-4, 7-dimethoxycoumarin (9) [5] except for the absence of a methoxy and an oxygenated methine in 9 being changed to be a methine in 1. In the HMBC spectrum, the correlations from the methyl (δH 2.10) to C-2 (δC 164.0) and C-4 (δC 166.9), and from the methoxy (δH 3.64) to C-4 (δC 166.9) affirmed the 3-methyl and 4-methoxy substitution. Taking the ROESY correlations of Me-3/OMe-4/Me-5/H-6 into consideration, this compound was characterized to be 7-hydroxy-4-methoxy-3, 5-dimethylcoumarin (1).
1H NMR data of compounds 1–7 (δ in ppm, J in Hz)
The molecular formula of 2 was assigned to be C13H14O5 by the positive HRESIMS ion at m/z 251.0897 ([M + H]+, calcd. for 251.0914). By comparing its 1H and 13C NMR data with those of 1, the 5-methyl in 1 was changed to be a hydroxymethyl (δH 4.89, δC 56.2) in 2, as well as an additional methoxyl group. The substitution of 5-hydroxymethyl and 7-methoxyl was confirmed by the HMBC correlations from H-10 (δH 4.89) to C-6 and C-4a, and from OMe-7 (δH 3.86) to C-7, as well as the ROESY correlations of H3-9/OMe-4/H2-10/H-6/OMe-7/H-8 (Fig. 2). Hence, compound 2 was defined as 5-hydroxymethyl-4, 7-dimethoxy-3-methylcoumarin.
Selected 2D NMR correlations of compounds 1–7
Compounds 3 and 4 were a pair of isomers with the same molecular formula of C14H16O6, indicating an additional CH2O moiety than 2. In their 1H and 13C NMR spectra, three methoxy groups (δH 3.86, 3.93, 4.01 and δC 56.2, 61.9, 62.1 for 3; δH 3.95, 3.97, 3.98 and δC 56.5, 61.67, 61.70 for 4) were obviously recognized (Table 2), suggesting compounds 3 and 4 should be the methoxylated derivatives of 2. The position of the additional methoxy in 3 and 4 were unambiguously determined by analyzing their ROESY experiments. In the ROESY spectrum of 3, the correlation peaks of H3-9/OMe-4/H2-10/OMe-6 and OMe-7/H-8 revealed the methoxy at C-6 position. Similarly, the ROESY signals of H3-9/OMe-4/H2-10/H-6/OMe-7 in 4 supported the methoxy at C-8 position. Thus, compounds 3 and 4 were concluded as 5-hydroxymethyl-4, 6, 7-trimethoxy-3-methylcoumarin (3) and 5-hydroxymethyl-4, 7, 8-trimethoxy-3-methylcoumarin (4), respectively.
13C NMR data of compounds 1–7 (δ in ppm, J in Hz)
Compound 5 had a chemical composition of C13H12O5 according to the protonated ion at m/z 249.0744 in the HRESIMS data. In the 1H NMR spectrum, the presence of a formyl group at δH 10.74, two meta-coupled aromatic protons at δH 7.24 (J = 2.7 Hz) and 6.99 (J = 2.7 Hz), two methoxys at δH 3.91 and 3.89, and a methyl at δH 2.19 were easily recognized. By comparing with 2, compound 5 had an additional formyl group at δH 10.74 and δC 192.3, but with the absence of a hydroxymethyl group (δH 4.89 and δC 64.6), indicating the dehydrogenated derivative of 2. The formyl group was assigned at C-5 by the ROESY correlations of H3-9/OMe-4/H-10, and HMBC correlation from H-6 to C-10 and from H-10 to C-5, C-6 and C-4a. Consequently, compound 5 was defined as 5-formyl-4, 7-dimethoxy-3-methylcoumarin.
The molecular formula of 6 was assigned as C14H14O6 by the [M + H]+ ion at m/z 279.0906 in positive HRESIMS spectrum. In the 1H NMR spectrum, one formal at δH 10.44, one aromatic singlet at δH 6.89, three methoxy groups at δH 3.81, 3.82, and 3.93, and one methyl group at δH 2.15 were observed, showing an extra methoxy than 5. The above deduction was consistent with that two meta-coupled protons at δH 7.24 and 6.99 in 5 was changed to be an aromatic singlet at δH 6.89 in 6. By analyzing the ROESY experiment, the correlations of H3-9/OMe-4/H-10/OMe-6 and OMe-7/H-8 demonstrated the structure of 5-formyl-4, 6, 7-trimethoxy-3-methylcoumarin (6).
Compound 7 was assigned with the chemical formula of C13H14O5 by the [M + H]+ ion at m/z 251.0898 in positive HRESIMS spectrum. In the 1H NMR spectrum, two aromatic protons at δH 7.07 and 6.85, three methoxy groups at δH 4.01, 3.95, and 3.94, and a methyl group at δH 2.17, were recognized. Compared with 4, 6, 7-trimethoxy-3, 5-dimethylcoumarin (10) [6], the 5-methyl in 10 was absent in 7 but with an extra aromatic singlet at δH 7.07. This proton (δH 7.07) was assigned to be H-5 by the HMBC correlation from H-5 to C-4, and ROESY correlations of H-5/OMe-6 and H-8/OMe-7. Thus, compound 7 was deduced as 4, 6, 7-trimethoxy-3-methylcoumarin, the demethylated derivative of 10. Although this compound has been synthesized by methylation of 4-hydroxy-6, 7-dimethoxy-3-methylcoumarin in 1949 [17], it is the first report of its natural occurrence and NMR spectroscopic data.
The known coumarins were determined to be 4, 7, 8-trimethoxy-3, 5-dimethylcoumarin (8) [7], 6-hydroxy-4, 7-dimethoxy-3, 5-dimethylcoumarin (9) [5], 4, 6, 7-trimethoxy-3, 5-dimethylcoumarin (10) [6], and 5-formyl-4, 7, 8-trimethoxy-3-methylcoumarin (11) [8] by comparing their 1H and 13C NMR data with those in the literatures.
In order to evaluate their antidiabetic potency, all the coumarins were assayed for their inhibitory activity on α-glucosidase, PTP1B, and TCPTP. As shown in Table 3, all the compounds showed only weak or no inhibition to three enzymes at the concentration of 200 μM. According to the previous study [5], this type of coumarins could enhance the glucose-triggered secretion of insulin from murine islets. Thus, further studies will be needed to reveal their targets and mechanisms in exerting hypoglycemic effects.
Inhibitory rates of the isolates (200 μM) on α-glucosidase, PTP1B, and TCPTP
3 Experimental Section
3.1 General Experimental Procedures
A Jasco model 1020 digital polarimeter (Jasco Corp., Tokyo, Japan) was used to measure optical rotations. UV and IR data were obtained using a Shimadzu UV2401PC spectrophotometer (Shimadzu, Kyoto, Japan) and a Nicolet iS10 spectrometer (Thermo Fisher Scientific, Madison, WI, USA), respectively. A Waters AutoSpec Premier P776 mass spectrometer (Waters, Milford, MA, USA) or a Shimadzu LCMS-IT-TOF mass spectrometer (Shimadzu, Kyoto, Japan) was used to acquire the high-resolution mass spectra. ECD spectra were recorded on an Applied Photophysics Chirascan apparatus (Applied Photophysics, Surrey, UK). NMR spectra were obtained by using DRX-500, Avance III-600, and Ascend™ 800 MHz spectrometers (Bruker, Karlsruhe, Germany). TLC detection was run on silica gel plates (60 F254). Silica gel (200–300 mesh, Qingdao Makall group Co. Ltd., Qingdao, China) and Sephadex LH-20 (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) were used for column chromatography. A Dr-Flash II apparatus was applied to accomplish the MPLC separations. HPLC purifications were conducted on a Shimadzu LC-CBM-20 system (Shimadzu, Kyoto, Japan), equipped with an Agilent Eclipse XDB-C18 column (5 μm, 9.4 × 250 mm).
3.2 Plant Materials
The aerial parts of three Chelonopsis plants were collected in October 2016 from Lijiang, Yunnan Province of China, which were authenticated to be Chelonopsis odontochila Diels, Chelonopsis pseudobracteata C. Y. Wu et H. W. Li, and Chelonopsis praecox Weckerle and F. Huber by Dr. Chun-Lei Xiang. Voucher specimens (Nos. 2016102101, 2016102102, 2016102103) were deposited in the Laboratory of Anti-virus and Natural Medicinal Chemistry, Kunming Institute of Botany, Chinese Academy of Sciences, China.
3.3 Extraction and Isolation
The air-dried plants of C. pseudobracteata (6 kg) were powdered and extracted three times with 90% aqueous EtOH (25 L × 3) at room temperature. The extract was evaporated under reduced pressure, and the residue was suspended in H2O and partitioned with CHCl3. The CHCl3 extraction (95 g) was subjected to silica gel column chromatography (Si CC) and eluted with an acetone-petroleum ether solvent system (from 10:90 to 50:50, v/v) to afford seven fractions (A–G). Fraction C (19.8 g) was subjected over MCI gel CHP 20P column (H2O–MeOH, 50:50–0:100) to provide five fractions, Frs. C1–C5. Fr. C3 (3.4 g) was purified via Si CC (EtOAc-petroleum ether, 10:90–50:50) to give three fractions, Frs. C3-1–C3-3. Fr. C3-1 (600 mg) was purified by Sephadex LH-20 CC (MeOH–CHCl3, 50:50) and semi-preparative HPLC (H2O–MeCN, 36:64) to give compounds 1 (16 mg) and 3 (18 mg). Compounds 2 (25 mg) and 4 (38 mg) were obtained from Fr. C3-2 (750 mg) by Sephadex LH-20 CC (MeOH–CHCl3, 50:50) and semi-preparative HPLC (H2O–MeCN, 50:50).
The air-dried plants of C. praecox (25 kg) were powdered and extracted three times with 90% aqueous EtOH (100 L × 3) at room temperature. The combined EtOH extract was concentrated and partitioned between H2O and CHCl3. The CHCl3 extraction (380 g) was subjected to Si CC (4.0 kg, 30 × 100 cm), using a gradient elution of EtOAc-petroleum ether (from 10:90 to 100:0) to afford seven fractions (A–G). MPLC separation of Fr. D (46 g) by using a CHP20P MCI gel column (H2O-MeOH, from 50:50 to 0:100) provided five fractions, Frs. D1–D5. Fr. D3 (1.3 g) was separated by Si CC (EtOAc-CHCl3, 2:98–50:50) to afford five fractions, Frs. D3-1–D3-5. Fr. D3-1 (265 mg) was purified by Si CC (acetone-petroleum ether, 5:95), and semi-preparative HPLC (H2O–MeCN, 42:58) to yield compounds 8 (25 mg), 5 (18 mg), and 6 (18 mg). Fr. E (17 g) was subjected to MPLC to give five fractions, Frs. E1–E5. Compounds 9 (5 mg) and 7 (25 mg) were obtained from Fr. E3 after repeated Si CC (acetone-petroleum ether, 10:90) and Sephadex LH-20 (MeOH–CHCl3, 50:50), and semi-preparative HPLC (MeCN–H2O, 40:60).
Air-dried and powdered plants of C. odontochila (8.0 kg) were extracted with 90% aqueous EtOH (35 L × 3) at room temperature. The combined EtOH extract was concentrated and partitioned between H2O and CHCl3. The CHCl3 extract (160 g) was chromatographed on a silica gel column (1.3 kg, 30 × 100 cm), and eluted with acetone-petroleum ether gradient (from 0:100 to 100:0) to yielded seven fractions, Frs. A–G. MPLC separation of Fr. C (15 g) with MCI gel CHP 20P column (H2O–MeOH, from 50:50 to 0:100) gave rise to five fractions, Frs. C1–C5. Fraction C3 (2.8 g) was chromatographed on a silica gel column (acetone-CHCl3, from 10:90 to 100:0) to provide four fractions, Frs. C3-1–C3-4. Compound 11 (12 mg) was purified from Fr. C3-2 (500 mg) by Sephadex LH-20 CC (MeOH–CHCl3, 50:50), and semi-preparative HPLC (H2O–MeCN, 64:36). After repeated separation over Si CC (acetone-petroleum ether, 15:85), Sephadex LH-20 CC (MeOH–CHCl3, 50:50), and semi-preparative HPLC (H2O–MeCN, 60:40), compounds 10 (27 mg) and 8 (21 mg) were obtained from Fr. C3-3 (470 mg).
3.4 Spectroscopic Data of Compounds
3.4.1 7-Hydroxy-4-Methoxy-3, 5-Dimethylcoumarin (1)
White amorphous powder; UV (MeOH) λmax (log ε): 321 (3.99) nm; IR (KBr) νmax: 3204, 1681, 1610, 1567, 1454, 1378, 1358, 1344, 1257, 1154, 1102, 1074, 1016 cm-1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 221.0809 [M + H]+ (calcd. for C12H13O4, 221.0808).
3.4.2 5-Hydroxymethyl-4, 7-Dimethoxy-3-Methylcoumarin (2)
White amorphous powder; UV (MeOH) λmax (log ε): 222 (3.91), 320 (3.90) nm; IR (KBr) νmax: 3398, 1663, 1616, 1592, 1558, 1453, 1432, 1369, 1336, 1251, 1197, 1151, 1083, 1047, 1012, 946 cm-1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 251.0897 [M + H]+ (calcd. for C13H15O5, 251.0914).
3.4.3 5-Hydroxymethyl-4, 6, 7-Trimethoxy-3-Methylcoumarin (3)
White amorphous powder; UV (MeOH) λmax (log ε): 294 (3.75), 328 (4.00) nm; IR (KBr) νmax: 3434, 1683, 1603, 1562, 1452, 1418, 1366, 1331, 1262, 1224, 1161, 1130, 1081, 1061, 1004, 987 cm-1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 281.1039 [M + H]+ (calcd. for C14H17O6, 281.1020).
3.3.4 5-Hydroxymethyl-4, 7, 8-Trimethoxy-3-Methylcoumarin (4)
White amorphous powder; UV (MeOH) λmax (log ε): 318 (4.07) nm; IR (KBr) νmax: 3435, 1687, 1595, 1571, 1457, 1421, 1337, 1274, 1136, 1096, 1052 1012 cm-1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 281.1014 [M + H]+ (calcd. for C14H17O6, 281.1020).
3.4.5 5-Formyl-4, 7-Dimethoxy-3-Methylcoumarin (5)
White amorphous powder; UV (MeOH) λmax (log ε): 218 (3.09), 291 (2.74), 328 (2.83) nm; IR (KBr) νmax: 3426, 1725, 1688, 1605, 1448, 1367, 1336, 1258, 1168, 1089, 953 cm-1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 249.0744 [M + H]+ (calcd. for C13H13O5, 249.0758).
3.4.6 5-Formyl-4, 6, 7-Trimethoxy-3-Methylcoumarin (6)
White amorphous powder; UV (MeOH) λmax (log ε): 207 (3.67), 224 (3.63), 290 (3.20), 329 (3.45) nm; IR (KBr) νmax: 3420, 1721, 1693, 1603, 1455, 1388, 1369, 1266, 1226, 1078, 1006, 959 cm-1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 279.0906 [M + H]+ (calcd. for C14H15O6, 279.0863).
3.4.7 4, 6, 7-Trimethoxy-3-Methylcoumarin (7)
Colorless gum; UV (MeOH) λmax (log ε): 208 (2.99), 222 (2.83), 287 (2.30), 333 (2.58) nm; IR (KBr) νmax: 3431, 1709, 1620, 1580, 1453, 1372, 1337, 1249, 1215, 1162, 1025, 994 cm-1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 251.0898 [M + H]+ (calcd. for C13H15O5, 251.0914).
3.5 In Vitro Enzyme Inhibition Assays
In this study, three enzymes closely related to diabetes, namely α-glucosidase, PTP1B, and TCPTP, were applied to assess the antidiabetic potency of compounds. Enzyme inhibition was assayed in accordance with the previous reports [18, 19]. Acarbose (for α-glucosidase) and Na3VO4 (for PTP1B and TCPTP) were used as the positive controls.
4 Supporting Information
1D and 2D NMR, HRMS, UV and IR spectra of compounds 1-7.
Notes
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.1007/s13659-021-00318-9.
Acknowledgements
This work was supported by the Yunnan Wanren Project (YNWR-QNBJ-2018-061), the Natural Science Foundation of Yunnan Province (2019FI017), and the Reserve Talents of Young and Middle-Aged Academic and Technical Leaders in Yunnan Province.
Declarations
Conflict of interest
The authors declare that they have no competing financial interest.
References
-
1.M.I. Hussain, Q.A. Syed, M.N.K. Khattak, B. Hafez, M.J. Rrigosa, A. El-Keblawy, Biologia 74, 863-888 (2019) PubMed Google Scholar
-
2.A.G. Estbvez-Braun, Nat. Prod. Rep. 14, 465-475 (1997) CrossRef PubMed Google Scholar
-
3.R.D.H. Murray, Nat. Prod. Rep. 12, 477-505 (1995) CrossRef PubMed Google Scholar
-
4.R.D.H. Murray, Nat. Prod. Rep. 6, 591-624 (1989) CrossRef PubMed Google Scholar
-
5.S. Ahmed, M. Nur-e-Alam, I. Parveen, S.J. Coles, R.M. Hafizur, A. Hameed, J.B. Orton, M.D. Threadgill, M. Yousaf, A.M. Alqahtani, A.J. Al-Rehaily, Phytochemistry 170, 112213 (2020) CrossRef PubMed Google Scholar
-
6.R.D. Waigh, B.M. Zerihun, D.J. Maitland, Phytochemistry 30, 333-335 (1991) CrossRef PubMed Google Scholar
-
7.J. de Pascual, A. San Feliciano, J.M. Miguel del Corral, A.F. Barrero, M. Rubio, L. Muriel, Phytochemistry 20, 2778-2779 (1981) CrossRef PubMed Google Scholar
-
8.M.H. Al Yousuf, A.K. Bashir, G. Blunden, M.-H. Yang, A.V. Patel, Phytochemistry 51, 95-98 (1999) CrossRef PubMed Google Scholar
-
9.L. Faiella, F. Dal Piaz, A. Bader, A. Braca, Phytochemistry 106, 164-170 (2014) CrossRef PubMed Google Scholar
-
10.Z.T. Deng, C.A. Geng, T.H. Yang, C.L. Xiang, J.J. Chen, Fitoterapia 132, 60-67 (2019) CrossRef PubMed Google Scholar
-
11.Z.T. Deng, J.J. Chen, C.A. Geng, Bioorg. Chem. 95, 103571 (2020) CrossRef PubMed Google Scholar
-
12.X.F. He, C.A. Geng, X.Y. Huang, Y.B. Ma, X.M. Zhang, J.J. Chen, Nat. Prod. Bioprospect. 9, 223-229 (2019) CrossRef PubMed Google Scholar
-
13.X.F. He, J.J. Chen, T.Z. Li, J. Hu, X.Y. Huang, X.M. Zhang, Y.Q. Guo, C.A. Geng, Chin. J. Chem. 39, 3051-3063 (2021) PubMed Google Scholar
-
14.X.F. He, J.J. Chen, T.Z. Li, J. Hu, X.M. Zhang, C.A. Geng, Bioorg. Chem. 108, 104683 (2021) PubMed Google Scholar
-
15.X.F. He, H.M. Wang, C.A. Geng, J. Hu, X.M. Zhang, Y.Q. Guo, J.J. Chen, Phytochemistry 177, 112418 (2020) CrossRef PubMed Google Scholar
-
16.Q. Huang, J.J. Chen, Y. Pan, X.F. He, Y. Wang, X.M. Zhang, C.A. Geng, J. Pharm. Biomed. Anal. 198, 113998 (2021) PubMed Google Scholar
-
17.G.H. Jones, J.B.D. Mackenzie, A. Robertson, W.B. Whalley, J. Chem. Soc. 0, 562-569 (1949) PubMed Google Scholar
-
18.D.X. Yan, C.A. Geng, T.H. Yang, X.Y. Huang, T.Z. Li, Z. Gao, Y.B. Ma, H. Peng, X.M. Zhang, J.J. Chen, Fitoterapia 128, 57-65 (2018) PubMed Google Scholar
-
19.C.C. Zhang, C.A. Geng, X.Y. Huang, X.M. Zhang, J.J. Chen, J. Agric. Food Chem. 67, 6765-6772 (2019) PubMed Google Scholar
Copyright information
© The Author(s) 2021
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.