Safflower (Carthamus tinctorius L.) is a highly branched, herbaceous and thistle-like plant that belongs to the family of Compositae. It is an economically important plant, being a commercial crop grown for oilseed, birdseed, spices, dyes and herbal medicines[1]. Plants of this crop grow to 30 to 150 cm tall, and most genotypes have many sharp spines on the leaves and bracts[2]. Safflower is one of the oldest known crops and is now grown in more than 60 countries worldwide, with cultivation in the New World and Australia being relatively recent[3]. In China, safflower is an important medicinal plant that grows all over the country, where its inflorescence is known as an important crude drug and has long been used in the prevention and treatment of cardiovascular diseases and thrombosis[4].
The inflorescence of the safflower is surrounded by spiny bracts, which results in difficulty in manual picking. Moreover, florets that incorporate spines indicate reduced medicinal quality. Spineless varieties of safflower are also the first choice for farmers due to convenience in general management, harvesting and threshing. Therefore, one of the most important characteristics of safflower is having no spines on the bracts and leaves. In addition, spininess has been found to be basically dominant over spinelessness in safflower and that four genes with epistatic effects are involved.
Although the inheritance of spininess has been investigated[2] and one SRAP marker linked to the spines in Carthamus tinctorius L. was found[5], the molecular mechanism of the spiny trait in safflower has not yet been reported at the transcriptional level. Screening for differentially expressed genes is one of the most straight forward approaches to unravel the molecular basis of a biological system. cDNA sequence-related amplified polymorphism (SRAP) is a sensitive, simple and reproducible method to identify novel genes without requiring any prior sequence information[6, 7], and it has already been widely used for genetic linkage map construction, genetic diversity analysis and comparative genetics of different species[7,8,9,10]. In this study, we attempted to use bulked segregant analysis (BSA)[11] together with cDNA-SRAP to screen differentially expressed genes correlated with spines, with the use of two safflower lines representing spininess and spinelessness. The recombination values showed that the two markers (GPY-1 and GPY-2) were closely associated with spininess. Analysis of transcript- derived gene revealed that GPY-2 is the ATP synthase CF1 alpha subunit gene and may be linked to the spines. By detailed analyses of the gene-specific mRNA accumulation and transcription, we showed that CTL-spn may directly take part in the formation of spininess. The following is our first report about this study.
Materials and methodsPlant material
Two parental strains, No.0016 (♀, spineless parent) and No.0025 (♂, spiny parent) (Figure 1), were selected from Chinese populations by our laboratory team. The former (P1) was spineless (n = 83), whereas the latter (P2) had sharp spines on the bracts (n = 89). The reciprocal crosses (P1×P2 and P2×P1) were made artificially by hand, which generated 87 and 93 F1 seeds, respectively, at the Medicinal Plant Garden of the Second Military Medical University, Shanghai, China. The F2 seeds of the crosses were produced in a field in Sanya, Hainan province, by placing F1 plants, which were all spiny, in paper bags before the flowering period. Then, a segregating F2 population was obtained by sifting a single F1 plant at the Medicinal Plant Garden of the Second Military Medical University, Shanghai, China. Two hundred and forty-three (243) segregating F2 individuals were obtained, and the spine type on the bracts of the plants at bloom stage was recorded. Plants with spines (from a few to many) on the bracts were considered as spiny. Plants with no spines on the bracts were considered as spineless, according to safflower descriptors.
 | Figure 1 A: ♀, No.0016, spineless parent. B: ♂, No.0025, spininess parent |
Total RNA was extracted from bracts by using TRIzol™ reagent according to the manufacturer’s instructions (Invitrogen, USA). The quality and concentration of RNA samples were examined by gel electrophoresis[12].
Construction of RNA poolsFor BSA, an equivalent amount of RNA (1 µg·µL−1) from ten spiny- and ten spineless-individuals in F2 populations were pooled to create H and H0 bulks, respectively. Both pools were analyzed by using the cDNA-SRAP methodology to identify putative markers linked to the spiny trait.
cDNA-SRAP PCR amplificationThe cDNA- SRAP procedure was done according to the method described in Li and Quiros[7], with some modifications. Sixty different combinations were applied with the use of 13 forward primers and 16 reverse primers (Table 1). RT-PCR (reverse transcription-polymerase chain reaction) was used in SRAP analysis. After establishing agreement between the OD values of the RNAs, an aliquot of 1 µg total RNA (1 µg·µL−1) was used as template in one-step RT-PCR analysis with one-step RT-PCR kit (TaKaRa, Japan). The template was reverse transcribed at 50 ℃ for 30 min and then denatured at 94 ℃ for 5 min and 5 cycles (94 ℃ for 1 min, 35 ℃ for 1 min, and 72 ℃ for 2 min), followed by 30 cycles of amplification (94 ℃ for 1 min, 50 ℃ for 1 min, and 72 ℃ for 2 min), and elongation at 72 ℃ for 5 min. The RT-PCR reaction products were resolved on 6% denaturing polyacrylamide gels and visualized with a silver staining system. The gels were then dried overnight.
|
|
Table 1 Forward and reverse sequence-related amplified polymorphism (SRAP) primer information |
Linkage analysis of cDNA-SRAP fragments was done with the use of the 243 plants of the F2 segregating population, of which 186 individuals were spiny and 57 were spineless type. Electrophoretic bands were documented with a digital camera (Sony DSC-F717), and only reliable fragments were considered. Recombination values are calculated in accordance with gene exchange and linkage law.
Cloning of SRAP markers by RT-PCRThe SRAP fragments that amplified specifically for spiny bulks were cloned for analysis. These SRAP bands were excised with utmost care from dried polyacrylamide gels, rehydrated in TE for 1 h at room temperature, and then transferred to 500 µL of elution buffer (0.5 mol·L−1 NH4Ac, 10 mmol·L−1 magnesium acetate, 1 mmol·L−1 EDTA; pH 8.0, 0.1% SDS) at 37 ℃, from which 1.0 µL of supernatant was taken and used as template for PCR amplification with primers and conditions similar to those used in the SRAP reaction. The amplified products were electrophoresed on 1% agarose gel. Then the bands were excised from the gel, and the DNA was cloned into the plasmid vector PMD18T (TaKaRa, Japan). Recombinant plasmid DNA was isolated and sequenced by using the dideoxy method.
Molecular cloning of GPY-2 full-length cDNAThe marker closely linked to spininess was termed as GPY-2. This band was present in the spinyparent No.0025, the spiny gene pool, and spiny F2 segregating individuals. Rapid amplification of cDNA ends (RACE) was used to clone of the full-length cDNA of GPY-2. The 3' and 5' cDNA libraries of safflower were constructed by use of the Clontech SmartTM RACE cDNA amplification kit (Clontech, USA) and all primers are designed in Table 2. The nested amplified PCR products were cloned into the PMD18T vector (TaKaRa, Japan), followed by sequencing. Based on the sequences of the 3'- and 5'-RACE products, GPY- FR and GPY-FF primers (Table 2) were designed and synthesized for the full-length GPY-2 cDNA. The PCR reaction is carried on by using TaKaRa LA Taq® according to the manufacturer’s instructions (TaKaRa Biotech, China). The PCR protocol is as the following: 94 ℃ for 5 min, followed by 30 cycles at 94 ℃ for 45 sec, 60 ℃ for 45 sec and 72 ℃ for 2 min and 30 sec, and then, by extension, at 72 ℃ for 10 min. The amplification of the full-length GPY-2 cDNA was repeated twice, and the amplified products were subcloned and sequenced. To detect whether introns existed within GPY-2, PCR amplification was carriedout by using the same reaction system as that in the cloning of the full-length cDNA, except that the template was substituted with 1.5 µg of total genomic DNA of safflower, and the extension time at 72 ℃ in the amplification cycles was extended up to 3.5 min.
|
|
Table 2 Primers used in the cloning of GPY-2 full-length cDNA by RACE |
Sequence alignments, and ORF translation were carried out on Vector NTI Suite 8. GenBank BLASTs were done on NCBI (http://www.ncbi.nlm.nih.gov/).
Results 1 Inheritance of spininess traitIn reciprocal crosses of two parental strains, all F1 individuals showed spininess. Therefore, the presence of spininess on bracts was considered dominant over the spineless type. A totle of 243 F2 segregating populations was obtained by selfing a single F1 plant. The 243 F2 segregating populations were divided into two groups: 186 spiny and 57 spineless individuals.
The spiny and spineless pools could be established from F2 populations to carry out BSA. Each pool contains individuals that are identical on a particular spininess trait but arbitrary at all unlinked regions. cDNA-SRAP analysis between two pools and sample verification based on BSA makes it easier to find polymorphismsat the expressional level. A useful material model for BSA is an essential and significant prerequisite for identification of specific fragments and further genetic mapping.
2 cDNA-SRAP analysisThere were 60 sets of primer combinations that combined 13 forward primers and 16 reverse primers (Table 1). The primer combinations were tested on cDNA from the two gene pools, of which, 19 primer pairs (data not shown) revealed polymorphisms. A total of 978 bands were obtained with the 19 different primer combinations, revealing 6%−20% polymorphism between the two pools. The primers amplified approximately 36−67 bands per assay. SRAP bands that were present in one pool and absent in the other were regarded as candidate markers linked to spininess. Of the 19 primer combinations tested from two gene pools and two parents, four primer combinations (Me3/ Em4, Me5/Em2, Me3/Em6, and Me2/Em1) produced fragments of SRAP-1, SRAP-2, SRAP-3, SRAP-4, SRAP-5, and SRAP-6 that were present in the spinyparent and spiny pool and absent in the spineless parent and spineless pool (Table 3).
|
|
Table 3 SRAP fragments specific for the parents and bulks |
To confirm the linkage of the candidate SRAP markers to spininess, 243 individuals in the F2 segregating population were screened for polymorphism. Four primer combinations amplified the polymorphism between the parents, the two gene pools, and F2 individuals. The four SRAPs (SRAP-1, SRAP-2, SRAP-3, and SRAP-4) showed relatively high recombination (more than 30%) and thus were not used for further analysis. In contrast, SRAP-5 and SRAP-6 showed relatively low recombination of 6.6% and 5.8%, respectively (Table 4).
|
|
Table 4 Data calculation for linkage analysis of GPY-1 and GPY-2 |
Figure 2 presents the amplification profile of the primer combination Me2/Em1, which shows that all theplants can be clearly distinguished by this single primer set. Two bands at 300 and 150 bp (Figure 2, arrow) were present in the spiny parent No.0025, the spiny pool, and spiny individuals. These two bands were designated as GPY-1 and GPY-2, respectively.
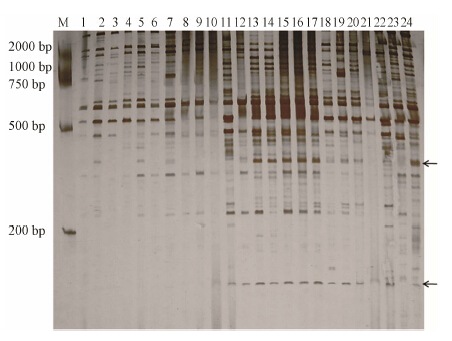 | Figure 2 A portion of the SRAP silver-staining generated with primer combinations Me2/Em1 for safflower cDNA.M: DL 2000 Marker. 1−10: Segregating spineless individuals; 11−20: Segregating spininess individuals; 21: ♀, No.0016, spineless parent; 22: ♂, No.0025, spininess parent; 23: Spineless pool; 24: Spininess pool. The SRAP markers linked to the spines are indicated by arrows |
GPY-1 and GPY-2, which were closely linked to spininess, were cloned with the objective of converting them into a simple PCR-based marker. RT-PCR analysis is sensitive, highly reproducible and able to reflect the complexity and relative abundance of the original RNA sample[13]. Suitable primer combinations were designed to test 12 random F2 populations, which generated GPY-1 and GPY-2 fragments[14]. The PCR products were checked on agarose gel to ensure that the inserts were of the same size as the SRAP fragment cloned. Figure 3 shows the resulting amplified products. The cloned cDNA was sequenced to facilitate the analysis of this marker.
 | Figure 3 The confirmed results of GPY-1 and GPY-2 fragments in Carthamus tinctorius L. individuals by RT-PCR.1-6: Results of spininess individuals; 7-12: Results of spineless individuals |
The exact length of the GPY-1 fragment was 278 bp:TGAGTCCAAACCGGAGCATCATCAGGTATCGGAAAATGACGATCTGCTGAAATCGTTGTATGATGAAATTAATGTATGATGATCTCCAT CTCCACCGGATCTGAATCGGTGCGAGAATCTTTACTATGTCGCATAAACATGCCCATGTGGCCACTGTTTTCTCTTGGAATTAATGC TTTAATCTTCTATTTTCTTTGTGTGGTGAAATTAATATAATGATAACATATTATTGAATTCGTTCTAATAGATTTATTTGTTCCA TTAATTCGTACGCAGTC.
The exact length of the GPY-2 fragment was 150 bp:TGAGTCCAAACCGGAGCCACAGAAGATGCTTTTTGACCAATAGCTACATAAACGCATATTACATTTTGGCCT TGTTGATTTAAAATTGTATCTGTTGCTACCGCTGTTTTACCGGTCTGTCTGTCCCCAATAATTAATTCGTACGCAGTC. 5 GPY-2 full-length cDNA
Rapid amplification of cDNA ends (RACE) was used clone the full-length cDNA of GPY-2. By using the RACE method, cDNA ends of 1 088 bp and 640 bp were amplified by 3' and 5' RACE, respectively. The 3'and 5' ends were assembled with Vector NTI Suite 8.0, and the deduced full-length GPY-2 cDNA was subsequently amplified by proof reading PCR amplification with the above-mentioned primers. The full- length cDNA sequence of GPY-2 was 1 679 bp (GenBank Accession No: EF104641),consisting of an 80 bp 5' untranslated region, a 75 bp 3'untranslated region, and a 1 524 bp ORF encoding a 508-aminoacid protein (Figure 4).
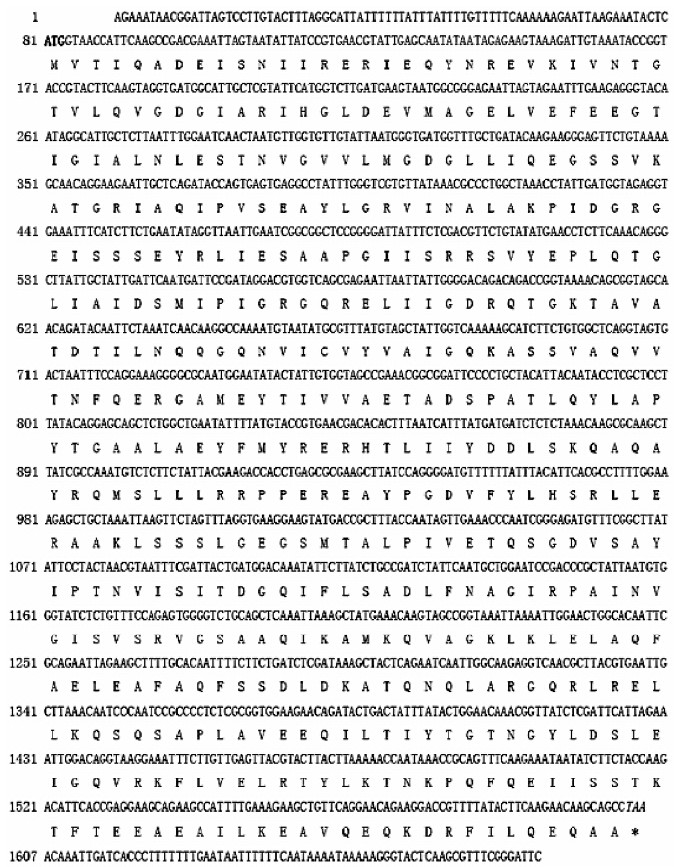 | Figure 4 The full-length cDNA sequence and the deduced amino acid sequence of GPY-2. The start codon (ATG) is in bold and the stop codon (TAA) is in italics |
The genomic sequence corresponding to the GPY-2full-length cDNA was 1 679 bp. The alignment result of the GPY-2 cDNA sequence and the genomic sequence indicated that the GPY-2 gene was intronless. The GPY-2 full-length cDNA is termed as CTL-spn originated from the closest linkage with spininess in safflower than other SRAP makers.
6 Sequence analysis and bioinformaticsProtein-protein BLAST showed that at the amino acid level, GPY-2 protein shared a high homology with ATP synthase CF1 alpha subunits from other plant species. Through Clustal W/X, the full-length alignment result showed that GPY-2 protein shared 97% identity with ATP synthase CF1 alpha subunit from Lactuca sativa chloroplast DNA of complete genome, 97% identity with ATP synthase CF1 alpha subunit from the Helianthus annuus cultivar line HA383 chloroplast of complete genome, 96% identity with ATPase alpha subunit from Panax ginseng chloroplast of complete genome and 95% identity with ATP synthase CF1 alpha subunit from Daucus carota chloroplast of complete genome (Figure 5).
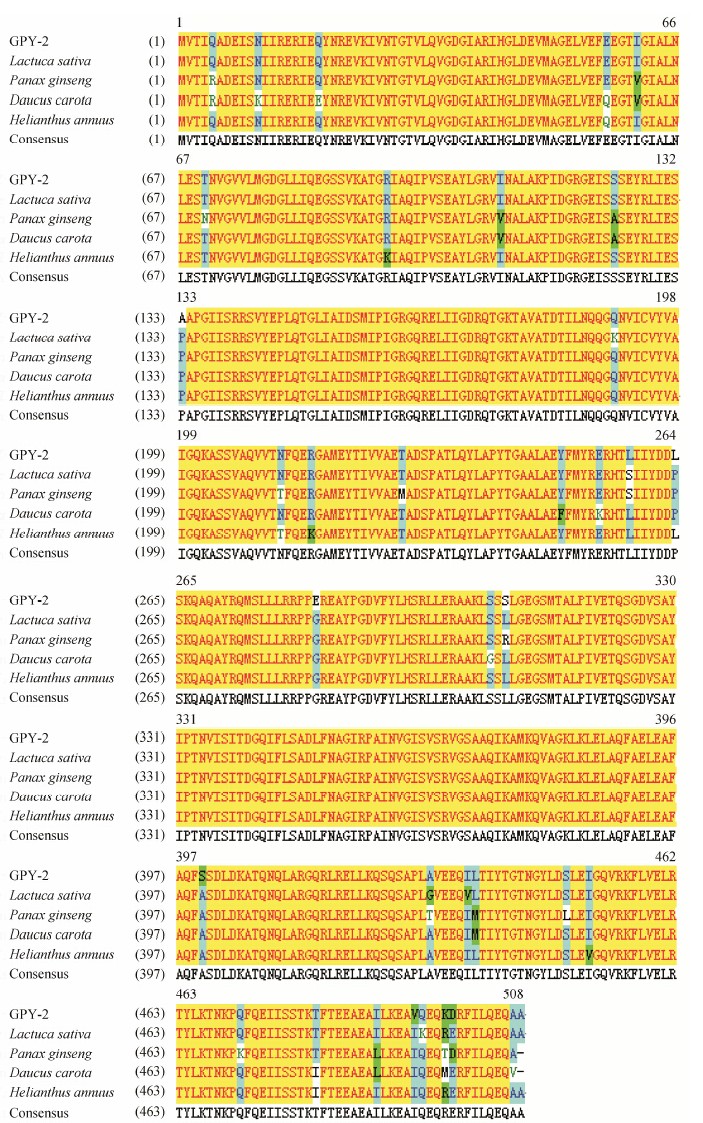 | Figure 5 Multiple sequence alignment of GPY-2 from Carthamus tinctorius L. with other ATP synthase CF1 alpha chain from Lactuca sativa, Panax ginseng, Daucus carota and Helianthus annuus. Amino acid residues identical at a given position are denoted with yellow backgrounds |
Although the inheritance of the spiny trait has been investigated, the biological characterization of this trait in safflower has not yet been reported. The present study investigated differentially expressed transcript-derived fragments of interest through BSA-SRAP analysis, applying RACE to get the full-length gene of GPY-2.
TheCTL-spn gene encodes the ATP synthase CF1 alpha subunit in safflower. ATPases are membrane- bound enzyme complexes that synthesize and/or hydrolyze ATP by the transport of protons across mitochondrial or chloroplast membranes[15]. As a kind of ATPase, ATP synthases are enzymes that can synthesize ATP from ADP and inorganic phosphate. The activity of ATPase plays an important role in promoting plant resistance to abiotic and biotic stress[16], such as high light, chilling stress, and virus infections [17, 18, 19, 20].Studies have proven that ATP synthase CF1 alpha subunit can trigger a series of resistance reactions by transporting protons and improving the cytoplasmic pH, or providing signal molecules, as well as by facilitating an increase in energy to enhance the resistance of plants[21].
Spines are known to evolve from leaves by metamorphosis, such as in cactuses, in which they protect the inner tissue structure against high light and other environmental injuries. This is consistent with the defense mechanism of ATP synthase CF1 alpha subunit during plant responses to environmental stimulation. Our findings suggest that CTL-spn may directly take part in the formation of spininess and in enhancing the resistance reaction of spiny safflower.
ConclusionThe study indicated that CTL-spn is related to the formation of spininess and enhance the resistance reaction of spiny safflower. Our work may help elucidate the molecular basis of the formation of spininess and may provide important insights for breeding spineless cultivars of safflower.
| [1] | Bowles VG, Mayerhofer R, Davis C, et al. A phylogenetic investigation of Carthamus combining sequence and microsatellite data [J]. Plant Syst Evol, 2010, 287: 85-97. |
| [2] | Pahlavani MH, Mirlohi AF, Saeidi G. Inheritance of flower color and spininess in safflower (Carthamus tinctorius L.) [J]. J Hered, 2004, 95: 265-267. |
| [3] | Johnston AM, Tanaka DL, Miller PR, et al. Oilseed crops for semiarid cropping systems in the northern Great Plains [J]. Agron J, 2002, 94: 231-240. |
| [4] | Siow YL, Choy PC, Leung WM, et al. Effect of Flos Carthami on stress-activated protein kinase activity in the isolated reperfused rat heart [J]. Mol Cell Biochem, 2000, 207: 41-47. |
| [5] | Guo QH, Guo ML. SRAP fragments linked to spines of Carthamus tinctorius [J]. Acta Pharm Sin (药学学报), 2007, 42: 94-797. |
| [6] | Huang CQ, Liu GD, Bai CJ, et al. Application of SRAP markers in the identification of stylosanthes guianensis hybrids [J]. Mol Biol Rep, 2014, 41: 5923-5929. |
| [7] | Li G, Quiros CF. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica [J]. Theor Appl Genet, 2001, 103: 455-461. |
| [8] | Ferriol M, Picó B, Nuez F. Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers [J]. Theor Appl Genet, 2003, 107: 271-282. |
| [9] | Lin ZX, Zhang XL, Nie YC. Evaluation of application of a new molecular marker SRAP on analysis of F2 segregation population and genetic diversity in cotton [J]. J Genet Genomics, 2004, 31: 622-626. |
| [10] | Li G, Gao M, Yang B, et al. Gene for gene alignment between the Brassica and Arabidopsis genomes by direct transcriptome mapping [J]. Theor Appl Genetics, 2003, 107: 168-180. |
| [11] | Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations [J]. Proc Natl Acad Sci USA, 1991, 88: 9828-9832. |
| [12] | Dongre AB, Raut MP, Bhandarkar MR, et al. Identification and genetic purity testing of cotton F1 hybrid using molecular markers [J]. Indian J Biotechnol, 2011, 10: 301-306. |
| [13] | Vernon SD, Unger ER, Rajeevan M, et al. Reproducibility of alternative probe synthesis approaches for gene expression profiling with arrays [J]. J Mol Diagn, 2000, 2: 124-127. |
| [14] | Feng N, Li YK, Tang J, et al. cDNA-AFLP analysis on transcripts associated with hydroxysafflor yellow A (HSYA) biosynthetic pathway in Carthamus tinctorius [J]. Biochem Syst Ecol, 2010, 38: 971-980. |
| [15] | Cross RL, Muller V. The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio [J]. FEBS Lett, 2004, 576: 1-4. |
| [16] | Mouekouba LD, Zhang LL, Guan X, et al. Analysis of Clonostachys rosea-induced resistance to tomato gray mold disease in tomato leaves [J]. PLoS One, 2014, 9: e102690. |
| [17] | Garavaglia BS, Thomas L, Zimaro T, et al. A plant natriuretic peptide-like molecule of the pathogen Xanthomonas axonopodis pv. citri causes rapid changes in the proteome of its citrus host [J]. BMC Plant Biol, 2010, 10: 51. |
| [18] | Wu LJ, Wang SX, Chen X, et al. Proteomic and phytohormone analysis of the response of Maize (Zea mays L.) seedlings to sugarcane mosaic virus [J]. PLoS One, 2013, 8: e70295. |
| [19] | Romanowska E, Powikrowska M, Zienkiewicz M, et al. High light induced accumulation of two isoforms of the CF1 alpha-subunit in mesophyll and bundle sheath chloroplasts of C4 plants [J]. Acta Biochim Pol, 2008, 55: 175-182. |
| [20] | Burkey KO, Mathis JN. Identification of a novel isoform of the chloroplast-coupling factor alpha-subunit [J]. Plant Physiol, 1998, 116: 703-708. |
| [21] | Shao HH, Cao Q, Tao X, et al. Cloning and characterization of ATP synthase CF1 α gene from sweet potato [J]. Afr J Biotechnol, 2011, 10: 19035-19042. |
 2015, Vol. 50
2015, Vol. 50


