Three new physalins from Physalis alkekengi var. franchetii
Abstract
Three new physalin steroids, physalin Ⅲ (1), physalin Ⅳ (2), 3-O-methylphysalin Ⅹ (3), together with five known physalins (4-8) were isolated from the 80% EtOH extract of calyces of Physalis alkekengi var. franchetii. The structures of the new compounds were revealed through 1D and 2D NMR and mass spectroscopic studies.Keywords
Physalis alkekengi var. franchetii steroid physalin neophysalinIntroduction
The genus of Physalis (Solanaceae), including about 120 species, is widely distributed throughout South and North America, which also includes five species, and two variations which also can be found in China1. The plant Physalis alkekengi var. franchetii, is used in traditional Chinese medicine2, and is chiefly used for the treatment and prevention of tumors, leishmaniasis, sore throat, cough, eczema, hepatitis and urinary problems3. The chemical constituents of P. alkekengi var. franchetii are mainly composed of alkaloids, flavonoids, sterols, fatty acids, amino acids, and steroids4.
Physalins, as one of the characteristic constituents from P. alkekengi var. franchetii, is a type of steroids with 16, 24-cyclo-13, 14-seco-ergostane skeleton which has been established by X-ray crystallographic analysis5-7. Neophysalins, are another characteristic structure, and are transformed from physalins through an acid-induced benzilic acid-type rearrangement reaction8. Until recently, more than 50 physalins and neophysalins have been identified from P. alkekegi var. franchetii, P. agulata, P. lancifolia and P. minima. Physalins and neophysalins have tremendous biological activities, including anti-tumor9, anti-microbial10, anti-malarial11, immunosuppressive12, anti-inflammatory13, 14, immunomodulatory15, cytotoxic16, and trypanocidal17, 18 effects.
In this study, five physalins (1, 2, 4-6) and three neophysalins (3, 7, 8) were isolated from P. alkekengi var. franchetii, three of which (1-3) were new. Their chemical structures and their elucidation are described herein.

|
Results and Discussion
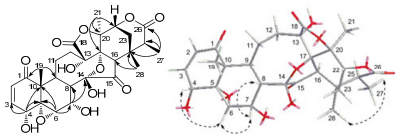
Compound 1 was obtained as a white powder. The molecular formula of 1 was determined to be C28H32O12 by HREIMS (m/z 560.1885, [M]+; calcd 560.1894) and NMR data (Table 1). The 1H NMR spectrum of 1 (in pyridine-d5) showed four methyl signals at δH 1.29, 1.47, 1.65, and 2.02, four oxygenated signals at δH 4.01 (H-4), 3.54 (H-6), 5.70 (H-7) and 4.73 (H-22), and two olefinic proton signals at δH 6.24 (H-2) and 7.19 (H-3). The 13C DEPT spectrum of 1 (in pyridine-d5) displayed the presence of twenty-eight carbons, including four methyls, three methylenes, ten methines (two olefinic carbons and four oxymethanes), eleven quaternary carbons (two ketone groups, two lactone groups, four oxygenated carbons, and one ketal carbon). These signals indicated the structural similarity of 1 and physalin J19. The chemical shift of the ketal carbon at C-14 (δC 103.4) of 1 differently from that (δC 107.8) of physalin J, together with one more methyl signal and less an oxygenated CH2 in 1 indicated that the C(27)-O-C(14) ether bridge was ruptured. This was further determined by the HMBC correlations (Figure 1) from Me-27 (δH 1.29) to C-25 (δC 42.4) and C-26 (δC 173.0). Moreover, the HMBC correlations from δH 1.65 (H-19), 6.24 (H-2), and 7.19 (H-3) to δC 202.9 (C-1) demonstrated the presence of an conjugated 2-en-1-one group, and the 1H-1H COSY correlations of δH 7.19 (H-3)/δH 4.01 (H-4) revealed that there existed an hydroxyl group at C-4. An oxymethine in 1 instead of a methene in physalin J suggests that one hydroxyl group could be located at C-7, which was confirmed by the 1H-1H COSY correlations of δH 3.54 (H-6)/δH 5.70 (H-7) and δH 5.70 (H-7)/δH 2.93 (H-8). The relative configurations of H-8 and Me-28 in the physalin skeleton were established to be β-oriented by X-ray.20, 21 Thus, the NOE correlations (Figure 1) of H-4/H-6, H-6/H-7, H-7/H-8β, and H-27/Me-28 established the relative configuration of OH-4 and OH-7 to be α-oriented and Me-27 to be β-oriented. Hence, the structure of compound 1 was identified to be physalin Ⅲ (1).
1H NMR (600 MHz) and 13C DEPT (150 MHz) data of compounds 1–3
Key HMBC (H→C) and ROESY(H 
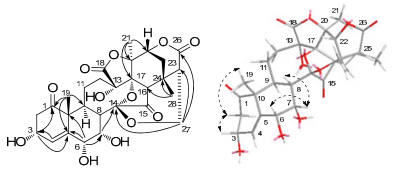
Compound 2 was obtained as a colorless needle crystal. The molecular formula of 2 was determined to be C28H32O12 on the basis of HREIMS (m/z 560.1854, [M]+; calcd 560.1894) and NMR data (Table 1). The NMR data of 2 were similar to those of physalin Z22, and the differences between them were two of the double-bond carbon signals in physalin Z were replaced by two oxygenated methine signals in 2 located at C-6 and C-7, which were deduced by 1H-1H COSY correlations of H-8 (δH 3.39)/H-7 (δH 5.23) and H-7 (δH 5.23)/H-6 (δH 4.79) and by HMBC correlations from H-6 to C-10, C-4, C-8, C-7, and C-5, from H-7 to C-6, C-5, C-8, and C-14. The ROESY correlations (Figure 2) of H-6/H-7 and H-7/H-8β revealed that OH-C(6) and OH-C(7) were α-oriented. So the structure of compound 2 was deduced and named as physalin Ⅳ (2).
Key HMBC (H→C) and ROESY(H 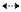
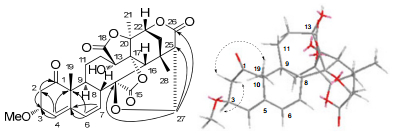
Compound 3 was obtained as an amorphous yellow powder. The molecular formula of 3 was determined as C29H32O10 using HREIMS and NMR data (Table 1). These NMR data of 3 were similar to those of physalin X23, except the presence of an additional methoxyl group. Comparing with physalin X, the chemical shift of C-3 (δC 76.7) at down field and the HMBC correlation (Figure 3) of OMe (δH 3.27) with C-3 in 3 indicated a methoxyl group at C-3. The relative configuration of Me-19 and H-16 in physalin skeleton were established to be β-oriented by X-ray.20, 21 So the relative configuration of 3-OMe could be determined as α-oriented by the NOE correlations of Me-19/H-3 and H-16/H-3. Hence, the structure of compound 3 was identified to be 3-O-methylphysalin X (3).
Key HMBC (H→C) and ROESY(H 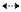
The structures of five known steroidal compounds, physalin Z (4)22, physalin C (5)24, physalin I (6)25, physalin P (7)26, 25, 27-dihydro-4, 7-dedehydro-7-deoxyneophysalin A (8)27, were elucidated in comparison to their spectroscopic data with those reported in the literature.
Experimental Section
General Experimental Procedures
Column chromatography (CC): silica gel (100-200 mesh, 200-300 mesh; Qingdao Marine Chemical Co., Ltd.), Sephadex LH-20 (Pharmacia). Thin-layer chromatography (TLC): silica gel 60F254 (Qingdao Marine Chemical Co., Ltd.). Optical rotations: Jasco P-1020 polarimeter. UV Spectra: Shimadzu UV-2401 PC spectraphotometer. IR Spectra: Bruker Tensor-27 infrared spectrophotometer, with KBr pellets in cm-1. NMR Spectra: Bruker Bruker Avance Ⅲ-600MHz spectrometer; chemical shifts δ were recorded as ppm relative to TMS, coupling constants J in Hz. ESIMS: API QSTAR Pulsar spectrometer; HREIMS: Waters Autospec Premier P776.
Plant Material
The calyces of Phsalis alkekengi var. franchetii were purchased in Kunming CTM market, Kunming city, Yunnan province at December 2011 and were identified by Prof. Xi-Wen Li who was botanic taxonomist in Kunming Institute of Botany.
Extraction and Isolation
Dried calyces (15 kg) of P. alkekengi var. franchetii were extracted with 80% EtOH under reflux. The extract was suspended in water (5 L) and then partitioned successively with PE (petroleum ether) (8 L × 4) and EtOAc (8 L × 5). The EtOAc-soluble layer (evaporated under vacuum to get 250 g) eluted successively with CHCl3, CHCl3-MeOH (100:1), CHCl3-MeOH (20:1), and CHCl3-MeOH (5:1). Then, the CHCl3-MeOH (100:1) portion (80 g) was chromatographed on a silica gel column using a gradient of CHCl3-CH3OH (100:0 to 0:100), which yielded five fractions (1-5). Fraction 2 was successively subjected to RP-18, Sephadex LH-20 and silica gel to derive subfractions Ⅰ-Ⅳ. Subfraction Ⅱ was further separated over silica gel with CHCl3-CH3OH (120:1) to yield compound 3 (5 mg), 6 (4 mg), 7 (10 mg) and 8 (3 mg). Subfraction Ⅲ was subjected to semipreparative HPLC (CH3CN-H2O, 50:50) to produce 4 (1 mg) and 5 (1 mg). Fraction 3 was successively subjected to RP-18, Sephadex LH-20 and separated over silica gel with CHCl3-CH3OH (80:1) to yield 1 (2 mg) and 2 (6 mg).
Physalin Ⅲ (1)
white powder; [α]D21-29.33 (c 0.2, CHCl3); UV (CHCl3): λmax (log ε): 240 (3.36) nm; IR (KBr): vmax 3430, 2924, 1787, 1758, 1728, 1462, 1376, 1218, 1073, 1041 cm-1; 1H and 13C NMR: see Table 1. Negative ESIMS: 559 ([M-H]-). HREIMS: 560.1885 [M]+ (calcd. for C28H32O12, 560.1894).
Physalin Ⅳ (2)
colorless needle crystal (CHCl3); [α]D16 -114.40 (c 0.1, MeOH); UV (MeOH): λmax (log ε): 201 (4.23) nm; IR (KBr): vmax 3431, 1781, 1729, 1632, 1383, 1374, 1167, 1134, 1065 cm-1; 1H and 13C NMR: see Table 1. Positive ESIMS: 583 ([M + Na]+). HREIMS: 560.1854 [M]+ (calcd. for C28H32O12, 560.1894).
3-O-Methylphysalin X (3)
yellow powder; [α]D18 -16.00 (c 0.1, pyridine); UV (MeOH): λmax (log ε): 225 (3.77), 201 (3.80) nm; IR (KBr): vmax 3432, 2923, 1786, 1629 cm-1; 1H and 13C NMR: see Table 1. Positive ESIMS: 563 ([M + Na]+). HREIMS: 540.1933 [M]+ (calcd. for C29H32O10, 540.1995).
Electronic Supplementary Material
Supplementary material is available in the online version of this article at http://dx.doi.org/10.1007/s13659-013-0021-z and is accessible for authorized users.
Notes
Acknowledgements
This project was financially supported by the National Major Basic Research Program (No. SB2007FY400) of the National Ministry of Science and Technology of China, the National Knowledge Innovation Program of Chinese Academy of Sciences (No. KSCX2-YW-G-038), and the Foundation of State Key Laboratory of Phytochemistry and Plant Resources in West China (P2010-ZZ14).
Compliance with Ethical Standards
References
-
1.K. R. Kuang, Flora Republicae Popularis Sinicae (Zhongguo Zhiwu Zhi) 67, 10 (1978) PubMed Google Scholar
-
2.N. P. Committee, Pharmacopeia of People's Republic of China (the 2010 edition) 1, 337-338 (2010) PubMed Google Scholar
-
3.H. Y. Wu, Q. L. Suo, J. Li, Neimenggu Shiyou Huagong 4, 5-8 (2007) PubMed Google Scholar
-
4.N. Zhang, Z. M. Bie, W. J. Qing, Y. Zhang, H. J. Ge, C. Shu, H. G. Shi, H. B. Ma, Journal of Jilin Medical College 29, 104-107 (2008) PubMed Google Scholar
-
5.M. Kawai, T. Matsuura, T. Taga, K. Osaki, J. Chem. Soc. B, 812-815 (1970) PubMed Google Scholar
-
6.M. Kawai, B. Makino, T. Taga, Y. Miwa, T. Yamamoto, T. Furuta, H. Yamamura, Y. Butsugan, K. Ogawa, M. Hayashi, Bull. Chem. Soc. Jpn. 67, 222-226 (1994) CrossRef PubMed Google Scholar
-
7.T. Taga, Y. Miwa, K. Machida, M. Kawai, Y. Buisugan, Acta Crystallogr. C 47, 2188-2191 (1991) CrossRef PubMed Google Scholar
-
8.M. Kawai, T. Ogura, Y. Butsugan, T. Taga, M. Hayashi, Tetrahedron 47, 2103-2110 (1991) CrossRef PubMed Google Scholar
-
9.H. Han, L. Qiu, X. Wang, F. Qiu, Y. Wong, X. Yao, Biol. Pharm. Bull. 34, 1584-1588 (2011) CrossRef PubMed Google Scholar
-
10.M. T. G. Silva, S. M. Simas, T. G. F. M. Batista, P. Cardarelli, T. C. B. Tomassini, Mem. Inst. Oswaldo Cruz 100, 779-782 (2005) CrossRef PubMed Google Scholar
-
11.M. S. Sa, M. N. de Menezes, A. U. Krettli, I. M. Ribeiro, T. C. B. Tomassini, D. S. R. Ribeiro, W. F. de Azevedo, M. B. P. Soares, J. Nat. Prod. 74, 2269-2272 (2011) CrossRef PubMed Google Scholar
-
12.Y. Yu, L. Sun, L. Ma, J. Li, L. Hu, J. Liu, Int. Immunopharmacol. 10, 290-297 (2010) CrossRef PubMed Google Scholar
-
13.N. B. Pinto, T. C. Morais, K. M. B. Carvalho, C. R. Silva, G. M. Andrade, G. A. C. Brito, M. L. Veras, O. D. L. Pessoa, V. S. Rao, F. A. Santos, Phytomedicine 17, 740-743 (2010) CrossRef PubMed Google Scholar
-
14.R. D. Budhiraja, S. Sudhir, K. N. Garg, Planta Med. 50, 134-136 (1984) CrossRef PubMed Google Scholar
-
15.Y. S. Lin, H. C. Chiang, W. S. Kan, E. Hone, S. J. Shih, M. H. Won, Am. J. Chin. Med. 20, 233-243 (1992) CrossRef PubMed Google Scholar
-
16.S. Habtemariam, Planta Med. 63, 15-17 (1997) CrossRef PubMed Google Scholar
-
17.F. Abe, S. Nagafuji, M. Okawa, J. Kinjo, Chem. Pharm. Bull. 54, 1226-1228 (2006) CrossRef PubMed Google Scholar
-
18.N. C. Vieira, L. S. Espindola, J. M. Santana, M. L. Veras, O. D. L. Pessoa, S. M. Pinheiro, d. A. R. Mendonca, M. A. S. Lima, E. R. Silveira, Bioorg. Med. Chem. 16, 1676-1682 (2008) CrossRef PubMed Google Scholar
-
19.L. R. Row, N. S. Sarma, K. S. Reddy, T. Matsuura, R. Nakashima, Phytochemistry 17, 1647-1650 (1978) CrossRef PubMed Google Scholar
-
20.M. Kawai, T. Matsuura, T. Taga, K. Osaki, J. Chem. Soc. B, 812-815 (1970) PubMed Google Scholar
-
21.M. Kawai, B. Makino, T. Taga, Y. Miwa, T. Yamamoto, T. Furuta, H. Yamamura, Y. Butsugan, K. Ogawa, M. Hayashi, Bull. Chem. Soc. Jpn. 67, 222-226 (1994) CrossRef PubMed Google Scholar
-
22.L. Qiu, F. Zhao, Z. H. Jiang, L. X. Chen, Q. Zhao, H. X. Liu, X. S. Yao, F. Qiu, J. Nat. Prod. 71, 642-646 (2008) CrossRef PubMed Google Scholar
-
23.R. Chen, J. Y. Liang, R. Liu, Helv. Chim. Acta. 90, 963-966 (2007) CrossRef PubMed Google Scholar
-
24.M. Kawai, T. Matsuura, Tetrahedron 26, 1743-1745 (1970) CrossRef PubMed Google Scholar
-
25.L. R. Row, K. S. Reddy, N. S. Sarma, T. Matsuura, R. Nakashima, Phytochemistry 19, 1175-1181 (1980) CrossRef PubMed Google Scholar
-
26.M. Kawai, A. Matsumoto, B. Makino, H. Mori, T. Ogura, Y. Butsugan, K. Ogawa, M. Hayashi, Bull. Chem. Soc. Jpn. 66, 1299-1300 (1993) CrossRef PubMed Google Scholar
-
27.R. Sunayama, M. Kuroyanagi, K. Umehara, A. Ueno, Phytochemistry 34, 529-533 (1993) CrossRef PubMed Google Scholar
Copyright information
© The Author(s) 2013
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.