Advances in Research on Chemical Constituents and Their Biological Activities of the Genus Actinidia
Abstract
Kiwi, a fruit from plants of the genus Actinidia, is one of the famous fruits with thousand years of edible history. In the past twenty years, a great deal of research has been done on the chemical constituents of the Actinidia species. A large number of secondary metabolites including triterpenoids, flavonoids, phenols, etc. have been identified from differents parts of Actinidia plants, which exhibited significant in vitro and in vivo pharmacological activities including anticancer, anti-inflammatory, neuroprotective, anti-oxidative, anti-bacterial, and anti-diabetic activities. In order to fully understand the chemical components and biological activities of Actinidia plants, and to improve their further research, development and utilization, this review summarizes the compounds extracted from different parts of Actinidia plants since 1959 to 2020, classifies the types of constituents, reports on the pharmacological activities of relative compounds and medicinal potentials.Keywords
Actinidia chemical constituents Isolation Biological activities1 Introduction
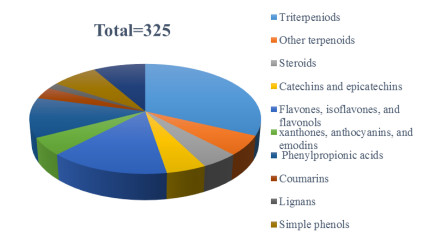
With the development of natural product research, a huge number of chemical constituents have been identified from natural resources. There is no doubt that the research on the chemical composition of fruits, including trace elements, has greatly improved the application prospects of these fruits. With no exception, it is the same to kiwifruit, one of the most prestigious fruits with a long history of eating [1, 2]. Kiwi belongs to plants of the genus Actinidia comprising more than 70 species around the world [3]. Some of these plants are proven to have a wide range of medicinal activities. For example, A. valvata, whose root is known as ''Mao-Ren-Shen" in traditional Chinese medicine, exhibits antitumor and anti-inflammatory activities and has been used for the treatment of hepatoma, lung carcinoma and myeloma for a long time [4, 5]. The roots of A. chinensis Planch, called "Teng-Li-Gen" usually, were used as a traditional Chinese medicine for the treatment of various cancers, such as esophagus cancer, liver cancer, and gastric cancer [6]. In the past two decades, great research had been accomplished about exploring the chemical composition of Actinidia plants. These studies have greatly promoted the understanding of the chemical components and functions of the Actinidia plant. According to literature survey, 12 Actinidia species including A. valvata, A. chinensis, A. argute, A. polygama, A. kolomikta, A. eriantha, A. macrosperma, A. deliciosa, A. chrysantha, A. rufa, A. indochinensis, and A. valvata were reported for their natural products. This review systematically summarizes the chemical components and their biological activities from different parts of 12 Actinidia species from 1959 to 2020. According to structure types, a total of 325 molecules have been collected including terpeniods, phenols, and other small groups (Fig. 1). Names and isolation information were listed in the tables, while the biological activities of the extracts or compounds were discussed in the text.
Constituents proportion of 12 Actinidia plants
2 Chemical Constituents
2.1 Terpenoids
In recent years, a large number of terpenoids were isolated from many Actinidia species. Among them, triterpenes account for the vast majority that are mainly composed by several normal frameworks including ursane-type, oleanane-type, and lupane-type. Of the total 325 compounds in this review, 104 are triterpenoids. From the literature review, ursolic acids and their saponins are undoubtedly the most abundant in Actinidia species.
2.1.1 Ursane Triterpenoids
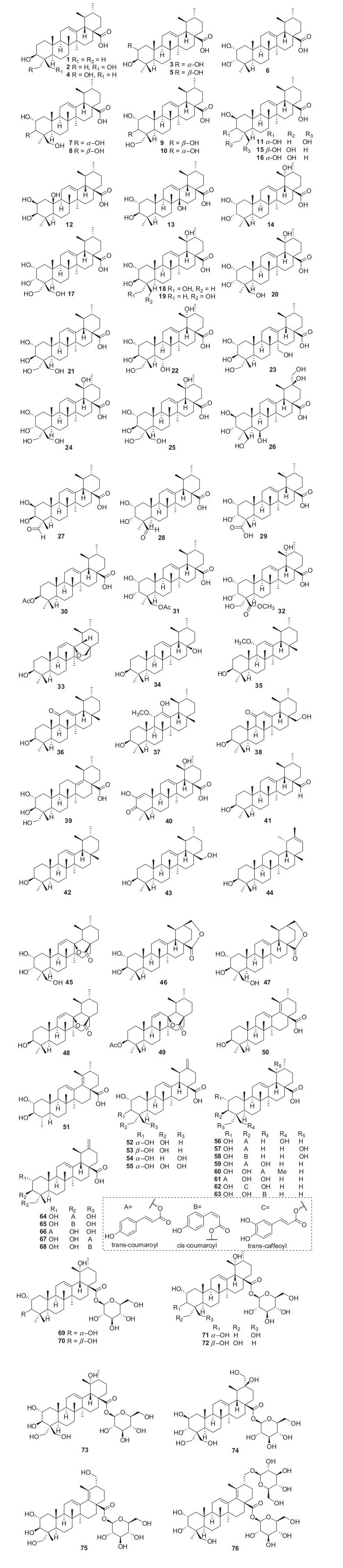
Ursane-type triterpenes are characterized of ursolic acid and its saponins, possessing a 6/6/6/6/6-fused carbon skeleton. A total of 76 ursane-type triterpenoids (1–76) have been identified from plants of the genus Actinidia (Fig. 2, Table 1). Ursolic acid (3β-Hydroxyurs-12-en-28-oic acid 1) [7], is one of the most frequently obtained compound in many kinds of kiwifruit plants with unique flavor. Great attention had been paid on biological activities about ursolic acid, attracting much interest in recent years. Ursolic acid exhibits different pharmacological activities, including anti-cancer, amylolytic enzyme inhibitors, cytotoxicity, downregulating thymic stromal lymphopoietin and others [7-11].
Structures of ursane triterpenoids 1‒76 from Actinidia plants
Information of ursane triterpenoids from Actinidia plants
Compounds 2‒7 are ursane triterpenoids featuring with two hydroxyl groups. Compound 3 (2α-Hydroxyursolic acid) was tested for its antiproliferative activity and cytotoxicity in MDA-MB-231 human breast cancer cells through the methylene blue assay. It significantly down-regulated expressions of TRAF2, PCNA, cyclin D1, and CDK4 and up-regulated the expressions of p-ASK1, p-p38, p-p53, and p-21. Furthermore, it induced apoptosis in MDA-MB-231 cell by significantly increasing the Bax/Bcl-2 ratio and inducing the cleaved caspase-3 [12]. Compound 4 exhibited inhibitory activity on pancreatic lipase with an IC50 value of 20.42 ± 0.95 μM [13]. It also showed cytotoxicity to human lung adenocarcinoma (A549), ovarian cancer (SK-OV-3), skin melanoma (SK-MEL-2), and colon cancer (HCT-15) cell lines with IC50 values ranging from 11.96 to 14.11 μM [14-17].
Compounds 7‒14 are trihydroxy-ursolic acid derivatives. In early 1992, Sashida et al. reported the isolation of 8‒10. Compounds 8 and 9 were evaluated for their cytotoxicity against A549 cells, LOVO cells, and HepG2 cells with IC50 values of 32.9, 31.6, 35.7 μg/mL respectively for 8 and 34.6, 13.9, 34.5 μg/mL respectively for 9 [18, 19]. Compounds 17‒26 are ursolic acids with four or five hydroxy groups. Xu et al. reported 17 from roots of A. valvata, this compound exhibited weak cytotoxicity against A549, LOVO and HepG2 cell lines with IC50 values of above 100 μg/mL [20]. 2α, 3α, 19α, 24-Tetrahydroxyurs-12-en-28-oic acid 20 was separated from the leaves of A. valvata which showed cytotoxicity against PLC, Hep3B, HepG2, HeLa, SW480, MCF-7 and Bel7402 in vitro [21]. A new polyoxygenated triterpenoid (2β, 3α, 6α)-2, 3, 6, 20, 23, 30-hexahydroxyurs-12-en-28-oic acid 26 was obtained from the roots of A. valvata DUNN, it exhibited moderate cytotoxic activity against BEL-7402 and SMMC-7721 tumor cell lines in vitro [22].
Compound 30 (3β-O-acetylursolic acid) was isolated from the fruit galls of A. polygama and the structure was elucidated on the basis of chemical and spectral evidence. It was reported to be a mixed-type protein tyrosine phosphatase 1B (PTP1B) inhibitor with an IC50 value of 4.8 ± 0.5 μΜ [23, 24]. Isolation of the antiviral active ingredient of A. chinensis root bark gave fupenzic acid 40, which showed moderate inactivity under the concentration of 100 μg/mL [25]. Callus tissue from the stems of A. arguta (Actinidiaceae) produced three ursane-type triterpenes including ursolaldehyde 41, α-amyrin 42, and uvaol 43 [26]. Of them, compound 43 showed anti-inflammatory, anticancer, and wound healing activities [27-29]. Anti-inflammatory properties of 43 on DSS-induced colitis and LPS-stimulated macrophages have been explored detailly and completely. It showed excellent potential of NO production inhibition. It could attenuate disease activity index (DAI), colon shortening, colon injury, and colonic myeloperoxidase activity in DSS-induced colitis mice. What's more, studies on LPS challenged murine macrophage RAW246.7 cells also revealed that uvaol reduces mRNA expression and production of pro-inflammatory cytokines and mediators. These results indicating that uvaol is a prospective anti-inflammatory agent for colonic inflammation [27]. Guided by the hepatoprotective activity, the phytochemical study on the roots of A. chinensis led to the isolation of two new compounds 2α, 3β-dihydroxyurs-12-en-28, 30-olide 46, 2α, 3β, 24-trihydroxyurs-12-en-28, 30-olide 47 and 3β-hydroxyurs-12, 18-dien-28-oic acid 50 [30]. Compounds 52‒54 showed antifungal activity against C. musae at 3 μg/mL [31]. A new compound 2α, 3α, 23, 24 -tetrahydroxyursa-12, 20(30)-dien-28-oic acid 55 was isolated from the roots of A. chinensis Planch. It exhibited moderate antitumor activities against five human cancer cell lines (HepG2, A549, MCF-7, SK-OV-3, and HeLa) with IC50 values of 19.62 ± 0.81, 18.86 ± 1.56, 45.94 ± 3.62, 62.41 ± 2.29, and 28.74 ± 1.07 μM, respectively [32].
Compounds 59‒63 are actinidic acid derivatives with a phenylpropanoid unit that were identified as 3-O-trans-p-coumaroylasiatic acid 59, 23-O-trans-p-coumaroylasiatic acid 60, actiniargupene E 61, actiniargupene F 62, and actiniargupene G 63 from the leaves of A. arguta. All the compounds showed inhibitory effects on α-glucosidase activity. Among them compound 59 showed most potentially inhibitory activity on α-glucosidase with an IC50 of 81.3 ± 2.7 μM, equal to that of the positive control (acarbose, 72.8 ± 3.1 μM) [33]. The structure–activity relationship suggested that triterpenoids with a phenylpropanoid moiety exhibited more potent effects than those without such a unit [34]. Compound 71 showed potent cytotoxic activity against human SKVO3 and TPC-1 cancer cell lines with IC50 values of 10.99 and 14.34 μM, respectively [19, 35]. Compound 74 exhibited moderate cytotoxic activity against BEL-7402 and SMMC-7721 tumor cell lines [22]. Compounds 75 and 76 were isolated from roots of A. valvata Dunn. They exhibited moderate cytotoxic activity in vitro against BEL-7402 and SMMC-7721 tumor cell line [36].
2.1.2 Oleanane Triterpenoids
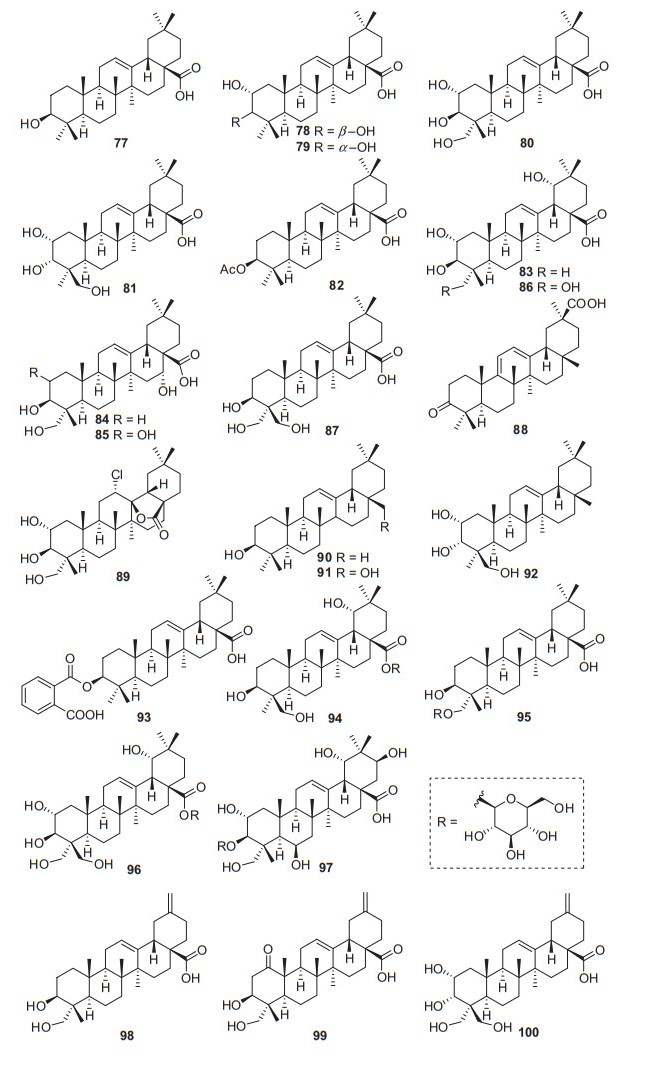
Oleanane-type triterpenoids also possessed a 6/6/6/6/6 pentacyclic carbon skeleton. Unlike ursane triterpenes, oleanane-type triterpenoids have two methyl groups at the C-20 position instead of each one at the C-19 and C-20, respectively. So far, a total of 24 oleanane-type triterpenoids have been identified from Actinidia plants (77‒100, Fig. 3, Table 2). The most representative compound is oleanolic acid 77. It was found from callus tissue from the stems of A. arguta, together with 2α, 3β-dihydroxyolean-12-en-28-oic acid 78 [26]. Oleanolic acid 77 is abundant in nature and exhibits a wide range of biological activities including anti-inflammatory [54], anti-hypertension [55], anti-tumor [56, 57], neuroprotection [58], and anti-cholesterol activities [59]. Oleanolic acid was performed to test the effect on apoptosis and autophagy of SMMC-7721 Hepatoma cells. It can significantly inhibit the growth of liver cancer SMMC-7721 cells and induce autophagy and apoptosis [57]. Compound 78 also showed anti-tumor and anti-inflammatory activities [60, 61]. Lim et al. have demonstrated that 78 showed very strong anti-tumor-promoting activity with an IC50 of 0.1 mg/mL [60].
Structures of oleanane triterpenoids 77‒100 from Actinidia plants
Information of oleanane triterpenoids from Actinidia plants
Bioassay- and 1H NMR-guided fractionation of the methanol extract afforded two oleanolic acids of 2α, 3β, 23-trihydroxyolean-12-en-28-oic acid 80 and 2α, 3α, 24-trihydroxyolean-12-en-28-oic acid 81, showing antifungal activity against C. musae at 3 μg/mL [31]. The EtOAc extract of the roots of A. eriantha Benth exhibited potent growth inhibitory activity against SGC7901 cells, CNE2 cells and HUVECs cells. From which, compound 87 (3β, 23, 24-trihydroxyl-12-oleanen-28-oic acid) was identified [62]. Compound 88 was extracted from the roots bark of A. chinensis, which showed anti-viral activity [25]. A new triterpenoid 12α-chloro-2α, 3β, 13β, 23 -tetrahydroxyolean-28-oic acid-13-lactone 89 was extracted from the roots of A. chinensis Planch (Actinidiaceae). It was tested for cytochrome P450 (CYPs) enzyme inhibitory activity in later years, which could significantly inhibit the catalytic activities of CYP3A4 to < 10% of its control activities [19, 52].
3β-(2-Carboxybenzoyloxy) oleanolic acid 93 and spathodic acid-28-O-β-D-glucopyranoside 94 were extracted from the root bark of A. chinensis. The anti-phytoviral activity test indicated that 94 showed potent activity on TMV, and CMV with inactivation effect of 46.67 ± 1.05, and 45.79 ± 2.23 (100 mg/L), compared to ningnanmycin with inactivation effect of 30.15 ± 1.16 and 27.18 ± 1.02 (100 mg/L) respectively [25]. 3β, 23-Dihydroxy- -30-norolean-12, 20(29)-dien-28-oic acid 98, 3β, 23-dihydroxy-1-oxo-30-norolean-12, 20(29)-dien-28-oic acid 99, and 2α, 3α, 23, 24-tetrahydroxy-30-norolean-12, 20(29)-dien-28-oicacid 100 are three one-carbon-degraded oleanane triterpenoids that were identified from A. chinensis Radix for the first time [50].
2.1.3 Lupane Triterpenoids
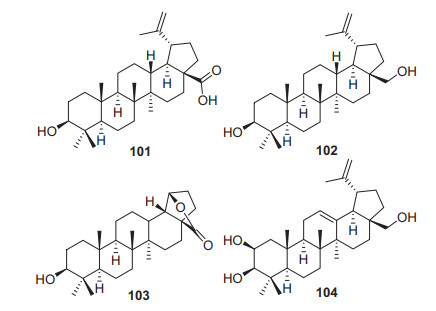
Lupane triterpenoids possess a 6/6/6/6/5-fused carbon skeleton. Compared with ursane and oleanane triterpenoids, the number of lupane triterpenoids in the Actinidia plants is much smaller, only four related compounds have been identified (101‒104, Fig. 4, Table 3). Three of them (101‒103) were identified from the rhizomes of A. kolomikta [51]. Betulinic acid 101 is one of the most representative compound of lupane triterpenes, it has been extensively studied in recent years based on the wide biological activities including anti-inflammatory, antitumor, anti-HIV, anti-diabetic and antimalarial activities [68-73]. Much attention as a molecular target about protein tyrosine phosphatase 1B had been paid to the treatment of insulin resistance diseases because of its critical roles in negatively regulating insulin- and leptin-signaling cascades. Betulinic acid showed significant PTP1B inhibitory activity, with IC50 values of 3.5 μM [24].
Structures of lupane triterpenoids 101‒104 from Actinidia plants
Information of lupane triterpenoids from Actinidia Plants
2.1.4 Other Terpenoids
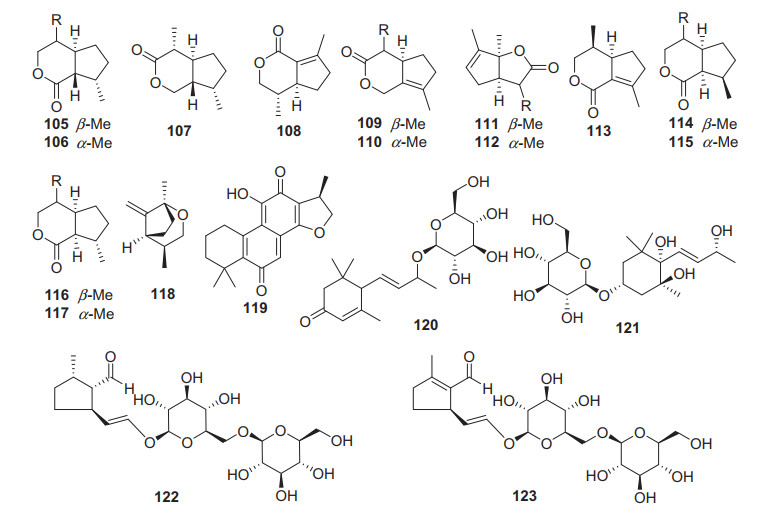
A total of 19 other terpenoids including iridoids, diterpenoids, and their glycosides have been found from Actinidia plants (105 − 123, Fig. 5, Table 4). None of these compounds have good biological activities, only compound 120 showed certain anti-angiogenesis activity [77].
Structures of other terpenoids 105‒123 from Actinidia plants
Information of other triterpenoids from Actinidia plants
2.2 Steroids
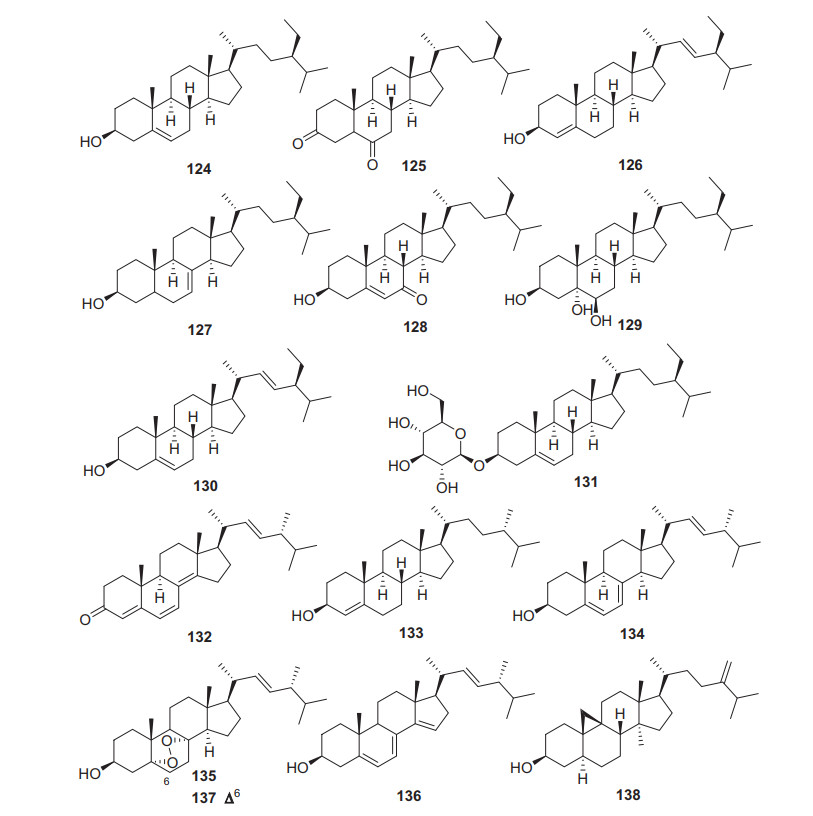
β-Sitosterol 124 is a very normal phytosterol almost distributed in all plants. Eight phytosterols have been obtained from the Actinidia plants (124‒131, Fig. 6, Table 5). Pharmacological studies on these steroids have demonstrated that β-sitosterol showed various bioactivities including anti-inflammatory, anti-cancer, antimicrobial and anti-diabetic properties [81-88]. A study suggested that β-sitosterol may serve as a potential therapeutic in the treatment of acute organ damages [82].
Structures of steroids 124‒138 from Actinidia plants
In addition to phytosterols, seven normal ergosterols (132‒137, Fig. 6, Table 5) were obtained from peel or rhizomes of kiwifruit plants. It is well known that ergosterols should be fungal products. Compounds 132‒137 may be produced by fungal infected kiwifruit plants.
Information of steroids from Actinidia plants
2.3 Phenols
2.3.1 Catechins and Epicatechins
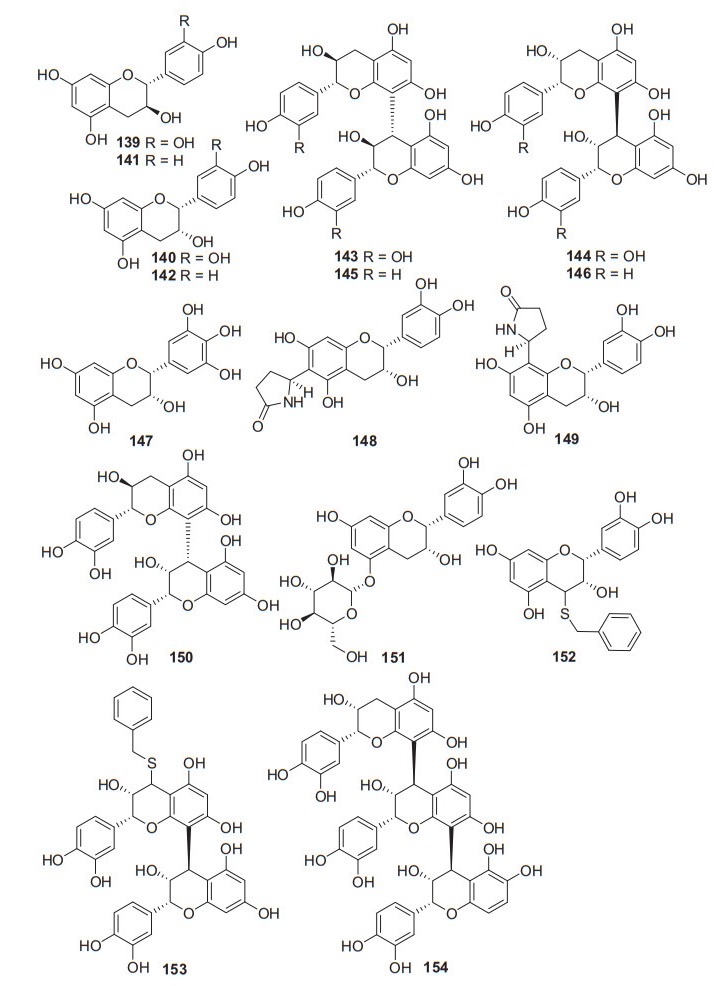
A total of 16 related compounds (139‒154) have been obtained from kiwifruit plants (Fig. 7, Table 6). Compounds 148 and 149 possessed a novel structure featuring with a pyrrolidin-2-one substituent at C-6 and C-8, respectively. Compounds 152 and 153 were two sulfur-containing catechins that was rare in nature. Pharmacological studies have revealed that (+)-catechin 139 and (−)-epi-catechin 140 showed nitric oxide inhibitory activity in LPS stimulated RAW 264.7 cell with IC50 values of 26.61 and 25.30 μg/mL, respectively [53, 91]. Compound 147 showed moderate radical scavenging and antioxidant capabilities by measuring their capacity to scavenge DPPH and anion superoxide radical and to reduce a Mo(VI) salt [89]. Two new flavan-3-ols, 6-(2-pyrrolidinone-5-yl)-(−)-epicatechin 148 and 8-(2-pyrrolidinone-5-yl)-(−)-epicatechin 149, as well as proanthocyanidin B-4 150, were isolated from an EtOAc-soluble extract of the roots of A. arguta. The isolates were tested in vitro for their inhibitory activity on the formation of advanced glycation end products (AGEs). All of them exhibited significant inhibitory activity against AGEs formation with IC50 values ranging from 10.1 to 125.2 μM [92].
Structures of catechins and epicatechins 139‒154 from Actinidia plants
Information of catechins and epicatechins from Actinidia plants
2.3.2 Flavones, Isoflavones, and Flavonols
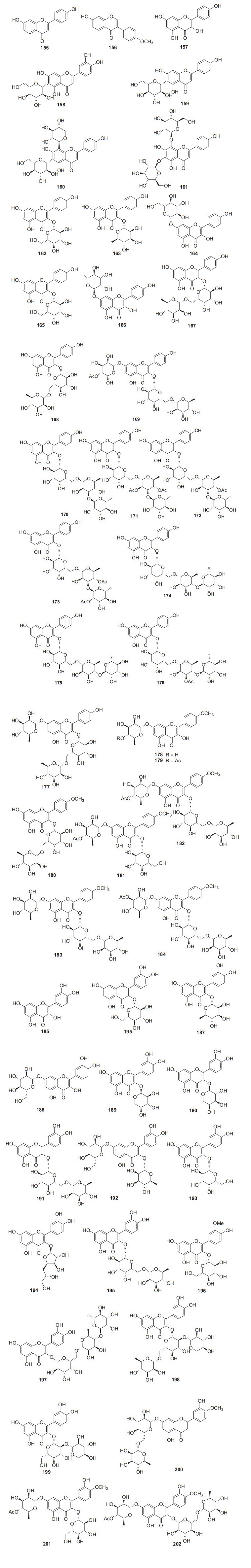
A total of 48 flavone derivatives have been identified from kiwifruit plants, most of which are glycosides (155‒202, Fig. 8, Table 7). Pharmacological studies indicated that these compounds, particularly kaempferol and its derivative, had a wide range of biological activities including antiproliferation, antioxidation, anti-inflammation, anticancer, anti-free radical, and neuroprotection activities [96-99]. Kaempferol 157 was found to prevent neurotoxicity by several ways which was able to completely block N-methyl-D-aspartate (NMDA)-induced neuronal toxicity and potently inhibited MAO (monoamine oxidase) with the IC50 of 0.8 μM [99]. Two novel flavonoids 171 and 172 were separated from the leaves of A. valvata Dunn. They exhibited dose-dependent activity in scavenging 1, 1-diphenyl-2-picrylhydrazyl (DPPH) free radicals, superoxide anion radicals, and hydroxyl radicals, and inhibited lipid peroxidation of mouse liver homogenate in vitro [100]. Compounds 178 [101] and 179 [102] were two new compounds obtained from the leaves of A. kolomikta. The latter was screened for its protective effect on human erythrocytes against AAPH-induced hemolysis, which could slow the hemolysis induced by AAPH [102]. Eerduna et al. evaluated the effects of compound 182 on acute myocardial infarction in rats, the groups treated with 182 showed a dose-dependent reduction in myocardial infarct size model, markedly inhibited the elevation of the activity of creatine kinase, troponin T level, and the content of malondialdehyde induced by AMI [103]. Compound 182 also showed a capacity to increase the activities of superoxide dismutase, catalase, and endothelial nitric oxide synthase [104]. Lim et al. tested the DPPH radical scavenging activity and nitric oxide production inhibitory activity in IFN-γ, LPS stimulated RAW 264.7 cell of quercetin 185, quercetin-3-O-β-D-glucoside 186, and quercetin 3-O-β-D-galactoside 193 with IC50 value of 20.41, 18.23, and 30.46 μg/mL, respectively [91].
Structures of flavones, isoflavones, and flavonols 155‒202 from Actinidia plants
Information of flavones, isoflavones, and flavonols from Actinidia plants
2.3.3 Xanthones
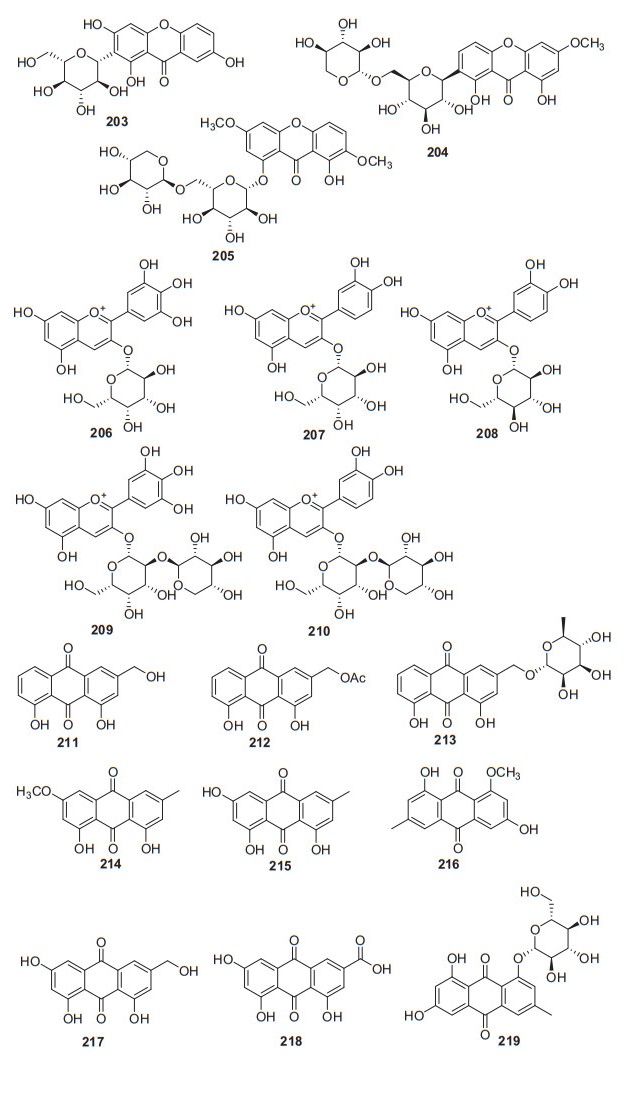
Three xanthones were isolated from n-butyl alcohol fraction of A. arguta (Sieb. & Zucc) Planch. ex Miq and identified as 2-β-D-glu-1, 3, 7-trihydrogen xanthone 203, 7-O-[β-D-xylose-(1 → 6)-β-D-glucopyranoside]-1, 8-dihydroxy-3-methoxy xanthone 204, and 1-O-[β-D-xylose-(1 → 6)-β-D-glucopyranside] -8-hydroxy-3, 7-dimethoxy xanthone 205 (Fig. 9, Table 8). They were isolated from this plant for the first time [108]. Compound 203 showed extensive biological activities, including inhibiting α-Glycosidase, NO production inhibition and NF-κB inhibition and PPAR activation [116, 117]. It has been demonstrated the inhibitory effects on NF-κB transcriptional activation in HepG2 cells stimulated with TNFα with an IC50 value of 0.85 ± 0.07 μM, which was more potent than the positive control of sulfasalazine (IC50 = 0.9 μM) [118].
Structures of xanthones, isoflavones, and flavonols 203‒219 from Actinidia plants
Information of xanthones, anthocyanins, and emodins from Actinidia plants
2.3.4 Anthocyanins
Five anthocyanins were obtained from the flesh of larger fruit of A. deliciosa and A. chinensis and identified as delphinidin 3-galactoside 206, cyanidin 3-galactoside 207, cyanidin 3-glucoside 208, delphinidin 3-[2-(xylosyl)galactoside] 209, and cyanidin 3-[2-(xylosyl)galactoside] 210, respectively (Fig. 9, Table 8) [119]. Cyanidin 3-glucoside 208 exhibited a wide range of pharmacological activities including anti-inflammatory, neuroprotective, anti-cancer, and antioxidant activities [120, 121, 122, 123, 124].
2.3.5 Emodins
A total of nine emodin derivatives were obtained (211‒219, Fig. 9, Table 8). Three emodin constituents were isolated from EtOAc fraction of the roots of A. deliciosa for the first time, and their structures were identified to be aloe-emodin 211, 11-O-acetyl-aloe-emodin 212, and aloe-emodin 11-O-α-L-rhamno -pyranoside 213 [125]. Compound 211 exhibited intriguing biological activities including inflammatory, antifungal, and anticancer activity [126-128]. Lipoxygenases (LOXs) are potential treatment targets in a variety of inflammatory conditions, enzyme kinetics showed that aloe emodin inhibited lipoxygenase competitively with an IC50 of 29.49 μM [126]. Compound 215 was reported to possess wide biological activities including anti-inflammatory, neuroprotection, anti-cardiovascular and α-glucosidase inhibitory activity [129-132]. It exhibited potent inhibition of α-glucosidase with an IC50 value of 19 ± 1 μM and lower cytotoxicity to the Caco-2 cell line [132].
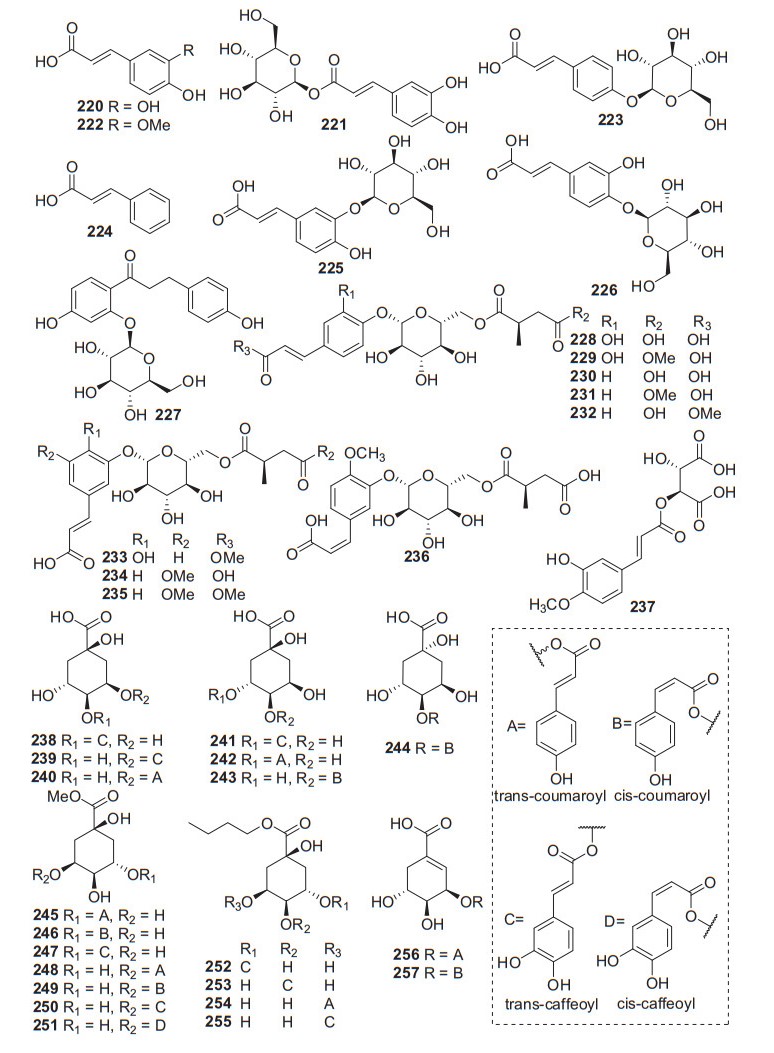
2.3.6 Phenylpropionic Acids
A total of 38 phenylpropionic acid derivatives have been identified from kiwifruit plants (220‒257. Fig. 10, Table 9), while most of them were glycosides or quinic acid derivatives. Phytochemical examination of the fruits of A. arguta led to the isolation of two organic acids including caffeic acid 220 and caffeoyl-β-D-glucopyranoside 221, which were tested for their nitric oxide production inhibitory activity in LPS-stimulated RAW 264.7 cells and DPPH radical scavenging activities. Compared with positive control (L-NMMA), they were potently reduced nitric oxide productions and showed anti-oxidative activities [135]. Nine succinic acid derivatives (228‒236), eleven quinic acid (245‒255) derivatives and two shikimic acid derivatives (256 and 257) were isolated from the fruits of A. arguta. The NF-κB transcriptional inhibitory activity of the compounds was evaluated using RAW 264.7 macrophages cells induced by lipopolysaccharide. Among the groups of different organic acid derivatives, the quinic acid derivatives inhibited NF-κB transcriptional activity with an IC50 value of 4.0 μM [136].
Structures of phenylpropionic acids 220‒257 from Actinidia plants
Information of phenylpropionic acids from Actinidia plants
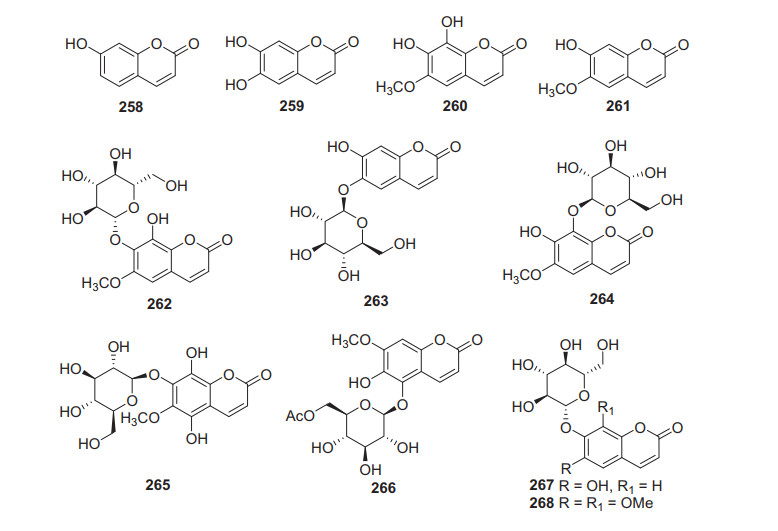
2.3.7 Coumarins
Coumarins are rarely identified from kiwifruit plants, and only eleven members have been reported (258‒267, Fig. 11, Table 10). Umbelliferone 258 was obtained from the leaves of A. polygama (Sieb. et Zucc.) Miq [109]. A number of studies demonstrate the pharmacological properties of 258 including antitumor, anti-inflammatory, antioxidant, antidiabetic, and immunomodulatory activities [143-149]. It showed cytotoxicity against MCF-7 and MDA-MB-231 cell lines with IC50 values of 15.56 and 10.31 μM, respectively [148]. Phytochemical examination of the fruits of A. arguta led to the isolation of esculetin 259 [135]. Two coumarins were isolated from the roots of A. deliciosa and identified as fraxetin 260 and isoscopoletin 261 [150]. Compound 260 showed potent inhibition against lipopolysaccharide (LPS)-induced nitric oxide (NO) generation with an IC50 value of 10.11 ± 0.47 µM [151]. Esculin 263 and fraxin 264 were characterized from the stems and fruits of A. deliciosa (kiwifruit) and A. chinensis [152]. Compound 264 showed inhibitory activity towards HepG2 with an IC50 value of 14.71 μM [153].
Structures of coumarins 258‒268 from Actinidia plants
Information of coumarins from Actinidia plants
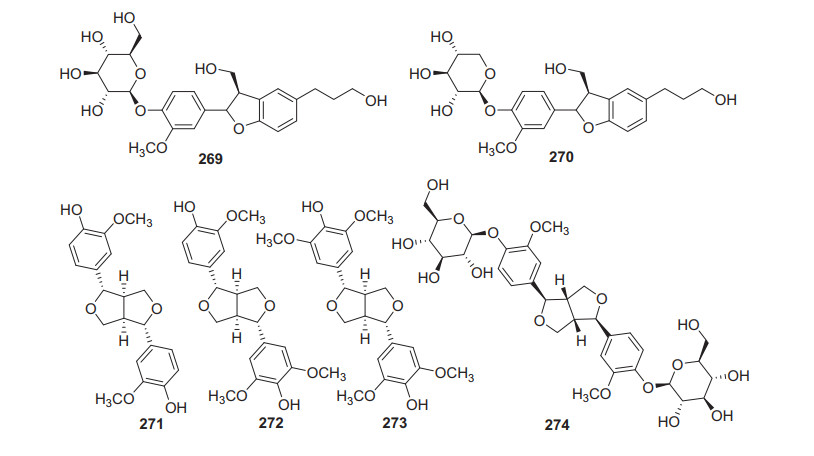
2.3.8 Lignans
Lignans also had a narrow distribution in kiwi plants, only six members have been identified from Actinidia plants (269‒274, Fig. 12, Table 11). (+)-Pinoresinol 271, (+)-medioresinol 272, and (−)-syringaresinol 273 were partitioned from the fraction of the roots of A. arguta [53]. Compound 271 is a biologically active lignan and widely found in many dietary plants. It was reported to possess antifungal, anti-inflammatory, antioxidant, hypoglycemic, and antitumor activities [158-162]. A study on this compound suggested that 271 displayed significant inhibition of fMLP/CB-induced superoxide anion generation and elastase release, with an IC50 value of 1.3 ± 0.2 μg/mL [159]. The 50% ethanol extract of A. arguta showed strong inhibitory effect on α-glucosidase (32.6%), while a bio-guided isolation on the extract gave a bioactive compound pinoresinol diglucoside 274 [138].
Structures of lignans 269‒274 from Actinidia plants
Information of of lignans from Actinidia plants
2.3.9 Simple Phenols
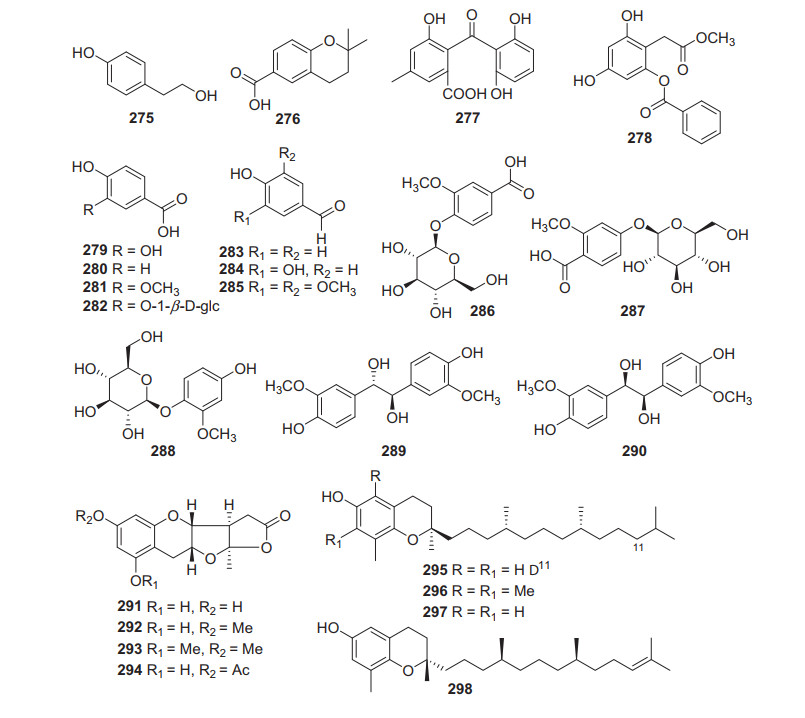
Simple benzene derivatives including glycosides and isoprenylated benzene products from Actinidia plants were collected (275‒298, Fig. 13, Table 12). Phytochemical examination of the fruits of A. arguta led to the isolation of protocatechuic acid 279 [135]. It showed anti-inflammatory [163], antioxidant [163], neuroprotective [164], and anti-proliferative activities [165]. Protocatechuic acid exhibited significant (p < 0.05) anti-inflammatory (83% and 88% inhibition for egg-albumin induced and xylene induced oedema, respectively), analgesic (56% inhibition and 22 s of pain suppression for acetic acid-induced and hot plate-induced pain, respectively), and antioxidant effects (97% inhibition and absorbance of 2.516 at 100 μg/mL for DPPH and FRAP assay, respectively) in the models [166]. Extraction of leaf tissue from the golden-fleshed kiwifruit cultivar A. chinensis "Hort16A" expressing genotype-resistance against the fungus Botrytis cinerea, a new phenolic compound, 3, 5-dihydroxy-2-(methoxycarbonylmethyl)phenyl 3, 4-dihydroxybenzoate 278 was therefore obtained [167].
Structures of simple phenols 275‒298 from Actinidia plants
Information of simple phenols from Actinidia plants
Four novel skeleton phenolic compounds planchols A‒D (291‒294) were isolated from the roots of A. chinensis Planch. Their structures were elucidated by spectroscopic analysis and chemical evidence. The structure of 291 was further confirmed by the single-crystal X-ray diffraction. Moreover, it was found that 291 and 292 showed remarkable cytotoxic activity against P-388 with IC50 of 2.50 and 3.85 μM, respectively, and against A-549 with IC50 of 1.42 and 2.88 μM, respectively [94].
2.4 Miscellaneous
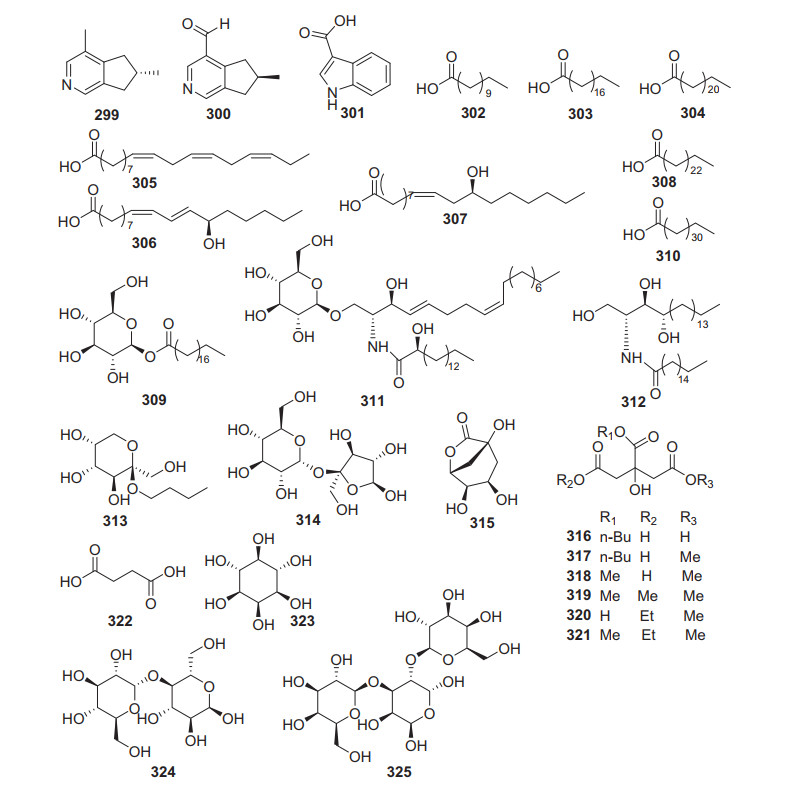
Three alkaloids (299‒301), eleven fatty acids and derivatives (302‒312), and other thirteen small molecules (313‒325) were obtained from Actinidia plants (Fig. 14, Table 13). Actinidine 299 and boschniakine 300 were isolated from the leaves and galls of A. polygama and also isolated from A. arguta which might be converted from iridoids [174, 175]. A bioassay-guided fractionation of the fruits of A. polygama led to the separation and identification of a polyunsaturated fatty acid, α-linolenic acid (ALA) 305 [176]. This compound was found to possess a broad biological properties including anti-inflammatory [177], anti-tumor [178], anti-hyperlipidemic [179], anti-diabetic [180], and anti-fungal [181] activities. By a bio-guided fractionation, a ceramide namely actinidiamide 312 was identified as an anti-inflammatory component from the EtOAc fraction of A. polygama Max. It potently inhibited nitric oxide production (30.6% inhibition at 1 μg/mL) in lipopolysaccharide (LPS)-stimulated RAW264.7 cells and β-hexosaminidase release (91.8% inhibition at 1 μg/mL) in IgE-sentized RBL-2H3 cells [182].
Structures of other molecules 299‒325 from Actinidia plants
Information other molecules from Actinidia plants
In summary, this review focused on the biological components and related pharmacological activities of various parts of Actinidia plants, including triterpenoids, steroids, flavonoids, catechins, coumarins, lignans, phenols, and other small organic molecules. A total of 325 molecules have been collected in this review. Most of the active molecules are derived from the roots of Actinidia plants, while triterpenes and flavonoids are the most important types regardless of the number of compounds and their biological activity significance. The stems, leaves, fruit galls, and other parts of kiwi are mainly rich in flavonoids, phenylpropionic acids, and other small molecule compounds. Currently, these chemical components are not structurally novel. In addition, there are few in-depth researches on pharmacological activities of the bioactive compounds. Therefore, research on the chemical constituents of Actinidia plants is still promising. We hope that this review can provide positive information for the further exploration of the chemical components and their biological activities of Actinidia plants.
Notes
Funding
This work was financially supported by the National Natural Science Foundation of China (22177139) and the Scientific Research Program of Hubei Provincial Department of Education, China (D20183001).
Copyright informatio
© The Author(s) 2021
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
References
-
1.S.J. Henare, S.M. Rutherfurd, L.N. Drummond, V. Borges, M.J. Boland, P.J. Moughan, Food Chem. 130, 67-72 (2012) CrossRef PubMed Google Scholar
-
2.G. Du, M. Li, F. Ma, D. Liang, Food Chem. 113, 557-562 (2009) CrossRef PubMed Google Scholar
-
3.C.V. Garcia, S.Y. Quek, R.J. Stevenson, R.A. Winz, J. Agric. Food Chem. 59, 8358-8365 (2011) CrossRef PubMed Google Scholar
-
4.X. Xiao, R. Sun, S. Jiang, T. Du, G. Yang, F. Ye, China Med. Hered. 11, 157-159 (2014) PubMed Google Scholar
-
5.Y.N. Zhang, L. Liu, C.Q. Ling, Chin J. Chin. Mater. Med. 31, 918-920 (2006) PubMed Google Scholar
-
6.T. Fang, M. He, J. Xia, J. Hou, L. Wang, M. Zheng, X. Wang, J. Xia, Cell Biol. Toxicol. 32, 499-511 (2016) CrossRef PubMed Google Scholar
-
7.C. Huang, Z. Zhang, G. Li, J. Zhou, Plant Divers. Resour. 10, 93-100 (1988) PubMed Google Scholar
-
8.A. Manayi, M. Nikan, N. Nobakht-Haghighi, M. Abdollahi, Curr. Med. Chem. 25, 4866-4875 (2018) PubMed Google Scholar
-
9.A.K. Singh, H. Pandey, P.W. Ramteke, S.B. Mishra, Lat. Am. J. Pharm. 38, 513-517 (2019) PubMed Google Scholar
-
10.A.B. Ramos-Hryb, N. Platt, A.E. Freitas, I.A. Heinrich, M.G. Lopez, R.B. Leal, M.P. Kaster, A.L.S. Rodrigues, Neurochem. Res. 44, 2843-2855 (2019) CrossRef PubMed Google Scholar
-
11.P.D. Moon, N.R. Han, J.S. Lee, H.M. Kim, H.J. Jeong, Int. J. Mol. Med. 43, 2252-2258 (2019) PubMed Google Scholar
-
12.X. Jiang, T. Li, R.H. Liu, J. Agric. Food Chem. 64, 1806-1816 (2016) CrossRef PubMed Google Scholar
-
13.D.S. Jang, G.Y. Lee, J. Kim, Y.M. Lee, J.M. Kim, Y.S. Kim, J.S. Kim, Arch. Pharmacal. Res. 31, 666-670 (2008) CrossRef PubMed Google Scholar
-
14.M. Takaya, M. Nomura, T. Takahashi, Y. Kondo, K.T. Lee, S. Kobayashi, Anticancer Res. 29, 995-1000 (2009) PubMed Google Scholar
-
15.J.H. Won, K.S. Chung, E.Y. Park, J.H. Lee, J.H. Choi, L.A. Tapondjou, H.J. Park, M. Nomura, A.H.E. Hassan, K.T. Lee, Molecules 23, 3306 (2018) CrossRef PubMed Google Scholar
-
16.K.W. Woo, S.U. Choi, K.H. Kim, K.R. Lee, J. Braz. Chem. Soc. 26, 1450-1456 (2015) PubMed Google Scholar
-
17.K.M. Shin, R.K. Kim, T.L. Azefack, L. David, S.B. Luc, M.I. Choudhary, H.J. Park, J.W. Choi, K.T. Lee, Planta Med. 70, 803-807 (2004) CrossRef PubMed Google Scholar
-
18.Y. Sashida, K. Ogawa, N. Mori, T. Yamanouchi, Phytochemistry 31, 2801-2804 (1992) CrossRef PubMed Google Scholar
-
19.Y.X. Xu, Z.B. Xiang, Y.S. Jin, Y. Shen, H.S. Chen, Fitoterapia 81, 920-924 (2010) CrossRef PubMed Google Scholar
-
20.Y.X. Xu, Z.B. Xiang, X.J. Chen, H.S. Chen, Acad. J. Second Mil. Med. Univ. 32, 749-753 (2011) PubMed Google Scholar
-
21.H.L. Xin, Y.C. Wu, Y.F. Xu, Y.H. Su, Y.N. Zhang, C.Q. Ling, Chin. J. Nat. Med. 8, 260-263 (2010) CrossRef PubMed Google Scholar
-
22.H.L. Xin, X.Q. Yue, Y.F. Xu, Y.C. Wu, Y.N. Zhang, Y.Z. Wang, C.Q. Ling, Helv. Chim. Acta. 91, 575-580 (2008) CrossRef PubMed Google Scholar
-
23.Y. Sashida, K. Ogawa, T. Yamanouchi, H. Tanaka, Y. Shoyama, I. Nishioka, Phytochemistry 35, 377-380 (1994) CrossRef PubMed Google Scholar
-
24.D. Li, W. Li, K. Higai, K. Koike, J. Nat. Med. 68, 427-431 (2014) CrossRef PubMed Google Scholar
-
25.X.Y. Zhang, Y. Zhou, Z.P. Wei, J. Shen, L.K. Wang, Z.Q. Ma, X. Zhang, Pest Manag. Sci. 74, 1630-1636 (2018) CrossRef PubMed Google Scholar
-
26.H. Takazawa, K. Yoshimura, A. Ikuta, K. Kawaguchi, Plant Biotechnol. 19, 181-186 (2002) CrossRef PubMed Google Scholar
-
27.S.Y. Du, H.F. Huang, X.Q. Li, L.X. Zhai, Q.C. Zhu, K. Zheng, X. Song, C.S. Xu, C.Y. Li, Y. Li, Z.D. He, H.T. Xiao, Chin. Med. 15, 43 (2020) CrossRef PubMed Google Scholar
-
28.G.C. Bonel-Perez, A. Perez-Jimenez, I. Gris-Cardenas, A.M. Parra-Perez, J.A. Lupianez, F.J. Reyes-Zurita, E. Siles, R. Csuk, J. Peragon, E.E. Rufino-Palomares, Molecules 25, 4254 (2020) CrossRef PubMed Google Scholar
-
29.J. Carmo, P. Cavalcante-Araujo, J. Silva, J. Ferro, A.C. Correia, V. Lagente, E. Barreto, Molecules 25, 4982 (2020) CrossRef PubMed Google Scholar
-
30.X.F. Zhou, P. Zhang, H.F. Pi, Y.H. Zhang, H.L. Ruan, H. Wang, J.Z. Wu, Chem. Biodivers. 6, 1202-1207 (2009) CrossRef PubMed Google Scholar
-
31.E.H. Lahlou, N. Hirai, T. Kamo, M. Tsuda, H. Ohigashi, Biosci. Biotechnol. Biochem. 65, 480-483 (2001) CrossRef PubMed Google Scholar
-
32.L.B. Wei, S.Y. Ma, H.X. Liu, C.S. Huang, N. Liao, Chem. Biodivers. 15, e1700454 (2018) CrossRef PubMed Google Scholar
-
33.N.A. Dangroo, J. Singh, A.A. Dar, N. Gupta, P.K. Chinthakindi, A. Kaul, M.A. Khuroo, P.L. Sangwan, Eur. J. Med. Chem. 120, 160-169 (2016) CrossRef PubMed Google Scholar
-
34.J.H. Ahn, Y. Park, S.W. Yeon, Y.H. Jo, Y.K. Han, A. Turk, S.H. Ryu, B.Y. Hwang, K.Y. Lee, M.K. Lee, J. Nat. Prod. 83, 1416-1423 (2020) CrossRef PubMed Google Scholar
-
35.C.S. Huang, S.Y. Ma, H.X. Liu, Q. Lu, C.S. Huang, H.X. Liu, L.B. Wei, L.G. Shi, N. Liao, L.B. Wei, China J. Chin. Mater. Med. 42, 2714-2718 (2017) PubMed Google Scholar
-
36.L.P. Qu, G.Y. Zheng, Y.H. Su, H.Q. Zhang, Y.L. Yang, H.L. Xin, C.Q. Ling, Int. J. Mol. Sci. 13, 14865-14870 (2012) CrossRef PubMed Google Scholar
-
37.Y. Shi, H. Wang, B. Ma, Chin. Tradit. Herb. Drugs. 24, 386-387 (1993) PubMed Google Scholar
-
38.S.C. Dong, T.Y. Shin, J.S. Eun, D.K. Kim, H. Jeon, Arch. Pharmacal. Res. 34, 425-436 (2011) CrossRef PubMed Google Scholar
-
39.D. Xu, A. Qiu, N. Tang, G. Liu, Y. Lai, CN101502542. PubMed Google Scholar
-
40.C. Huang, G. Li, H. Fan, Z. Zhang, J. Zhou, Plant Divers. Resour. 8, 489-491 (1986) PubMed Google Scholar
-
41.C. Huang, X. Chen, Nat. Prod. Res. Dev. 4, 27-30 (1992) PubMed Google Scholar
-
42.L. Meng, C. Huang, H. Liu, X. Chen, Chin. Tradit. Herb. Drugs 32, 1544-1546 (2009) PubMed Google Scholar
-
43.L. Wei, C. Huang, H. Liu, X. Chen, Technol. Dev Chem. Ind. 38, 1-3 (2009) PubMed Google Scholar
-
44.Y. Cui, X.M. Zhang, J.J. Chen, Y. Zhang, X.K. Lin, L. Zhou, China J. Chin. Mater. Med. 32, 1663-1665 (2007) PubMed Google Scholar
-
45.W.J. Zhu, D.H. Yu, M. Zhao, M.G. Lin, Q. Lu, Q.W. Wang, Y.Y. Guan, G.X. Li, X. Luan, Y.F. Yang, X.M. Qin, C. Fang, G.H. Yang, H.Z. Chen, Anti-Cancer Agents Med. Chem. 13, 195-198 (2013) CrossRef PubMed Google Scholar
-
46.Y. Qin, C. Huang, X. Chen, M. Cai, H. Liu, Chin. Tradit. Herb. Drugs. 30, 323-326 (1999) PubMed Google Scholar
-
47.F. Lu, L. Zhao, L. Zheng, L. Lu, Cent. S. Pharm. 12, 165-168 (2014) PubMed Google Scholar
-
48.Y.D. Xu, L. Yin, Chin. Tradit. Herb. Drugs. 44, 935-937 (2013) PubMed Google Scholar
-
49.S. Bei, C. Huang, X. Chen, Nat. Prod. Res. Dev. 9, 15-18 (1997) PubMed Google Scholar
-
50.C. Nie, J. Yang, D. Wu, L. Wan, G. Liang, Chem. Res. Chin. Univ. 35, 823-829 (2019) CrossRef PubMed Google Scholar
-
51.C. Ye, M. Jin, Y. Zhou, W. Zhou, G. Li, Chem. Nat. Compd. 55, 975-977 (2019) CrossRef PubMed Google Scholar
-
52.Y.X. Xu, Z.B. Xiang, Y.S. Jin, W. Xu, L.N. Sun, W.S. Chen, HSJC Chem. Biodivers. 13, 1454-1459 (2016) CrossRef PubMed Google Scholar
-
53.J.I. Whang, H.I. Moon, O.P. Zee, Saengyak Hakhoechi. 31, 357-365 (2000) PubMed Google Scholar
-
54.Y. Han, Z. Tong, C. Wang, X. Li, G. Liang, Eur. J. Pharmacol. 893, 173811 (2021) CrossRef PubMed Google Scholar
-
55.S. Zhang, Y. Liu, X. Wang, Z. Tian, D. Qi, Y. Li, H. Jiang, Int. J. Mol. Med. 46, 2019-2034 (2020) CrossRef PubMed Google Scholar
-
56.P.M. Edathara, S. Chintalapally, V.K.K. Makani, C. Pant, S. Yerramsetty, M.D. Rao, M.P. Bhadra, Gene 771, 145370 (2021) CrossRef PubMed Google Scholar
-
57.W. Zhou, X. Zeng, X. Wu, Med. Sci. Monit. 26, e921606 (2020) PubMed Google Scholar
-
58.J.M. Castellano, S. Garcia-Rodriguez, J.M. Espinosa, M.C. Millan-Linares, M. Rada, J.S. Perona, Biomolecules 9, 683 (2019) CrossRef PubMed Google Scholar
-
59.W.J.A. Musa, B. Situmeang, J. Sianturi, Int. J. Food Prop. 22, 1439-1444 (2019) CrossRef PubMed Google Scholar
-
60.L.Y. Mooi, N. Abdul Wahab, N.H. Lajis, A.M. Ali, Chem. Biodivers. 7, 1267-1275 (2010) CrossRef PubMed Google Scholar
-
61.L. Huang, T. Guan, Y. Qian, M. Huang, X. Tang, Y. Li, H. Sun, Eur. J. Pharmacol. 672, 169-174 (2011) CrossRef PubMed Google Scholar
-
62.J.G. Wu, L. Ma, S.H. Lin, Y.B. Wu, J. Yi, B.J. Yang, J.Z. Wu, K.H. Wong, J. Ethnopharmacol. 203, 1-10 (2017) CrossRef PubMed Google Scholar
-
63.F. A. Ma, D. L. Wu, F. Q. Xu, W. Zhang, S. R Y., Chin. Tradit. Pat. Med. 38, 591-593 (2016) PubMed Google Scholar
-
64.H. Zhao, B.Z. Wang, B.R. Ma, J.Y. Sun, Chin. Pharm. J. 29, 523-524 (1994) PubMed Google Scholar
-
65.Y. Lai, D. Xu, J. Chin. Med. Mater. 30, 166-168 (2007) PubMed Google Scholar
-
66.L. Ding, S. Wang, Z. Wang, China J. Chin. Mater. Med. 32, 1893-1895 (2007) PubMed Google Scholar
-
67.J. Lu, R. Yang, M. Gui, Y. Jin, J. Dong, X. Li, Chin. Pharm. J. 44, 1215-1217 (2009) PubMed Google Scholar
-
68.L.J. Zhu, S.T. Xiang, X.H. Wang, J. Zhao, Z.I. Tan, J.E. Yi, J. Tradit. Chin. Vet. Med. 35, 18-22 (2016) PubMed Google Scholar
-
69.C.G. Farcas, C. Dehelean, I.A. Pinzaru, M. Mioc, V. Socoliuc, E.A. Moaca, S. Avram, R. Ghiulai, D. Coricovac, I. Pavel, P.K. Alla, O.M. Cretu, C. Soica, F. Loghin, Int. J. Nanomed. 15, 8175-8200 (2020) CrossRef PubMed Google Scholar
-
70.L. Kun, J.Y. Wang, L. Zhang, Y.Y. Pan, X.Y. Chen, Y. Yuan, Int. J. Immunopathol. Pharmacol. 34, 2058738420945078 (2020) PubMed Google Scholar
-
71.H. Wang, F. Dong, Y. Wang, X.A. Wang, D. Hong, Y. Liu, J. Zhou, Acta Biochim. Biophys. Sin. 52, 200-206 (2020) CrossRef PubMed Google Scholar
-
72.Q. Wang, Y. Li, L. Zheng, X. Huang, Y. Wang, C.H. Chen, Y.Y. Cheng, S.L. Morris-Natschke, K.H. Lee, A.C.S. Med, Chem. Lett. 11, 2290-2293 (2020) PubMed Google Scholar
-
73.G.A. Birgani, A. Ahangarpour, L. Khorsandi, H.F. Moghaddam, Braz. J. Pharm. Sci. 54, e17171 (2018) PubMed Google Scholar
-
74.Z. Zhou, C. Zhu, Z. Cai, F. Zhao, L. He, X. Lou, X. Qi, Oncol. Lett. 15, 7319-7327 (2018) PubMed Google Scholar
-
75.Y.H. Han, J.G. Mun, H.D. Jeon, J.Y. Kee, S.H. Hong, Nutrients 12, 66 (2019) CrossRef PubMed Google Scholar
-
76.F. Yin, F. Feng, L. Wang, Z. Li, X. Wang, Y. Cao, Cell Death Dis. 10, 672 (2019) CrossRef PubMed Google Scholar
-
77.F. Murai, M. Tagawa, Planta Med. 37, 234-240 (1979) CrossRef PubMed Google Scholar
-
78.T. Sakai, K. Nakajima, T. Sakan, Bull. Chem. Soc. Jpn. 53, 3683-3686 (1980) CrossRef PubMed Google Scholar
-
79.S. Isoe, T. Ono, S.B. Hyeon, T. Sakan, Tetrahedron Lett. 9, 5319-5323 (1968) CrossRef PubMed Google Scholar
-
80.F. Murai, M. Tagawa, H. Ohishi, Planta Med. 58, 112-113 (1992) CrossRef PubMed Google Scholar
-
81.S. Jain, A. Ganeshpurkar, N. Dubey, Pharmacogn. Commun. 10, 134-135 (2020) CrossRef PubMed Google Scholar
-
82.K. Koc, F. Geyikoglu, O. Cakmak, A. Koca, Z. Kutlu, F. Aysin, A. Yilmaz, H. Askin, Naunyn-Schmiedeberg's Arch. Pharmacol. 394, 469-479 (2021) CrossRef PubMed Google Scholar
-
83.J.Y. Ye, L. Li, Q.M. Hao, Y. Qin, C.S. Ma, Korean J. Physiol. Pharmacol. 24, 39-46 (2020) CrossRef PubMed Google Scholar
-
84.F. Zhang, Z. Liu, X. He, Z. Li, B. Shi, F. Cai, Drug Deliv. 27, 1329-1341 (2020) CrossRef PubMed Google Scholar
-
85.L. Karthik, B. Vijayakumar, Int. J. Pharm. Phytopharm. Res. 10, 8-21 (2020) PubMed Google Scholar
-
86.A. Sen, P. Dhavan, K.K. Shukla, S. Singh, G. Tejovathi, Sci. Secure J. Biotechnol. 1, 9-13 (2012) PubMed Google Scholar
-
87.S. Babu, S.J.B. Jayaraman, Pharmacotherapy 131, 110702 (2020) CrossRef PubMed Google Scholar
-
88.S. Babu, M. Krishnan, P. Rajagopal, V. Periyasamy, V. Veeraraghavan, R. Govindan, S. Jayaraman, Eur. J. Pharmacol. 873, 173004 (2020) CrossRef PubMed Google Scholar
-
89.A. Fiorentino, B.D. Abrosca, S. Pacifico, C. Mastellone, M. Scognamiglio, P. Monaco, J. Agric. Food Chem. 57, 4148-4155 (2009) CrossRef PubMed Google Scholar
-
90.M. Ahmad Khan, A.H.M.G. Sarwar, R. Rahat, R.S. Ahmed, S. Umar, Int. Immunopharmacol. 85, 106642 (2020) CrossRef PubMed Google Scholar
-
91.H.W. Lim, J.G. Shim, H.K. Choi, M.W. Lee, Saengyak Hakhoechi. 36, 245-251 (2005) PubMed Google Scholar
-
92.D.S. Jang, G.Y. Lee, Y.M. Lee, Y.S. Kim, H. Sun, D.H. Kim, J.S. Kim, Chem. Pharm. Bull. 57, 397-400 (2009) CrossRef PubMed Google Scholar
-
93.B. Riyana, D.H. Putri Huspa, M.H. Satari, D. Kurnia, Lett. Drug Des. Discov. 17, 1531-1537 (2020) CrossRef PubMed Google Scholar
-
94.J. Chang, R. Case, Planta Med. 71, 955-959 (2005) CrossRef PubMed Google Scholar
-
95.J. Michaud, M. Ane-Margail, Bull. Soc. Pharm. Bordeaux. 116, 52-64 (1977) PubMed Google Scholar
-
96.J. Wang, X. Fang, L. Ge, F. Cao, L. Zhao, Z. Wang, W. Xiao, PLoS ONE 13(5), e0197563 (2018) CrossRef PubMed Google Scholar
-
97.S. Bakhshii, S. Khezri, R. Ahangari, A. Jahedsani, A. Salimi, Drug Dev. Res. (2021). https://doi.org/10.1002/ddr.21790 PubMed Google Scholar
-
98.M.A. Arowosegbe, O.T. Amusan, S.A. Adeola, O.B. Adu, I.A. Akinola, B.F. Ogungbe, O.I. Omotuyi, G.M. Saibu, A.J. Ogunleye, R.I. Kanmodi, N.E. Lugbe, O.J. Ogunmola, D.C. Ajayi, S.O. Ogun, F.O. Oyende, A.O. Bello, P.G. Ishola, P.E. Obasieke, Curr. Drug Discov. Technol. 17, 682-695 (2020) CrossRef PubMed Google Scholar
-
99.B.D. Sloley, L.J. Urichuk, P. Morley, J. Durkin, J.J. Shan, P.K.T. Pang, R.T. Coutts, J. Pharm. Pharmacol. 52, 451-459 (2000) PubMed Google Scholar
-
100.H.L. Xin, Y.C. Wu, Y.H. Su, J.Y. Sheng, C.Q. Ling, Planta Med. 77, 70-73 (2011) CrossRef PubMed Google Scholar
-
101.X. Chang, B. Ma, L. He, Y. Xiao, X. Li, Chin. Tradit. Herb. Drugs. 24, 283-285 (1993) PubMed Google Scholar
-
102.J. Lu, Y. Jin, G. Liu, N. Zhu, M. Gui, A. Yu, X. Li, Chem. Nat. Compd. 46, 205-208 (2010) CrossRef PubMed Google Scholar
-
103.M. Horiuch, C. Murakami, N. Fukamiya, D. Yu, K.H. Lee, J. Nat. Prod. 69, 1271-1274 (2006) CrossRef PubMed Google Scholar
-
104.G. Eerduna, D. Wei, X. Yu, S. Qu, D. Sui, Pharmazie 68, 453-458 (2013) PubMed Google Scholar
-
105.G.N. He, B.C. Wang, H. Wang, L. Fan, X.M. Hu, Chin. Arch. Tradit. Chin. Med. 31, 2353-2355 (2016) PubMed Google Scholar
-
106.C. Liu, D. Weir, P. Busse, N. Yang, Z. Zhou, C. Emala, X.M. Li, Phytother. Res. 29, 925-932 (2015) CrossRef PubMed Google Scholar
-
107.R.F. Webby, N. Z. J. Crop Hortic. Sci. 18, 1-4 (1990) CrossRef PubMed Google Scholar
-
108.M. Xiang, C. Jin, R. Kou, G. Yang, J. Li, J. Huazhong, Norm. Univ. Nat. Sci. 49, 397-401 (2015) PubMed Google Scholar
-
109.J. Lu, G. Cui, X. Wang, N. Zhu, G. Liu, X. Li, Y. Jin, Chin. Pharm. J. 44, 328-330 (2009) PubMed Google Scholar
-
110.A.S. Syed, J.S. Jeon, C.Y. Kim, Nat. Prod. Res. 31, 1501-1508 (2017) CrossRef PubMed Google Scholar
-
111.J. Lu, X.W. Li, M.Y. Gui, G.Y. Liu, N. Zhu, A.M. Yu, T. Okuyama, B. Masaki, Y.R. Jin, Chem. J. Chin. Univ. 30, 468-473 (2009) PubMed Google Scholar
-
112.R.F. Webby, K.R. Markham, Phytochemistry 29, 289-292 (1990) CrossRef PubMed Google Scholar
-
113.Y. Jin, M. Gui, X. Li, CN1566127A. PubMed Google Scholar
-
114.A. Kalandiya, M. Vanidze, S. Papunidze, I. Chkhikvishvili, A. Shalashvili, Bull. Georgian Acad. Sci. 163, 157-159 (2001) PubMed Google Scholar
-
115.R.F. Webby, Phytochemistry 30, 2443-2444 (1991) CrossRef PubMed Google Scholar
-
116.C.T. Luo, H.H. Zheng, S.S. Mao, M.X. Yang, C. Luo, H. Chen, Planta Med. 80, 201-208 (2014) PubMed Google Scholar
-
117.T.X. Shi, S. Wang, K.W. Zeng, P.F. Tu, Y. Jiang, Bioorg. Med. Chem. Lett. 23, 5904-5908 (2013) CrossRef PubMed Google Scholar
-
118.W. Li, Y. Ding, T.H. Quang, T.T.N. Nguyen, Y.N. Sun, X.T. Yan, S.Y. Yang, C.W. Choi, E.J. Lee, K.Y. Paek, Y.H. Kim, Bull. Korean Chem. Soc. 34, 1407-1413 (2013) CrossRef PubMed Google Scholar
-
119.D.J. Comeskey, M. Montefiori, P.J.B. Edwards, T.K. McGhie, J. Agric. Food Chem. 57, 2035-2039 (2009) CrossRef PubMed Google Scholar
-
120.D. Ferrari, F. Cimino, D. Fratantonio, M.S. Molonia, R. Bashllari, R. Busa, A. Saija, A. Speciale, Mediat. Inflamm. 2017, 3454023 (2017) PubMed Google Scholar
-
121.S.R. Pereira, L.M. Almeida, T.C.P. Dinis, J. Funct. Foods. 63, 103586 (2019) CrossRef PubMed Google Scholar
-
122.G.C. Di, R. Acquaviva, R. Santangelo, V. Sorrenti, L. Vanella, V.G. Li, N.D. Orazio, A. Vanella, F. Galvano, J. Evid. Based Complement. Altern. Med. 20, 285750 (2012) PubMed Google Scholar
-
123.X. Ma, S. Ning, Phytother. Res. 33, 81-89 (2019) CrossRef PubMed Google Scholar
-
124.P. Zhang, S. Liu, Z. Zhao, L. You, M.D. Harrison, Z. Zhang, Food Chem. 343, 128482 (2021) CrossRef PubMed Google Scholar
-
125.W. Fu, C. Tan, X. Meng, L. Lu, S. Jiang, D. Zhu, Chin. J. Med. Chem. 20, 116-118 (2010) PubMed Google Scholar
-
126.C.S. Sharanya, K.G. Arun, A. Sabu, M. Haridas, Prostaglandins Other Lipid Mediat. 150, 106453 (2020) CrossRef PubMed Google Scholar
-
127.W. Ma, C. Liu, J. Li, M. Hao, Y. Ji, X. Zeng, Photochem. Photobiol. Sci. 19, 485-494 (2020) CrossRef PubMed Google Scholar
-
128.S. Jangra, B. Sharma, S. Singh, Mater. Res Innov. 25, 264-275 (2020) PubMed Google Scholar
-
129.M.R. De Oliveira, I.C.C. De Souza, F.B. Brasil, Neurochem. Res. 46, 482-493 (2020) PubMed Google Scholar
-
130.S.W. Leung, J.H. Lai, J.C.C. Wu, Y.R. Tsai, Y.H. Chen, S.J. Kang, Y.H. Chiang, C.F. Chang, K.Y. Chen, Int. J. Mol. Sci. 21, 2899 (2020) CrossRef PubMed Google Scholar
-
131.Q. Li, J. Gao, X. Pang, A. Chen, Y. Wang, Front. Pharmacol. 11, 559607 (2020) CrossRef PubMed Google Scholar
-
132.C. Wang, L. Guo, J. Hao, L. Wang, W. Zhu, J. Nat. Prod. 79, 2977-2981 (2016) CrossRef PubMed Google Scholar
-
133.Z. Ji, X. Liang, Acta Pharm Sin. 20, 778-781 (1985) PubMed Google Scholar
-
134.M.Y. Ali, S. Jannat, H.A. Jung, B.S. Min, P. Paudel, J.S. Choi, J. Food Biochem. 42 (2018) PubMed Google Scholar
-
135.H.W. Lim, S.J. Kang, M. Park, J.H. Yoon, B.H. Han, S.E. Choi, M.W. Lee, Nat. Prod. Sci. 12, 221-225 (2006) PubMed Google Scholar
-
136.J.H. Ahn, Y. Park, Y.H. Jo, S.B. Kim, S.W. Yeon, J.G. Kim, A. Turk, J.Y. Song, Y. Kim, B.Y. Hwang, M.K. Lee, Food Chem. 308, 125666 (2020) CrossRef PubMed Google Scholar
-
137.J. He, B.Z. Ma, X.X. Wang, F. Liu, W.J. Qin, X.I. Zhang, T. Zhao, Chin. Pharm. J. 50, 1960-1963 (2015) PubMed Google Scholar
-
138.D. Kwon, G.D. Kim, W. Kang, J.E. Park, S.H. Kim, E. Choe, J.I. Kim, J.H. Auh, J. Korean Soc. Appl. Biol. Chem. 57, 473-479 (2014) CrossRef PubMed Google Scholar
-
139.X.H. Gao, S.D. Zhang, L.T. Wang, L. Yu, X.I. Zhao, H.Y. Ni, Y.Q. Wang, J.D. Wang, C.H. Shan, Y.J. Fu, Molecules 25, 1385 (2020) CrossRef PubMed Google Scholar
-
140.J. Hu, X. Han, X. Li, B. Huang, Med. Plant. 9, 9-13 (2018) PubMed Google Scholar
-
141.A. Zeng, X. Liang, S. Zhu, C. Liu, S. Wang, Q. Zhang, J. Zhao, D.L. Song, Oncol. Rep. 45, 717-727 (2021) PubMed Google Scholar
-
142.D. Wang, L. Tian, H. Lv, Z. Pang, D. Li, Z. Yao, S. Wang, Biomed. Pharmacother. 132, 110773 (2020) CrossRef PubMed Google Scholar
-
143.J.S. Lopez-Gonzalez, H. Prado-Garcia, D. Aguilar-Cazares, J.A. Molina-Guarneros, J. Morales-Fuentes, J.J. Mandoki, Lung Cancer 43, 275-283 (2004) CrossRef PubMed Google Scholar
-
144.V.M. Navarro-Garcia, G. Rojas, M. Aviles, M. Fuentes, G. Zepeda, Mycoses 4, e569-e571 (2011) PubMed Google Scholar
-
145.J.R.S. Hoult, M. Paya, Gen. Pharmacol. 27, 713-722 (1996) CrossRef PubMed Google Scholar
-
146.H. Li, Y. Yao, L. Li, J. Pharm. Pharmacol. 69, 1253-1264 (2017) CrossRef PubMed Google Scholar
-
147.J.F. Vasconcelos, M.M. Teixeira, J.M. Barbosa-Filho, M.F. Agra, X.P. Nunes, A.M. Giulietti, R. Ribeiro-dos-Santos, M.B.P. Soares, Eur. J. Pharmacol. 609, 126-131 (2009) CrossRef PubMed Google Scholar
-
148.R. Rashmi, N. Prakash, D. Rathnamma, S. Rao, A. Sahadev, C.R. Santhosh, U. Sunilchandra, K.S. Naveen, R.S. Wilfred, G.P. Kalmath, K.R.A. Kumar, H.M. Yathish, P. Waghe, Pharma Innov. 8, 29-35 (2019) PubMed Google Scholar
-
149.R. Rashmi, N. Prakash, D. Rathnamma, S. Rao, A. Sahadev, C.R. Santhosh, U. Sunilchandra, N.S. Kumar, W.S. Ruban, G.P. Kalmath, H. Dhanalakshmi, G L., A.R. Gomes, K.R.A. Kumar, P. Waghe, Pharma Innov. 8, 36-40 (2019) PubMed Google Scholar
-
150.W.W. Fu, C.H. Tan, L.L. Lu, X.X. Meng, H.F. Luo, D.Y. Zhu, Chin. J. Nat. Med. 8, 247-249 (2010) CrossRef PubMed Google Scholar
-
151.H.C. Chang, S.W. Wang, C.Y. Chen, T.L. Hwang, M.J. Cheng, P.J. Sung, K.W. Liao, J.J. Chen, Molecules 25, 5911 (2020) CrossRef PubMed Google Scholar
-
152.A.M. Hirsch, A. Longeon, M. Guyot, Biochem. Syst. Ecol. 30, 55-60 (2002) CrossRef PubMed Google Scholar
-
153.Y. Li, C. Ma, J. Huang, Chin. Pharm. J. 44, 1294-1297 (2009) PubMed Google Scholar
-
154.Y. Kimura, M. Sumiyoshi, Eur. J. Pharmacol. 746, 115-125 (2015) CrossRef PubMed Google Scholar
-
155.B. Aouey, A.M. Samet, H. Fetoui, M.S.J. Simmonds, M. Bouaziz, Biomed. Pharmacother. 84, 1088-1098 (2016) CrossRef PubMed Google Scholar
-
156.S. Ren, Y. Xing, C. Wang, F. Jiang, G. Liu, Z. Li, T. Jiang, Y. Zhu, D. Piao, Int. J. Biochem. Cell Biol. 125, 105777 (2020) CrossRef PubMed Google Scholar
-
157.P. Wu, W. He, Y. Fu, J. Wu, J. Li, L. Xiao, Immunol. J. 36, 22-28 (2020) PubMed Google Scholar
-
158.B. Hwang, J. Lee, Q.H. Liu, E.R. Woo, D.G. Lee, Molecules 15, 3507-3516 (2010) CrossRef PubMed Google Scholar
-
159.P.C. Kuo, H.Y. Hung, C.W. Nian, T.L. Hwang, J.C. Cheng, D.H. Kuo, E.J. Lee, S.H. Tai, T.S. Wu, J. Nat. Prod. 80, 1055-1064 (2017) CrossRef PubMed Google Scholar
-
160.X.L. Ouyang, L.X. Wei, H.S. Wang, Y.M. Pan, S. Afr, J. Bot. 98, 162-166 (2015) PubMed Google Scholar
-
161.A. Wikul, T. Damsud, K. Kataoka, P. Phuwapraisirisan, Bioorg. Med. Chem. Lett. 22, 5215-5217 (2012) CrossRef PubMed Google Scholar
-
162.Y. Zhang, H. Zhao, Y. Di, Q. Li, D. Shao, J. Shi, Q. Huang, J. Funct. Foods. 45, 206-214 (2018) CrossRef PubMed Google Scholar
-
163.I. Paterniti, D. Impellizzeri, M. Cordaro, R. Siracusa, C. Bisignano, E. Gugliandolo, A. Carughi, E. Esposito, G. Mandalari, S. Cuzzocrea, Nutrients 9, 915 (2017) CrossRef PubMed Google Scholar
-
164.A.N. Winter, M.C. Brenner, N. Punessen, M. Snodgrass, C. Byars, Y. Arora, D.A. Linseman, Oxid. Med. Cell. Longevity. 2017, 6297080 (2017) PubMed Google Scholar
-
165.Y.H. Wang, Y. Gao, Z. Li, D.I. Wang, W.H. Ling, Acta Nutr. Sin. 36, 53-57 (2014) PubMed Google Scholar
-
166.P. Thomas, E. Essien, A. Udoh, B. Archibong, O. Akpan, E. Etukudo, M. Leo, O. Eseyin, G. Flamini, K. Ajibesin, J. Ethnopharmacol. 269, 113737 (2021) CrossRef PubMed Google Scholar
-
167.K.V. Wurms, J.M. Cooney, Asian J. Biochem. 1, 325-332 (2006) CrossRef PubMed Google Scholar
-
168.X. Qin, C.H. Zhang, D.L. Yao, J.M. Cui, G. Li, J. Yanbian Med. Coll. 36, 187-189 (2013) PubMed Google Scholar
-
169.D.A. Sumilat, H. Yamazaki, K. Endo, H. Rotinsulu, D.S. Wewengkang, K. Ukai, M. Namikoshi, J. Nat. Med. 71, 776-779 (2017) CrossRef PubMed Google Scholar
-
170.M.H. Farah, G. Samuelsson, Planta Med. 58, 14-18 (1992) CrossRef PubMed Google Scholar
-
171.J. He, B.Z. Ma, T. Zhao, W. Wang, F.L. Wei, J. Lu, X.L. Zhang, Chin. Pharm. J. 49, 184-186 (2014) PubMed Google Scholar
-
172.Z.Q. Chang, E. Gebru, S.P. Lee, M.H. Rhee, J.C. Kim, H. Cheng, S.C. Park, J. Nutr. Sci. Vitaminol. 57, 118-122 (2011) CrossRef PubMed Google Scholar
-
173.A. Fiorentino, C. Mastellone, B.D. Abrosca, S. Pacifico, M. Scognamiglio, G. Cefarelli, R. Caputo, P. Monaco, Food Chem. 115, 187-192 (2009) CrossRef PubMed Google Scholar
-
174.T. Sakan, A. Fujino, F. Murai, Y. Butsugan, A. Suzui, Bull. Chem. Soc. Jpn. 32, 315-316 (1959) CrossRef PubMed Google Scholar
-
175.D. Gross, W. Berg, H.R. Schuette, Phytochemistry 11, 3082-3083 (1972) CrossRef PubMed Google Scholar
-
176.J. Ren, E.J. Han, S.H. Chung, Arch. Pharmacal. Res. 30, 708-714 (2007) CrossRef PubMed Google Scholar
-
177.J. Kim, M. Ahn, Y. Choi, T. Kang, J. Kim, N.H. Lee, G.O. Kim, T. Shin, Inflammation 43, 1876-1883 (2020) CrossRef PubMed Google Scholar
-
178.M. Vara-Messler, M.E. Pasqualini, A. Comba, R. Silva, C. Buccellati, A. Trenti, L. Trevisi, A.R. Eynard, A. Sala, C. Bolego, M.A. Valentich, Eur. J. Nutr. 56, 509-519 (2017) CrossRef PubMed Google Scholar
-
179.Y. Mounika, R.M. Naik, Int. J. Pharm. Pharm. Res. 17, 306-328 (2019) PubMed Google Scholar
-
180.T. Suanarunsawat, G. Anantasomboon, C. Piewbang, Exp. Ther. Med. 11, 832-840 (2016) CrossRef PubMed Google Scholar
-
181.P.U.M. Devi, P.S. Reddy, N.R.U. Rani, K.J. Reddy, M.N. Reddy, P. Reddanna, Eur. J. Plant Pathol. 106, 857-865 (2000) CrossRef PubMed Google Scholar
-
182.M.H. Bang, I.G. Chae, E.J. Lee, N.I. Baek, Y.S. Baek, D.Y. Lee, I.S. Lee, S.P. Lee, S.A. Yang, Biosci. Biotechnol. Biochem. 76, 289-293 (2012) CrossRef PubMed Google Scholar
-
183.K. Kono, A. Yamashita, T. Ishihara, JP2008120772A. PubMed Google Scholar
-
184.G.N. He, X.M. Hu, H. Wang, L. Fan, B.C. Wang, China J. Chin. Mater. Med. 30, 498-500 (2015) PubMed Google Scholar
-
185.Y. Lai, D.P. Xu, Chin. Tradit. Herb. Drugs. 30, 166-168 (2007) PubMed Google Scholar
-
186.P. Roger, J. P. Fournier, A. Martin, J. Choay, EP238401A2. PubMed Google Scholar
-
187.X. Chen, S. Yang, S. Bai, Chin. Tradit. Herb. Drugs. 42, 841-843 (2011) PubMed Google Scholar
-
188.J. Liang, H. Zhen, S. Li, W. Zhang, X. Wang, C. Liang, China J. Chin. Mater. Med. 33, 1275-1277 (2008) PubMed Google Scholar
-
189.X. Chang, B. Ma, J. Shan, L. Chen, Chin. Tradit. Herb. Drugs. 27, 395 (1996) PubMed Google Scholar
-
190.P. Li, A. Lu, B. Ma, J. Wei, China J. Chin. Mater. Med. 17, 420-421 (1992) PubMed Google Scholar