New Secodaphnane-Type Alkaloids with Cytotoxic Activities from Daphniphyllum angustifolium Hutch
Abstract
One new Daphniphyllum alkaloid, daphnioldhanol A (1), together with three known ones, were isolated from the stem part of Daphniphyllum angustifolium Hutch. Their structures were elucidated by spectroscopic methods and comparing with the literature data. Compound 2 is a new natural product, but known by synthesis as a racemate. Compound 1 exhibited week cytotoxic activity against Hela cell line with IC50 of 31.9 μM.Graphical Abstract

Keywords
Daphniphyllum angustifolium hutch Secodaphnane-type Daphnioldhanol A Cytotoxic activity1 Introduction
Alkaloids are a class of compounds with significant activities (with a variety of novel skeletons) that are widely found in nature [1-7]. Daphniphyllum alkaloids are a structurally diversified group of complex polycyclic natural products isolated from the Daphniphyllum genus [8]. Since these unique, versatile and complex nitrogen heterocyclic compounds exhibit a wide range of biological activities and are extremely challenging, they have aroused great interest in total synthesis and biosynthetic studies [9-16]. In recent years, quite a number of new Daphniphyllum alkaloids have been isolated and identified, and some of them possessed novel skeletons [17-20]. In our continued search for Daphniphyllum alkaloids with interesting skeletons [21-24], one new Daphniphyllum alkaloid, daphnioldhanol A (1), together with three known ones, were isolated from the stems of Daphniphyllum angustifolium Hutch (Fig. 1). Herein, the isolation, structural elucidation, and bioactivities of these compounds are reported.
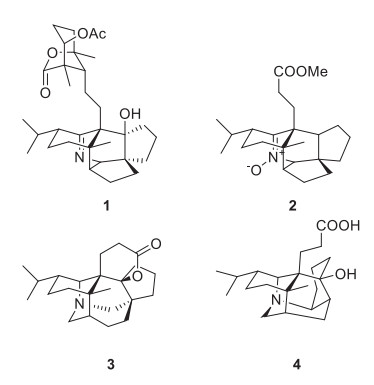
The structures of compounds 1–4
2 Results and Discussion
2.1 Structure Elucidation of the Compounds
Daphnioldhanol A (1) was obtained as white amorphous powder. Its molecular formula, C32H48NO5, was established by positive HRESIMS at m/z 526.3534 [M+ H]+ (calcd 526.3534), with 10 degrees of unsaturation. The IR absorptions implied the presence of hydroxyl (3441 cm−1), and an imine moiety (1631 cm−1). The 13C NMR and DEPT data of 1 displayed 32 carbon signals (Table 1), due to three tetrasubstituted sp2 carbon atoms at lower field and 29 sp3 carbon atoms (5 × C, 6 × CH, 11 × CH2, 6 × CH3) at higher field. According to the molecular formula and relative NMR data, one CH group (δC 65.4, δH 2.95) was ascribed to those bearing an N-atom, while one quaternary C-atom (δC 84.4) were attributed to those bearing an O-atom. Additionally, three sp2 quaternary carbons were attributable to one lactone carbonyl (δC 180.2), one ester carbonyl (δC 172.2), and one iminium group (δC 168.4), while taking into account the three degrees of unsaturation. The remaining seven degrees of unsaturation were accounted for the presence of the heptacyclic system of 1.
1H and 13C NMR spectroscopic data for compound 1a (δ in ppm and J in Hz)
The 1H and 13C NMR spectra of 1 were closely related to those of the known compound Daphnioldhanine Ⅰ [25], with the exception of the loss of signal for a CH in the latter and the addition of signal for quaternary carbon with a hydroxyl (δC 84.4), which were supported by the HMBC correlations of H-11/C-9, H-13/C-9, H-15/C-9, H-16a/C-9, H-17/C-9.
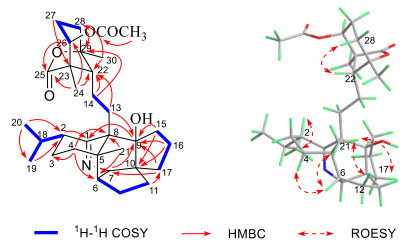
To determine the orientation of the hydroxyl at C-9, we compared the 13C NMR data of both 1 and daphnioldhanine Ⅰ, which revealed that 9-OH substituent significantly shields the C-21 (5 ppm decrease) in the former. This indicated that the 9-OH in 1 should take a β-orientation. Moreover, the remaining relative configuration of 1 was elucidated from ROESY correlations as shown in computer-generated 3D drawing, which was the same as that of the daphnioldhanine Ⅰ (Fig. 2).
1H-1H COSY, key HMBC and ROESY correlations of 1
The known compounds were identified as (−)-nitrone 17 (2) [26], daphnilactone A (3) [27], dapholdhamine B (4) [28], respectively, by comparison of their spectroscopic data with those reported in the literature (Fig. 1). Compound 2 was obtained as white amorphous powder. MS analysis of 2 revealed a [2 M + H]+ peak at m/z 747. By comparison of its 1H and 13C NMR data with those of (±)-nitrone 17 in the literature, high similarity between them indicated that 2 shared the same structure as the latter. However, the compound 2 is a new chiral natural product with OR at − 31.75°, but known by synthesis is racemate.
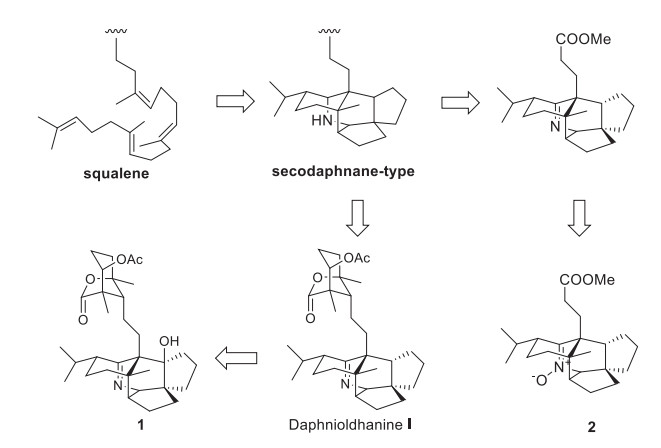
A plausible biogenetic pathway for 1 and 2 was proposed as shown in Scheme 1. Biogenetically, both 1 and 2 should be the derivatives of secodaphnane-type alkaloid [13, 29], which might be originated from sequalene, as proposed from Heathcock [26]. Then, 1 and 2 might be formed via different pathway.
Plausible Biosynthetic Pathway of 1
2.2 Cytotoxic Activity
Both compounds 1 and 2 have been tested for their cytotoxicity against Hela, MCF-7, A549, MGC-803 and COLO-205 human cancer cell lines in vitro. The results indicated that 1 exhibited weak cytotoxic activity against Hela cell line with IC50 of 31.9 μM (Table 2).
Cytotoxic activity of Compound 1 against Hela, MCF-7, A549, MGC-803 and COLO-205 human cancer cell lines in vitro
3 Experimental
3.1 General Experimental Procedures
Optical rotations were measured with a Jasco P-1020 polarimeter. UV spectra were obtained using a Shimadzu UV-2401A spectrophotometer. A Tenor 27 spectrophotometer was used for IR spectra as KBr pellets. 1D and 2D NMR spectra were recorded on Bruker spectrometer with TMS as internal standard. HRESIMS was performed on a triple quadrupole mass spectrometer. Semi-preparative HPLC was performed on an Agilent 1100 liquid chromatograph with a Waters X-Bridge Prep Shield RP18 (10 × 150 mm) column. Column chromatography (CC) was performed using silica gel (100–200 mesh and 300–400 mesh, Qingdao Marine Chemical, Inc., Qingdao, P. R. China) and Sephadex LH-20 (40–70 μm, Amersham Pharmacia Biotech AB, Uppsala, Sweden).
3.2 Plant Material
The stems of Daphniphyllum Angustifolium used in this study was collected from Jinfo mountain, Chongqing, P. R. China, in October 2013, and botanically authenticated by professor Deng Hong-ping. A voucher specimen (KIBHAO2014012) was deposited in State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences.
3.3 Extraction and Isolation
Air-dried stems of Daphniphyllum Angustifolium (20 kg) were powdered and extracted with MeOH (24 h × 3) at room temperature, and the solvent was evaporated in vacuo. The MeOH extract was then partitioned between EtOAc and TFA/H2O at pH 3.0. Water-soluble materials, after being adjusted at pH 10.0 with saturated Na2CO3, were partitioned with CHCl3. CHCl3-soluble materials (100.6 g) was subjected to silica gel column chromatography (CC) and eluted with gradient CHCl3/MeOH to yield five fractions F-1–F-5. F-4 was repeatedly submitted to silica gel CC and Sephadex LH-20, then purified by HPLC to afford compounds 1 (1.2 mg) and 2 (2.0 mg). Accordingly, 3 (18.0 mg) was obtained from F-1; 4 (2.5 mg) was obtained from F-5.
3.4 Daphnioldhanol A (1)
Daphnioldhanol A (1): White amorphous powder; C32H47NO5; Positive HR-EI-MS at m/z 526.3534 [M + H] + (calcd. for C32H48NO5, 526.3527); [α] D20 = + 9.88° (c = 0.54, MeOH); UV (MeOH) λmax (log ε) 265 (3.43) nm, 242 (3.56) nm, 215 (3.93) nm; IR: νmax (KBr) cm–1: 3440, 2928, 2869, 1772, 1743, 1713, 1631, 1383, 1226, 1057, 1028 cm–1.
3.5 Nitrone 17 (2)
(−)-Nitrone 17 (2): Colorless oil; C23H35NO3; ESI-MS (positive): m/z 747 [2 M + H] +; 1H NMR (CDCl3, 400 MHz) δH: 3.72 (3H, s), 1.58 (3H, s), 1.02 (1H, d, 6.16), 0.94 (3H, s), 0.85 (3H, d, 6.48); 13C NMR (CDCl3, 100 MHz) δC: 157.2 (C-1), 48.8 (C-2), 27.0 (C-3), 39.0 (C-4), 51.7 (C-5), 52.5 (C-6), 84.1 (C-7), 50.9 (C-8), 52.5 (C-9), 52.9 (C-10), 33.4 (C-11), 22.7 (C-12), 26.2 (C-13), 31.6 (C-14), 25.7 (C-15), 37.0 (C-16), 38.9 (C-17), 31.5 (C-18), 21.0 (C-19), 20.6 (C-20), 23.3 (C-21), 174.1 (C-22), 51.9 (C-23).
3.6 Cytotoxicity Assays
Cytotoxic activity of compound 1 against Hela, MCF-7, A549, MGC-803, and COLO-205 human cancer cell lines in vitro were measured using methylthiazoletetrazolium (MTT) assay [30]. Doxorubicin was used as a positive control.
4 Concluding Remarks
In conclusion, one new Daphniphyllum alkaloid, daphnioldhanol A (1), together with three known ones, were isolated from the stem part of D. angustifolium Hutch. Compound 1 exhibited week cytotoxic activity against Hela cell line.
Notes
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.1007/s13659-021-00309-w.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 31770392 to YTD), and the Science and Technology Program of Yunnan Province (2018ZF013 to YTD). The authors are grateful to the analytical group of the Laboratory of Phytochemistry, Kunming Institute of Botany, Chinese Academy of Sciences, for recorded spectra.
References
-
1.Z.Q. Huo, Q. Zhao, W.T. Zhu, X.J. Hao, Y. Zhang, Nat. Prod. Bioprospect. 11, 207-213 (2021) PubMed Google Scholar
-
2.M.M. Cao, H.P. He, Y.C. Gu, Q. Zhang, X.N. Li, G.Y. Zuo, Y.T. Di, C.M. Yuan, S.L. Li, Y. Zhang, X.J. Hao, Nat. Prod. Bioprospect. 3, 29-32 (2013) PubMed Google Scholar
-
3.Y.L. Zhao, M. Su, J.H. Shang, X. Wang, G.S.S. Njateng, G.L. Bao, J. Ma, Q.D. Sun, F. Yuan, J.K. Wang, X.D. Luo, Nat. Prod. Bioprospect. 10, 77-88 (2020) PubMed Google Scholar
-
4.Y. Zhang, Y.T. Di, H.Y. Liu, C.S. Li, C.J. Tan, Q. Zhang, X. Fang, S.L. Li, X.J. Hao, Helv. Chim. Acta 91, 2153-2158 (2008) PubMed Google Scholar
-
5.X.W. Ye, W.Y. Chai, X.Y. Lian, Z.Z. Zhang, Nat. Prod. Res. 31, 1390-1396 (2017) PubMed Google Scholar
-
6.A.K. Chattopadhyay, S. Hanessian, Chem. Rev. 117, 4104-4146 (2017) PubMed Google Scholar
-
7.S. Liu, J.H. Zhang, Y.T. Di, J.Y. Dong, X.J. Hao, Nat. Prod. Res. 32, 2165-2170 (2017) PubMed Google Scholar
-
8.J. Kobayashi, S. Ueno, H. Morita, J. Org. Chem. 67, 6546-6549 (2002) PubMed Google Scholar
-
9.J. Kobayashi, T. Kubota, Nat. Prod. Rep. 26, 936-962 (2009) PubMed Google Scholar
-
10.H.F. Wu, X.P. Zhang, L.S. Ding, S.L. Chen, J.S. Yang, X.D. Xu, Planta Med. 79, 1589-1598 (2013) PubMed Google Scholar
-
11.X.H. Tang, R.C. Luo, M.S. Ye, H.Y. Tang, Y.L. Ma, Y.N. Chen, X.M. Wang, Q.Y. Lu, S. Liu, X.N. Li, Y. Yan, J. Yang, X.Q. Ran, X. Fang, Y. Zhou, Y.G. Yao, Y.T. Di, J. Hao, Org. Lett. 23, 262-267 (2021) PubMed Google Scholar
-
12.S.E. Denmark, R.Y. Baiazitov, J. Org. Chem. 71, 593-605 (2006) PubMed Google Scholar
-
13.G.A. Wallace, C.H. Heathcock, J. Org. Chem. 66, 450-454 (2001) PubMed Google Scholar
-
14.Y. Zhang, Y.T. Di, Q. Zhang, S.Z. Mu, C.J. Tan, Org. Lett. 11, 5414-5417 (2009) PubMed Google Scholar
-
15.Y.H. Li, Y. Zhang, L.Y. Peng, X.N. Li, Q.S. Zhao, R.T. Li, X.D. Wu, Nat. Prod. Bioprospect. 6, 291-296 (2016) PubMed Google Scholar
-
16.Y.C. Liu, Z.J. Zhang, J. Su, L.Y. Peng, L.T. Pan, X.D. Wu, Q.S. Zhao, Nat. Prod. Bioprospect. 7, 405-411 (2017) PubMed Google Scholar
-
17.C.R. Zhang, H.B. Liu, T. Feng, J.Y. Zhu, M.Y. Geng, J.M. Yue, J. Nat. Prod. 72, 1669-1672 (2009) PubMed Google Scholar
-
18.Q. Zhang, Y.T. Di, C.S. Li, X. Fang, C.J. Tan, Z. Zhang, H.P. He, S.L. Li, X.J. Hao, Org. Lett. 11, 2357-2359 (2009) PubMed Google Scholar
-
19.Y. Zhang, Y.T. Di, H.P. He, S.F. Li, Y. Lu, N.B. Gong, X.J. Hao, Eur. J. Org. Chem. 2011, 4103-4107 (2011) PubMed Google Scholar
-
20.M.M. Cao, L. Wang, Y. Zhang, H.P. He, Y.C. Gu, Q. Zhang, Y. Li, C.M. Yuan, S.L. Li, Y.T. Di, Fitoterapia 89, 205-209 (2013) PubMed Google Scholar
-
21.T.Q. Yang, Y.T. Di, H.P. He, Q. Zhang, Y. Zhang, X.J. Hao, Helv. Chim. Acta 94, 397-403 (2011) PubMed Google Scholar
-
22.T. He, Y. Zhou, Y.H. Wang, S.Z. Mu, X.J. Hao, Helv. Chim. Acta 94, 1019-1023 (2011) PubMed Google Scholar
-
23.C.J. Tan, Y.H. Wang, Y.T. Di, H.P. He, S.Z. Mu, S.F. Li, Y. Zhang, X.J. Hao, Tetrahedron Lett. 53, 2588-2591 (2012) PubMed Google Scholar
-
24.M.M. Cao, Y. Zhang, H.P. He, S.F. Li, S.D. Huang, D.Z. Chen, G.H. Tang, S.L. Li, Y.T. Di, X.J. Hao, J. Nat. Prod. 75, 1076-1082 (2012) PubMed Google Scholar
-
25.S.Z. Mu, J.S. Wang, X.S. Yang, H.P. He, C.S. Li, Y.T. Di, Y. Wang, Y. Zhang, X. Fang, L.J. Huang, X.J. Hao, J. Nat. Prod. 71, 564-569 (2008) PubMed Google Scholar
-
26.C.H. Heathcock, D. Joe, J. Org. Chem. 60, 1131-1142 (1995) PubMed Google Scholar
-
27.K. Sasaki, Y. Hirata, Tetrahedron Lett. 13, 1275-1278 (1972) PubMed Google Scholar
-
28.Y. Zhang, Y.T. Di, S.Z. Mu, C.S. Li, Q. Zhang, C.J. Tan, Z. Zhang, X. Fang, X.J. Hao, J. Nat. Prod. 72, 1325-1327 (2009) PubMed Google Scholar
-
29.C.H. Heathcock, S. Piettre, R.B. Ruggeri, J.A. Ragan, J.C. Kath, J. Org. Chem. 57, 2554-2566 (1992) PubMed Google Scholar
-
30.A. Monks, D. Scudiero, P. Skehan, R. Shoemaker, K. Paull, D. Vistica, C. Hose, J. Langley, P. Cronise, A. VaigroWolff, J. Natl. Cancer Ⅰ(83), 757-766 (1991) PubMed Google Scholar
Copyright information
© The Author(s) 2021
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.