Structures and cytotoxicities of three new sesquiterpenes from cultures of Armillaria sp.
Abstract
Three new sesquiterpene aryl esters, named 10-dehydroxy-melleoliede B(1), 1-O-formyl-10-dehydroxy-melleoliede B(2) and 10-oxo-melleoliede B(3) together with six known ones(4-9), were isolated from the cultures of Armillaria sp. The structures of the new compounds were elucidated based on the extensive spectroscopic methods. Compounds 1, 2, and 5-9 exhibited moderate cytotoxicities.Keywords
basidiomycete Armillaria sp. sesquiterpene aryl ester cytotoxicitiesIntroduction
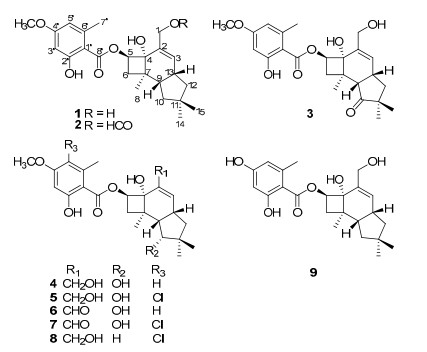
The basidiomycete genus Armillaria is recognized for the production of rich secondary metabolites with structure diversities as well as interesting biological activities.1 Among those metabolites, sesquiterpene aryl esters are quite a big family which showed antibacterial, antifungal activities,1c, 1j and cytotoxicity.1o, 1p As a part of our search for naturally occurring secondary metabolites with diverse structures from fungi in China,2 we have carried out the chemical investigation of Armillaria sp. cultures, which led to the isolation of three new sesquiterpene aryl esters, named 10-dehydroxy-melleoliede B (1), 1-O-formyl-10-dehydroxy-melleoliede B (2), and 10-oxo-melleoliede B (3), together with melleolide B (4),1g 5'-chloromelleolide (5),1g armillarigin (6),1g armillarikin (7),1g A52a (8),3 and armillane (9).1p Those new compounds were elucidated by means of spectroscopic methods, while the known compounds were identified by comparison with data in the literature. All of these compounds were evaluated for cytotoxicity against five human cancer cell lines.
Results and Discussion
|
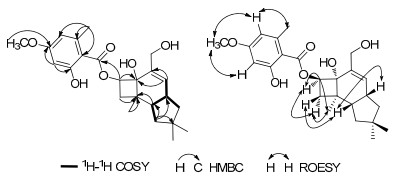
Compound 1, obtained as a colorless oil, had the molecular formula of C24H32O6 based on the negative-ion HRESIMS at m/z 415.2118 [M - H]- (calcd for C24H31O6, 415.2120). The IR data exhibited absorption bands for a hydroxy at 3430 cm-1, an ester group at 1646 cm-1 and a substituted aromatic ring at 1617 cm-1 and 1578 cm-1,1 which consisted with the UV spectra. The 1H NMR spectrum (Table 1) of 1 exhibited chelated phenolic hydroxy proton at δH 11.63, two aromatic protons at δH 6.31, one methoxy group at δH 3.80, four methyls including an aromatic one at δH 2.41. The 13C NMR spectrum revealed 24 carbons resonances (Table 2), which were classified into five methyl groups (three aliphatic, one oxygenated and one aromatic), four aliphatic methylenes (one oxygenated), six methines (three olefinic), and nine quaternary carbons (one carbonyl), indicating the presence of a tetrasubstituted benzene unit as well as one ester carbonyl signal at δC 171.8. The above data were quite closely related to those of melleolide B (4).1g The significant differences of the 13C NMR data between 1 and 4 were the lack of a secondary alcohol signal and the presence of a methylene signal at δC 41.9 in 1, revealing the absence of HO-10 in 1. The above assumption was further confirmed by the 1H-1H COSY correlations of H-10/H-9/H-13/H-12, and the HMBC correlations from H-10 to C-9, C-11, and C-13 (Figure 1).
1H NMR data of 1-3 (1, 3 in Me2CO-d6; 2 in CDCl3)
13C NMR data of 1-3 (1, 3 in Me2CO-d6; 2 in CDCl3)
Key HMBC, 1H-1H COSY, and ROESY correlations for 1
The ROESY correlations (Figure 1) of H-9/H-13, H-9/H-6β, H-5/H-6α, H-5/H-8, and H-6α/H-8 of 1, as well as the similar chemical shifts of the chiral carbons comparing with those of melleolide B, suggested the cis-anti-cis stereochemistry of the three rings.4 Therefore, the structure of compound 1 was established as 10-dehydroxy-melleoliede B.
Compound 2, purified as a colorless oil, had a molecular formula of C25H32O7 according to its negative-ion HRESIMS at m/z 443.2070 ([M - H]-, calcd, 443.2069). The IR spectrum showed the presence of a hydroxy group at 3432 cm-1 and an ester moiety at 1629 cm-1.The 1D NMR (Tables 1 and 2) data were quite similar with those of 1 except for an additional signal for a formyl group (δH 7.87, δC 160.8). The formyl group attached at C-1 was confirmed by the HMBC of H-1 to C-CHO. Detailed analysis of 1D and 2D NMR data suggested that the other parts of 2 were the same to those of 1. Therefore, compound 2 was established as 1-O-formyl-10-dehydroxy-melleoliede B.
Compound 3, was obtained as a colorless oil. The negative-ion HRESIMS afforded the molecular formula of C24H30O7 at m/z 429.1917 ([M - H]-, calcd, 429.1913). The IR data exhibited absorption bands for a hydroxy at 3431 cm-1, an ester group at 1642 cm-1 and a substituted aromatic ring at 1618 cm-1 and 1579 cm-1, which were similar with those of compound 1. Detailed analysis of the 13C NMR suggested that the main difference between 3 and 1 was that a methine was oxidized into a keto carbonyl group at δC 220.6 (C-10), which resulted in the downfield shift of C-9 (Δδ 7.1 ppm), C-11 (Δδ 6.2 ppm) in compound 3. The above conclusion was supported by the HMBC correlations of H-9, H-13, and H-14 to C-10, and H-3 (δH 5.98) to C-12 (δC 41.9). Further analysis of 1D and 2D NMR data and coupling constants suggested that the other parts of the molecular was the same to compound 1. Thus, compound 3 was established as 10-oxo-melleoliede B.
All compounds were evaluated for their cytotoxicity against five human cancer cell lines using the MTT method as reported previously.5 Compounds 1, 2, and 5-9 showed cytotoxicities as shown in Table 3. It is notable that compound 2 showed stronger cytotoxicities than that of the positive control in MCF-7 and SW480 cell lines.
Cytotoxicity for compounds 1-9 (IC50, μM)
Experimental Section
General Experimental Procedures. Optical rotations (OR) were recorded on a Jasco P-1020 digital polarimeter. UV and the IR spectra were obtained on a Shimadzu UV2401PC and a Bruker Tensor 27 FT-IR spectrometer with KBr pellets. Nuclear Magnetic Resonance (NMR) spectra were obtained on a BRUKER AM-400 and a BRUKER DRX-500 MHz spectrometer with tetramethylsilane (TMS) as an internal standard at room temperature. High-resolution (HR) ESIMS were recorded on an API QSTAR Pulsar spectrometer. Silica gel (200-300 mesh, Qingdao Marine Chemical Ltd., China) and Sephadex LH-20 (Amersham Biosciences, Sweden) were used for open column chromatography (CC). Preparative HPLC was performed on an Agilent 1100 liquid chromatography system equipped with a Zorbax SB-C18 column (9.4 mm × 150 mm). Fractions were monitored by TLC. Spots were visualized by heating silica gel plates immersed in Vanillin-H2SO4 in ethanol.
Fungal Material and Cultivation Conditions. The fungus Arimillaria sp. was collected from Wuding, in Yunnan Province, China, in 2005. The culture medium consisted of glucose (5%), peptone from porcine meat (0.15%), yeast powder (0.5%), KH2PO4 (0.5%) and MgSO4. Fermentation was carried out on a shaker at 160RPM for 25 days.
Extraction and Isolation. The culture broth (21 L) was filtered, and the filtrate was extracted three times with ethyl acetate, while the mycelium was extracted three times with CHCl3-MeOH (1:1). The EtOAc layer together with the mycelium extraction was concentrated under reduced pressure to give a crude extract (20 g), and this residue was chromato-graphed on silica gel (200-300 mesh) column eluted with a gradient of CHCl3-MeOH (1:0 → 0:1) to obtain six fractions (1-6). Fraction 4 was subjected to a silica gel CC (petroleum ether-EtOAc, 6:1) to give 4 sub fractions (4a-4d). Subfraction 4c was further purified by Sephadex LH-20 column (eluted with acetone) to give 2 (6 mg) while 4d was subjected to Sephadex LH-20 column (eluted with CHCl3-MeOH, 1:1) followed by semipreparative HPLC (MeCN-H2O, eluting from 55:45 to 100:0 for 40 min with a flow rate of 10 mL/min) to yield 1 (14 mg) and 3 (11 mg).
10-Dehydroxy-melleoliede B (1): colorless oil; [α]D15 + 41.4 (c 1.08, MeOH); UV (MeOH) λmax (log ε) 302 (3.73), 264 (4.13), 215 (4.35) nm; IR (KBr) νmax 3430, 2949, 2864, 1646, 1617, 1578, 1444, 1257, 1160 cm-1; 1H (400 MHz) and 13C NMR (100 MHz) data (Me2CO-d6), see Tables 1 and 2; negative ion HRESIMS m/z 415.2118 [M - H]- (calcd for C24H31O6, 415.2120).
1-O-Formyl-10-dehydroxy-melleoliede B (2): colorless oil; [α]D15 + 29.4 (c 0.29, MeOH); UV (MeOH) λmax (log ε) 302 (3.72), 264 (4.12), 215 (4.35) nm; IR (KBr) νmax 3432, 2920, 1629 cm-1; 1H (400 MHz) and 13C NMR (125 MHz) data (CDCl3), see Tables 1 and 2; negative ion HRESIMS m/z 443.2070 ([M - H]-, calcd 443.2069).
10-Oxo-melleoliede B (3): colorless oil; [α]D15 - 42.5 (c 0.28, MeOH); UV (MeOH) λmax (log ε) 302 (3.70), 265 (4.10), 215 (4.33) nm; IR (KBr) νmax 3431, 2928, 1730, 1642, 1618, 1579, 1256, 1160 cm-1; 1H (400 MHz) and 13C NMR (100 MHz) data (Me2CO-d6), see Tables 1 and 2; negative ion HRESIMS m/z 429.1917 ([M - H]-, calcd 429.1913).
Cytotoxicity Assay. Five human cancer cell lines, breast cancer MCF-7, hepatocellular carcinoma SMMC-7721, human myeloid leukemia HL-60, colon cancer SW480, and lung cancer A-549 cells, were used in the cytotoxic assay. All the cells were cultured in RPMI-1640 or DMEM medium (Hyclone, USA), supplemented with 10% fetal bovine serum (Hyclone, USA) in 5% CO2 at 37 ℃. The cytotoxicity assay was performed according to the MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide) method in 96-well microplates.5 Briefly, 100 μL adherent cells were seeded into each well of 96-well cell culture plates and allowed to adhere for 12 h before drug addition, while suspended cells were seeded just before drug addition with initial density of 1 × 105 cells/mL. Each tumor cell line was exposed to the test compound dissolved in DMSO at concentrations of 0.0625, 0.32, 1.6, 8, and 40 μmol in triplicates for 48 h, with cisplatin (Sigma, USA) and vinorelbine (National Institute for the Control of Pharmaceutical and Biological Products, China) as positive controls. After compound treatment, cell viability was detected and a cell growth curve was graphed. IC50 values were calculated by Reed and Muench's method.6
Notes
Electronic Supplementary Material
Supplementary material is available in the online version of this article at http://dx.doi.org/10.1007/s13659-012-0077-1 and is accessible for authorized users.
Acknowledgments
This project was supported by the National Basic Research Program of China (973 Program, 2009CB522300), the National Natural Sciences Foundation of China (30830113, U1132607).
References
-
1.(a) Ikeguchi, T. J. Bio. Chem. 1919, 40, 175-182. (b) Yang, J. ; Chen, Y. ; Feng, X. ; Yu, D. ; Liang, X. Planta Med. 1984, 50, 288-290. (c) Donnelly, D. M. ; Abe, F. ; Coveney, D. ; Fukuda, N. ; O'Reilly, J. ; Polonsky, J. ; Prange, T. J. Nat. Prod. 1985, 48, 10- 16. (d) Donnelly, D. M. X. ; Coveney, D. J. ; Fukuda, N. ; Polonsky, J. J. Nat. Prod. 1986, 49, 111-116. (e) Donnelly, D. M. X. ; Quigley, P. F. ; Coveney, D. J. ; Polonsky, J. Phytochemistry 1987, 26, 3075-3077. (f) Yang, J. S. ; Chen, Y. W. ; Feng, X. Z. ; Yu, D. Q. ; He, C. H. ; Zheng, Q. T. ; Yang, J. ; Liang, X. T. Planta Med. 1989, 55, 564-565 (g) Yang, J. S. ; Su, Y. L. ; Wang, Y. L. ; Feng, X. Z. ; Yu, D. Q. ; Cong, P. Z. ; Tamai, M. ; Obuchi, T. ; Kondoh, H. ; Liang, X. T. Planta Med. 1989, 55, 479-481. (h) Donnelly, D. M. X. ; Hutchinson, R. M. Phytochemistry 1990, 29, 179-182. (i) Donnelly, D. M. X. ; Hutchinson, R. M. ; Coveney, D. ; Yonemitsu, M. Phytochemistry 1990, 29, 2569-2572. (j) Obuchi, T. ; Kondoh, H. ; Watanabe, N. ; Tamai, M. ; Omura, S. ; Yang, J. ; Liang, X. Planta Med. 1990, 56, 198-201. (k) Watanabe, N. ; Obuchi, T. ; Tamai, M. ; Araki, H. ; Omura, S. ; Yang, J. S. ; Yu, D. Q. ; Liang, X. T. ; Huan, J. H. Planta Med. 1990, 56, 48-52. (l) Yang, J. S. ; Su, Y. L. ; Wang, Y. L. ; Feng, X. Z. ; Yu, D. Q. ; Liang, X. T. Planta Med. 1991, 57, 478-480. (m) Donnelly, D. M. X. ; Konishi, T. ; Dunne, O. ; Cremin, P. Phytochemistry 1997, 44, 1473-1478. (n) Guo, W. J. ; Guo, S. X. Chem. Nat. Compd. 2008, 44, 403. (o) Misiek, M. ; Williams, J. ; Schmich, K. ; Huttel, W. ; Merfort, I. ; Salomon, C. E. ; Aldrich, C. C. ; Hoffmeister, D. J. Nat. Prod. 2009, 72, 1888-1891. (p) Bohnert, M. ; Miethbauer, S. ; Dahse, H. M. ; Ziemen, J. ; Nett, M. ; Hoffmeister, D. Bioorg. Med. Chem. Lett. 2011, 21, 2003-2006. (q) Guo, W. J. ; Guo, S. X. Chem. Nat. Compd. 2011, 46, 995-996. PubMed Google Scholar
-
2.(a) Ding, J. H. ; Feng, T. ; Li, Z. H. ; Yang, X. Y. ; Guo, H. ; Yin, X. ; Wang, G. Q. ; Liu, J. K. Org. Lett. 2012, 14, 4976-4978. (b) Zhou, Z. Y. ; Liu, J. K. Nat. Prod. Rep. 2010, 27, 1531-1570; (c) Jiang, M. Y. ; Feng, T. ; Liu, J. K. Nat. Prod. Rep. 2011, 28, 783-808; (d) Jiang, M. Y. ; Li, Y. ; Wang, F. ; Liu, J. K. Phytochemistry 2011, 72, 923-928. (e) Yin, X. ; Feng, T. ; Li, Z. H. ; Su, J. ; Li, Y. ; Tan, N. H. ; Liu, J. K. Nat. Prod. Bioprospect. 2011, 1, 75-80. (f) Liu, L. Y. ; Li, Z. H. ; Dong, Z. J. ; Li, X. Y. ; Su, J. ; Li, Y. ; Liu, J. K. Nat. Prod. Bioprospect. 2012, 2, 130-132. (g) Wang, F. ; Zhou, D. S. ; Wei, G. Z. ; Ren, F. C. ; Liu, J. K. Phytochemistry 2012, 77, 312- 317. (h) Zhou, Z. Y. ; Shi, G. Q. ; Fontaine, R. ; Wei, K. ; Feng, T. ; Wang, F. ; Wang, G. Q. ; Qu, Y. ; Li, Z. H. ; Dong, Z. J. ; Zhu, H. J. ; Yang, Z. L. ; Zeng, G. ; Liu, J. K. Angew. Chem., Int. Ed. 2012, 51, 2368-2370. PubMed Google Scholar
-
3.J. G. J. J. Sonnenbichler, H. Peipp, D. Schwarz, Eur. J. For. Path. 27, 241 (1997) CrossRef PubMed Google Scholar
-
4.A. C.R Arnone, G. Nasini, Phytochemistry 25, 471-474 (1986) CrossRef PubMed Google Scholar
-
5.T. Mosmann, J. Immunol. Method 65, 55-63 (1983) CrossRef PubMed Google Scholar
-
6.L. J. Reed, H. Muench, Am. J. Hygiene 27, 493-497 (1938) PubMed Google Scholar
Copyright information
© The Author(s) 2012
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.