New amide alkaloids from Piper longum fruits
Abstract
Three new amide alkaloids piperlongumamides A-C (1-3), together with 12 known ones (4-15), were isolated from the fruits of Piper longum.The structures of the new isolates were determined using spectroscopic data analyses.Cytotoxic activity of these amides against HL-60 (human leukemia), A-549 (human lung cancer), MCF-7 (human breast cancer), SMMC-7721 (human liver cancer) and SW480 (human rectal cancer) cell lines were evaluated.Piperchabamide B (11) exhibited weak inhibitory activity against HL-60 (IC50=21.32 μM), A-549 (IC50=23.82 μM) and MCF-7 (IC50=16.58 μM) cell lines.Keywords
Piperaceae Piper longum amide alkaloids piperlongumamides piperchabamide B cytotoxicityIntroduction
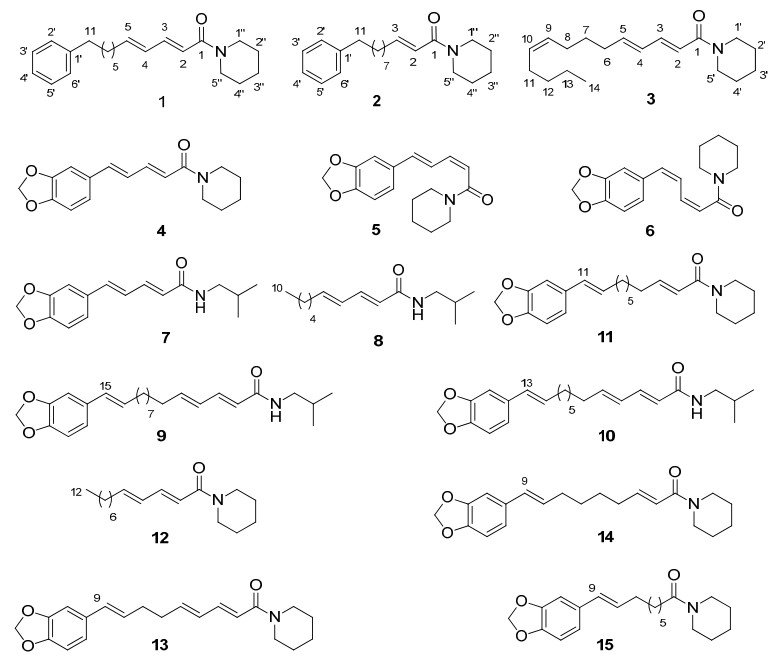
Piper longum L.(Piperaceae) is a slender aromatic climber with perennial woody roots which grows primarily in tropical regions.Its fruits and roots are used to treat various disease and ailments in traditional Chinese medicinal and ethnomedicinal practice.Uses include expectorant, curing dyspepsia, sleep problems, asthma, nausea, diarrhea, lumbarleg pain and arthralgia.1 The plant mainly contains amide alkaloids which have been used with anti-hepatitis B virus (antiHBV), 2, 3 apoptotic, 4 leishmanicidal, 5 cytotoxic, 6 mosquito larvicidal, 7, 8 phytotoxic, 9 anti-inflammatory, 10 antihyperlipidemic, 11, 12 cell adhesion inhibitory, 13 antiplatelet, 11, 14 acylCoA:cholesterol acyltransferase (ACAT) inhibitory, 15 antifungal16 and coronary vasorelaxant activities.17 In addition to amide alkaloids, phytochemicals present also include prenylated phenolic compounds18 and aromatic esters19 are found in the plant.In our continuing research on bioactive constituents of Piper species, 20-22 three new amide alkaloids piperlongumamides A–C (1–3), along with 12 previously identified ones (Figure 1) were isolated from the fruits of P.longum.The structural elucidation of the new compounds and the bioassay results are reported.
Structures of amide alkaloids 1–15 from Piper longum
Results and Discussion
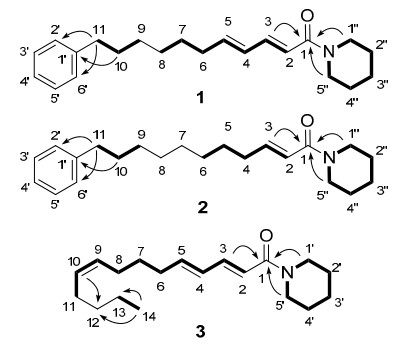
The molecular formula of compound 1, C22H31NO, was determined using the HREIMS (m/z 325.2413 [M]+), indicating eight degrees of unsaturation.Its IR spectrum showed strong absorption bands at 1722, 1636 and 1452 cm–1 implying the existence of unsaturated amide and aromatic functionalities. The 1H NMR and 13C NMR spectra of 1 (Table 1) showed signals for one monosubstituted benzene ring [δH 7.27 (2H, m) and 7.17 (3H, m)], two conjugated trans double bonds [δH 7.30 (dd, J = 14.8, 10.8 Hz), 6.24 (d, J = 14.8 Hz), 6.17 (dd, J = 15.2, 10.8 Hz) and 6.08 (dt, J = 15.2, 7.0 Hz)], one carbonyl group (δC 166.0) and several methylene groups.Moreover, amide alkaloids are major constituents of Piper plants, and the compound might be a phenylalkenoyl derivative.A piperidine ring was determined by the 1H-1H COSY spectrum (Figure 2). The length of the alkenoyl group was determined as 11 carbon atoms according to the molecular formula of compound 1. Finally, on the basis of the HMBC correlations (Figure 2) from H-3 to C-1, H2-1″ and H2-5″ to C-1, H2-10 to C-1′ and H2-11 to C-2′ and C-6′, the structure of 1 was determined to be (2E, 4E)-N-(11-phenylundecadienoyl)piperidine and was given the common name piperlongumamide A.
1H and 13C NMR spectral data of 1 and 2 in CDCl3 (δ in ppm)
Key 1H-1H COSY (bold) and HMBC (arrows, H→C) correlations of compounds 1–3
The molecular formula of compound 2, C22H33NO, was determined by HREIMS at m/z 327.2554 [M]+.Its IR spectrum showed absorption bands at 1723, 1657, 1618 and 1439 cm–1 indicating the existence of unsaturated amide and aromatic groups.Comparison of the MS and NMR data of 2 with those of 1 (Table 1), the difference was that a trans double bond at C-4(5) disappeared in compound 2.Based on the 1H-1H COSY and HMBC correlations of 2 (Figure 2), the structure of 2 was deduced to be (2E)-N-(11-phenylundecenoyl)piperidine and was given the common name piperlongumamide B.
Compound 3 has the molecular formula C19H31NO based on HREIMS at m/z 289.2414 [M]+.The IR spectrum indicated the presence of an unsaturated amide group (1720, 1713, 1630 and 1447 cm–1).The 1H and 13C NMR spectra of 3 (Table 2) displayed signals for two trans-conjugated diene moiety [δH 6.24 (1H, d, J = 15.0 Hz, H-2), 7.29 (1H, dd, J = 15.0, 11.0 Hz, H-3), 6.18 (1H, dd, J = 15.0, 11.0 Hz, H-4) and 6.08 (1H, dt, J = 15.0, 7.2 Hz, H-5)], one methyl group [δH 0.89 (3H, t, J = 6.8 Hz, H-14)], one carbonyl group (δH 166.1) and several methylene groups.Also, signals at δH 5.32 (1H, m, H-9) and 5.38 (1H, m, H-10) were due to an additional double bond, which was deduced as Z configuration by the chemical shifts of the allylic carbons [δC 26.8 (C-8) and 27.1 (C-11)].21, 22 Based on above analysis and by comparison of its NMR data with those of compound 1, compound 3 was deduced as an alkenoyl piperidine.According to 1H-1H COSY correlations of 3 (Figure 2), the connections from C-1 to C-11, C-13 to C-14 and C-1′ to C-5′ were confirmed.Finally, based on the HMBC correlations (Figure 2) from H-3, H2-1′ and H2-5′ to C-1, H-10 to C-12, Me-14 to C-12 and C-13, the structure of 3 was determined to be (2E, 4E, 9Z)-N-tetradecatrienoylpiperidine and was given the common name piperlongumamide C.
1H (600 MHz) and 13C NMR (150 MHz) spectral data of 3 in CDCl3 (δ in ppm)
The known compounds were identified as piperine (4), 23 isopiperine (5), 24 chavicine (6), 24 piperlonguminine (7), 25 pellitorine (8), 26 brachystamide B (9), 27 guineensine (10), 25 piperchabamide B (11), 28 (2E, 4E)-N-dodecadienoylpiperidine (12), 29 dehydropipernonaline (13), 12 pipernonaline (14)16 and piperolein B (15)12 by comparison of their NMR and MS data with those reported in the literature.
In a previous study, we found that an amide alkaloid 1-[(9E)-10-(3, 4-methylenedioxyphenyl)-9-decenoyl]pyrrolidine from P.boehmeriaefolium was cytotoxic.21 Therefore, all of the amides from P.longum were evaluated for their inhibitory activities against HL-60 (human leukemia), A-549 (human lung cancer), MCF-7 (human breast cancer), SMMC-7721 (human liver cancer) and SW480 (human rectal cancer) cell lines.Piperchabamide B (11) exhibited weak inhibitory activity against HL-60 cell line (IC50 = 21.32 μM), A-549 (IC50 = 23.82 μM) and MCF-7 (IC50 = 16.58 μM) (Table 3).Other tested compounds were inactive.
Cytotoxicity of piperchabamide B (11) from Piper longum*
Experimental Section
General Experimental Procedures.UV spectra were recorded on a Shimadzu double-beam 210A spectrometer (Shimadzu Co., Shimadzu, Japan).IR spectra were recorded on a Bruker Tensor 27 Fourier transform infrared spectrometer (Bruker, Karlsruhe, Germany) with KBr pellets.ESIMS and HREIMS analyses were carried out on an API Qstar-Pulsar-1 mass spectrometer (Applied Biosystems/MDS Sciex, Ontario, Canada) and Waters AutoSpec Premier P776 (Waters, Milford, USA), respectively.1H and 13C NMR spectra were collected on a Bruker AM-400, DRX-500 and Avance Ⅲ-600 spectrometers (Bruker Bio-Spin GmbH, Rheinstetten, Germany) with TMS as an internal standard.Semi-preparative HPLC was performed on an Agilent 1200 series pump (Agilent Technologies, Santa Clara, USA) equipped with a diode array detector and a Zorbax SB-C18 column (5.0 μm, φ 9.4 × 250 mm).Both analytical and preparative TLC conductedusing silica gel plates (GF254, Yantai Institute of Chemical Technology, Yantai, China).The spots were initially visualized using UV light (254 and 366 nm) and subsequently visualised by spraying a solution of 5% H2SO4 onto the TLC plate, which was subsequently heated.Column chromatography was performed using silica gel (80–100 mesh and 300–400 mesh; Qingdao Makall Group Co., Ltd., Qingdao, China), C18 silica gel (40–75 μm, Fuji Silysia Chemical, Ltd., Kasugai, Japan) and Sephadex LH-20 (GE Healthcare Bio-Xciences AB, Uppsala, Sweden).
Plant Material.The fruits of P.longum were purchased from Yikan Chinese Herbal Medicine Ltd., Qujing, China, in October 2011.The plant material was identified by Dr. Guang-Wan Hu, at Kunming Institute of Botany, Chinese Academy of Sciences.A voucher specimen (QJ1101) has been deposited at Key Laboratory of Economic Plants and Biotechnology, Kunming Institute of Botany.
Extraction and Isolation.The dried powdered fruits (20 kg) of P.longum were extracted using MeOH (4, 3 and 3 h, resp.) under reflux.The combined MeOH extracts were evaporated under reduced pressure to yield a residue, which was suspended in H2O and then partitioned successively with petroleum ether and CHCl3 to produce two corresponding portions.After TLC testing, the two portions were combined as both contained alkaloids.The combined extract (1377 g) was subjected to column chromatography over silica gel G (80–100 mesh) using petroleum ether/EtOAc (1׃0→0׃1) to yield ten fractions (Fr.A–J) and also compound 4 (1.8 g).
Fr.D (19 g) was separated using column chromatography (C18, MeOH/H2O, 80׃20) to give 8 (56.2 mg).Fr.E (12 g) was partitioned by column chromatography (C18, MeOH/H2O, 60׃40→95׃5) to produce fractions (E1–E7).Fr.E1 (1.1 g) was fractionated by column chromatography (Sephadex LH-20, MeOH; silica gel, petroleum ether/Me2CO, 1׃1) and prep. TLC (petroleum ether/EtOAc, 10׃1) to give 5 (20.0 mg), 6 (27.0 mg) and 7 (8.2 mg).
Fr.E3 (2.5 g) was fractionated by column chromatography (Sephadex LH-20, MeOH; silica gel, petroleum ether/Me2CO, 3׃1) and semi-preparative HPLC (MeOH/Me3CN/H2O, 50׃35׃15, 4 mL/min) to obtain 13 (23.8 mg, tR = 5.995 min) and 14 (37.3 mg, tR = 6.818 min).Fr.E4 was fractionated by column chromatography (Sephadex LH-20, MeOH) to give two parts (E4a and E4b).Fr.E4a (233.6 mg) was subjected to by column chromatography (silica gel, petroleum ether/Et2NH, 50׃1) and semi-preparative HPLC (MeCN/H2O, 90׃10, 3 ml/min) to give 12 (1 mg, tR = 11.984 min), 1 (2.1 mg, tR = 12.858 min), 2 (4 mg, tR = 16.182 min), 3 (8.4 mg, tR = 13.959 min).Fr.E4b (743.7 mg) was isolated repeatedly by column chromatography (silica gel, petroleum ether/Me2CO, 5׃1; petroleum ether/EtOAc, 4׃1;petroleum ether/Et2NH, 10׃1) to give 10 (17.2 mg) and 11 (18.0 mg), and a remaining fraction, which was purified by semi-preparative HPLC (MeCN/H2O, 60׃40, 4 mL/min) to give 15 (3.5 mg, tR = 17.027 min) and 11(4 mg, tR = 28.529 min).Fr.E5 (1.3 g) was fractionated by column chromatography (Sephadex LH-20, MeOH) and prep. TLC (CHCl3/EtOAc, 10׃1) to give 9 (23.9 mg).
Piperlongumamide A (1):pale yellow oil; UV (MeOH): λmax (log ε) 205 (2.77) nm.IR (KBr) νmax 1722, 1636, 1452, 1254, 748, 699, 571 cm–1.1H and 13C NMR spectral data see Table 1.ESIMS (positive) m/z 326 [M + H]+; HREIMS:m/z 325.2413 [M]+ (calcd for C22H31NO, 325.2406).
Piperlongumamide B (2):pale yellow oil; UV (MeOH): λmax (log ε) 209 (2.93) nm.IR (KBr) νmax 1723, 1657, 1618, 1439, 1219, 1069, 1023, 974, 903, 637 cm–1.1H and 13C NMR spectral data see Table 1.ESIMS (positive) m/z 350 [M + Na]+; HREIMS:m/z 327.2554 [M]+ (calcd for C22H33NO, 327.2562).
Piperlongumamide C (3):pale yellow oil; UV (MeOH): λmax (log ε) 207 (2.33) nm.IR (KBr) νmax 1720, 1713, 1630, 1447, 1253, 1229, 1166, 1082, 1022, 978, 800, 588 cm–1.1H and 13C NMR spectral data see Table 2.ESIMS (positive) m/z 290 [M + H]+; HREIMS:m/z 289.2414 [M]+ (calcd for C19H31NO, 289.2406).
MTS Assay for Cytotoxicity.The isolated amide alkaloids were tested in vitro for their cytotoxicity against proliferation of human leukemia HL-60 cell line, human lung cancer A-549, human breast cancer MCF-7, human liver cancer SMMC-7721 and human rectal cancer SW480 using the MTS assay.
The 3-(4, 5-dimethylthiazol-2-yl)-5(3-carboxymethoxyphenyl)-2-(4-sulfopheny)-2H-tetrazolium (MTS) (Promega, Beijing, China) is a mix-based cell titer assay and was performed as previous described.30 Initially, cells in their log-phase of their cycle were seeded in 96-well plates (5000– 10000 cells/well, NEST Biotechnology, Wuxi, China) using a standard 100 μL volume, with paired single cell suspension containing 10% fetal bovine serum (DMEM or RMPI1640, Thermo Fisher Scientific, Beijing, China).The cell were further treated with indicated concentrations of the compounds dissolved in DMSO, which were set the regular thickness 40 μM for the initial screening and five concentrations of each compound were fixed in the concentration compounds inhibited tumor cell growth by approximately 50% to achieve a total culture medium in a volume of 200 μL.After incubation for 48 h at 37 ℃, a 20 μL of MTS solution and 100 μL DMEM were added into the well, incubation was continued for another 1–4 h.The absorbance was measured at the detection wavelength of 490 nm (L1) and the reference wavelength of 680 nm (L2)31 and cytotoxicity for each compound was expressed as IC50 values.
Notes
Electronic Supplementary Material
Supplementary material is available in the online version of this article at http://dx.doi.org/10.1007/s13659-013-0073-0 and is accessible for authorized users.
Acknowledgments
This work was funded by the Natural Science Foundation of Yunnan Province, China (No.2011FZ205), the National Natural Science Foundation of China (Nos.31070288, 31161140345), the Ministry of Education of China through its 111 and 985 projects (Nos.B08044, MUC98506-01000101 & MUC985-9), and the Japan Society for the Promotion of Science (No.JSPS/AP/109080).
References
-
1.Editorial Board of 'Zhonghua Bencao', State Administration of Traditional Chinese Medicine of the People's Republic of China, Zhonghua Bencao[M]. Shanghai: Shanghai Scientific and Technical Publishing House, 1999, 434. PubMed Google Scholar
-
2.Z.Y. Jiang, W.F. Liu, X.M. Zhang, J. Luo, Y.B. Ma, J.J. Chen, Bioorg.Med.Chem.Lett. 23, 2123-2127 (2013) CrossRef PubMed Google Scholar
-
3.Z.Y. Jiang, W.F. Liu, C.G. Huang, X.Z. Huang, Fitoterapia 84, 222-226 (2013) CrossRef PubMed Google Scholar
-
4.W. Lee, K.Y. Kim, S.N. Yu, S.H. Kim, S.S. Chun, J.H. Ji, H.S. Yu, S.C. Ahn, Biochem.Biophys.Res.Commun. 430, 406-412 (2013) CrossRef PubMed Google Scholar
-
5.S. Ghosal, A. Deb, P. Mishra, R. Vishwakarma, Planta Med. 78, 906-908 (2012) CrossRef PubMed Google Scholar
-
6.P. Mishra, S. Sinha, S. Guru, S.K. Bhushan, R.A. Vishwakarma, S. Ghosal, J.Asian Nat.Prod.Res. 13, 143-148 (2011) CrossRef PubMed Google Scholar
-
7.S.K. Madhu, V.A. Vijayan, A.K. Shaukath, Asian Pac.J.Tropical Med. 4, 112-116 (2011) CrossRef PubMed Google Scholar
-
8.Y.C. Yang, S.G. Lee, H.K. Lee, M.K. Kim, S.H. Lee, H.S. Lee, J.Agric.Food Chem. 50, 3765-3767 (2002) CrossRef PubMed Google Scholar
-
9.H.Z. Huang, C.M. Morgan, R.N. Asolkar, M.E. Koivunen, P.G. Marrone, J.Agric.Food Chem. 58, 9994-10000 (2010) CrossRef PubMed Google Scholar
-
10.S.W. Lee, M.S. Kim, M.H. Park, S.J. Park, W.S. Lee, J.S. Chang, M.C. Rho, Bull.Korean Chem.Soc. 31, 921-924 (2010) CrossRef PubMed Google Scholar
-
11.Z. Jin, G. Borjihan, R. Zhao, Z. Sun, G.B. Hammond, T. Hammond, Phytother.Res. 23, 1194-1196 (2009) CrossRef PubMed Google Scholar
-
12.S.W. Lee, M.C. Rho, H.R. Park, J.H. Choi, J.Y. Kang, J.W. Lee, K. Kim, H.S. Lee, Y.K. Kim, J.Agric.Food Chem. 54, 9759-9763 (2006) CrossRef PubMed Google Scholar
-
13.S.W. Lee, Y.K. Kim, K. Kim, H.S. Lee, J.H. Choi, W.S. Lee, C.D. Jun, J.H. Park, J.M. Lee, M.C. Rho, Bioorg.Med.Chem.Lett. 18, 4544-4546 (2008) CrossRef PubMed Google Scholar
-
14.M. Iwashita, N. Oka, S. Ohkubo, M. Saito, N. Nakahata, Eur.J.Pharmacol. 570, 38-42 (2007) CrossRef PubMed Google Scholar
-
15.S.W. Lee, M.C. Rho, J.Y. Nam, E.H. Lim, O.E. Kwon, Y.H. Kim, H.S. Lee, Y.K. Kim, Planta Med. 70, 678-679 (2004) CrossRef PubMed Google Scholar
-
16.S.E. Lee, B.S. Park, M.K. Kim, W.S. Choi, H.T. Kim, K.Y. Cho, S.G. Lee, H.S. Lee, Crop Prot. 20, 523-528 (2001) CrossRef PubMed Google Scholar
-
17.N. Shoji, A. Umeyama, N. Saito, T. Takemoto, A. Kajiwara, Y. Ohizumi, J.Pharm.Sci. 75, 1188-1189 (1986) CrossRef PubMed Google Scholar
-
18.O. Ohno, T. Watabe, K. Nakamura, M. Kawagoshi, N. Uotsu, T. Chiba, M. Yamada, K. Yamaguchi, K. Yamada, K. Miyamoto, Biosci.Biotechnol.Biochem. 74, 1504-1506 (2010) CrossRef PubMed Google Scholar
-
19.S. Kumar, P. Arya, C. Mukherjee, B.K. Singh, N. Singh, V.S. Parmar, A.K. Prasad, B. Ghosh, Biochem. 44, 15944-15952 (2005) CrossRef PubMed Google Scholar
-
20.H.X. Liu, K. Chen, Q.Y. Sun, F.M. Yang, G.W. Hu, Y.H. Wang, C.L. Long, J.Nat.Prod. 76, 732-736 (2013) CrossRef PubMed Google Scholar
-
21.G.H. Tang, D.M. Chen, B.Y. Qiu, L. Sheng, Y.H. Wang, G.W. Hu, F.W. Zhao, L.J. Ma, H. Wang, Q.Q. Huang, J.J. Xu, C.L. Long, J. Li, J.Nat.Prod. 74, 45-49 (2011) CrossRef PubMed Google Scholar
-
22.S.X. Yang, Q.Y. Sun, F.M. Yang, G.W. Hu, J.F. Luo, Y.H. Wang, C.L. Long, Planta Med. 79, 693-696 (2013) CrossRef PubMed Google Scholar
-
23.R.A. Olsen, G.O. Spessard, J.Agric.Food Chem. 29, 942-944 (1981) CrossRef PubMed Google Scholar
-
24.K. Hashimoto, T. Yaoi, H. Koshiba, T. Yoshida, T. Maoka, Y. Fujiwara, Y. Yamamoto, K. Mori, Food Sci.Technol.Int. 2, 24-29 (1996) PubMed Google Scholar
-
25.S.A. Lee, J.S. Hwang, X.H. Han, C. Lee, M.H. Lee, S.G. Choe, S.S. Hong, D. Lee, M.K. Lee, B.Y. Hwang, Arch.Pharmacal Res. 31, 679-683 (2008) CrossRef PubMed Google Scholar
-
26.I. Yasuda, K. Takeya, H. Itokawa, Chem.Pharm.Bull. 29, 564-566 (1981) CrossRef PubMed Google Scholar
-
27.M.J. Abad Martínez, P.B. Benito, Stud.Nat.Prod.Chem. 30, 393-418 (2005) CrossRef PubMed Google Scholar
-
28.T. Morikawa, H. Matsuda, I. Yamaguchi, Y. Pongpiriyadacha, M. Yoshikawa, Planta Med. 70, 152-159 (2004) CrossRef PubMed Google Scholar
-
29.R.N. Shakhmaev, A.U. Ishbaeva, V.V. Zorin, Russ.J.Org.Chem. 48, 908-913 (2012) CrossRef PubMed Google Scholar
-
30.X. Yang, D.D. Yuan, X.J. Jiang, Z.J.J. Xi, Peking Univ.(Health Sci.). 45, 221-226 (2013) PubMed Google Scholar
-
31.S.A.A. Ghafar, M. Ismail, L.S. Yazan, S. Fakurazi, N. Ismail, K.W. Chan, P.M. Tahir, Evid-Based Complement.Altern.Med. 2013, 1-8 (2013) PubMed Google Scholar
Copyright information
© The Author(s) 2013
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.